Transcriptome Profile of a New Mouse Model of Spinocerebellar Ataxia Type 14 Implies Changes in Cerebellar Development
Abstract
1. Introduction
2. Materials and Methods
2.1. The SCA14 Mouse Model
2.2. Total RNA Extraction and Total RNA Sequencing (RNA-Seq)
2.3. RNA-Seq Data Collection and Analysis
3. Results
3.1. Overview of Gene Expression Changes
3.2. Highlighted Genes
3.3. Cluster Analysis
3.4. Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chelban, V.; Wiethoff, S.; Fabian-Jessing, B.K.; Haridy, N.A.; Khan, A.; Efthymiou, S.; Becker, E.B.; O’Connor, E.; Hersheson, J.; Newland, K.; et al. Genotype-phenotype correlations, dystonia and disease progression in spinocerebellar ataxia type 14. Mov. Disord. 2018, 33, 1119–1129. [Google Scholar] [CrossRef]
- Verbeek, D.S.; Knight, M.A.; Harmison, G.G.; Fischbeck, K.H.; Howell, B.W. Protein kinase C gamma mutations in spinocerebellar ataxia 14 increase kinase activity and alter membrane targeting. Brain 2005, 128, 436–442. [Google Scholar] [CrossRef]
- Chopra, R.; Wasserman, A.H.; Pulst, S.M.; De Zeeuw, C.I.; Shakkottai, V.G. Protein kinase C activity is a protective modifier of Purkinje neuron degeneration in cerebellar ataxia. Hum. Mol. Genet. 2018, 27, 1396–1410. [Google Scholar] [CrossRef]
- Hirai, H. Protein Kinase C in the Cerebellum: Its Significance and Remaining Conundrums. Cerebellum 2018, 17, 23–27. [Google Scholar] [CrossRef] [PubMed]
- White, J.J.; Sillitoe, R.V. Development of the cerebellum: From gene expression patterns to circuit maps. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 149–164. [Google Scholar] [CrossRef]
- Schrenk, K.; Kapfhammer, J.P.; Metzger, F. Altered dendritic development of cerebellar Purkinje cells in slice cultures from protein kinase Cgamma-deficient mice. Neuroscience 2002, 110, 675–689. [Google Scholar] [CrossRef]
- Kapfhammer, J.P. Cellular and molecular control of dendritic growth and development of cerebellar Purkinje cells. Prog. Histochem. Cytochem. 2004, 39, 131–182. [Google Scholar] [CrossRef]
- Adachi, N.; Kobayashi, T.; Takahashi, H.; Kawasaki, T.; Shirai, Y.; Ueyama, T.; Matsuda, T.; Seki, T.; Sakai, N.; Saito, N. Enzymological analysis of mutant protein kinase Cgamma causing spinocerebellar ataxia type 14 and dysfunction in Ca2+ homeostasis. J. Biol. Chem. 2008, 283, 19854–19863. [Google Scholar] [CrossRef]
- Shimobayashi, E.; Kapfhammer, J.P. A New Mouse Model Related to SCA14 Carrying a Pseudosubstrate Domain Mutation in PKCgamma Shows Perturbed Purkinje Cell Maturation and Ataxic Motor Behavior. J. Neurosci. 2021, 41, 2053–2068. [Google Scholar] [CrossRef]
- Uesaka, N.; Kano, M. Presynaptic Mechanisms Mediating Retrograde Semaphorin Signals for Climbing Fiber Synapse Elimination During Postnatal Cerebellar Development. Cerebellum 2018, 17, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Kano, M.; Abeliovich, A.; Chen, L.; Bao, S.; Kim, J.J.; Hashimoto, K.; Thompson, R.F.; Tonegawa, S. Impaired motor coordination correlates with persistent multiple climbing fiber innervation in PKC gamma mutant mice. Cell 1995, 83, 1233–1242. [Google Scholar] [CrossRef]
- Kano, M.; Hashimoto, K.; Chen, C.; Abeliovich, A.; Aiba, A.; Kurihara, H.; Watanabe, M.; Inoue, Y.; Tonegawa, S. Impaired synapse elimination during cerebellar development in PKC gamma mutant mice. Cell 1995, 83, 1223–1231. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Lund, S.P.; Nettleton, D.; McCarthy, D.J.; Smyth, G.K. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat. Appl. Genet. Mol. Biol. 2012, 11. [Google Scholar] [CrossRef]
- Lawrence, M.; Gentleman, R.; Carey, V. rtracklayer: An R package for interfacing with genome browsers. Bioinformatics 2009, 25, 1841–1842. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Gaidatzis, D.; Lerch, A.; Hahne, F.; Stadler, M.B. QuasR: Quantification and annotation of short reads in R. Bioinformatics 2015, 31, 1130–1132. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Wu, Q.W.; Kapfhammer, J.P. Serine/threonine kinase 17b (STK17B) signalling regulates Purkinje cell dendritic development and is altered in multiple spinocerebellar ataxias. Eur. J. Neurosci. 2021, 54, 6673–6684. [Google Scholar] [CrossRef]
- Cui, W.Q.; Zhang, W.W.; Chen, T.; Li, Q.; Xu, F.; Mao-Ying, Q.L.; Mi, W.L.; Wang, Y.Q.; Chu, Y.X. Tacr3 in the lateral habenula differentially regulates orofacial allodynia and anxiety-like behaviors in a mouse model of trigeminal neuralgia. Acta Neuropathol. Commun. 2020, 8, 44. [Google Scholar] [CrossRef]
- Izzi, L.; Charron, F. Midline axon guidance and human genetic disorders. Clin. Genet. 2011, 80, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.A.; Tymanskyj, S.; Yuan, R.C.; Leung, H.C.; Lefebvre, J.L.; Sanes, J.R.; Chedotal, A.; Ma, L. Dendrite self-avoidance requires cell-autonomous slit/robo signaling in cerebellar purkinje cells. Neuron 2014, 81, 1040–1056. [Google Scholar] [CrossRef]
- Liao, C.; Sarayloo, F.; Vuokila, V.; Rochefort, D.; Akçimen, F.; Diamond, S.; Houle, G.; Laporte, A.D.; Spiegelman, D.; He, Q.; et al. Transcriptomic Changes Resulting From STK32B Overexpression Identify Pathways Potentially Relevant to Essential Tremor. Front. Genet. 2020, 11, 813. [Google Scholar] [CrossRef]
- Cao, J.; O’Day, D.R.; Pliner, H.A.; Kingsley, P.D.; Deng, M.; Daza, R.M.; Zager, M.A.; Aldinger, K.A.; Blecher-Gonen, R.; Zhang, F.; et al. A human cell atlas of fetal gene expression. Science 2020, 370, eaba7721. [Google Scholar] [CrossRef]
- Saywell, V.; Cioni, J.M.; Ango, F. Developmental gene expression profile of axon guidance cues in Purkinje cells during cerebellar circuit formation. Cerebellum 2014, 13, 307–317. [Google Scholar] [CrossRef]
- Paulhus, K.; Ammerman, L.; Glasscock, E. Clinical Spectrum of KCNA1 Mutations: New Insights into Episodic Ataxia and Epilepsy Comorbidity. Int. J. Mol. Sci. 2020, 21, 2802. [Google Scholar] [CrossRef]
- Pizzagalli, M.D.; Bensimon, A.; Superti-Furga, G. A guide to plasma membrane solute carrier proteins. FEBS J. 2021, 288, 2784–2835. [Google Scholar] [CrossRef]
- Smagin, D.A.; Babenko, V.N.; Redina, O.E.; Kovalenko, I.L.; Galyamina, A.G.; Kudryavtseva, N.N. Reduced Expression of Slc Genes in the VTA and NAcc of Male Mice with Positive Fighting Experience. Genes 2021, 12, 1099. [Google Scholar] [CrossRef] [PubMed]
- Rask-Andersen, M.; Masuram, S.; Fredriksson, R.; Schioth, H.B. Solute carriers as drug targets: Current use, clinical trials and prospective. Mol. Asp. Med. 2013, 34, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Tsumagari, R.; Kakizawa, S.; Kikunaga, S.; Fujihara, Y.; Ueda, S.; Yamanoue, M.; Saito, N.; Ikawa, M.; Shirai, Y. DGKgamma Knock-Out Mice Show Impairments in Cerebellar Motor Coordination, LTD, and the Dendritic Development of Purkinje Cells through the Activation of PKCgamma. eNeuro 2020, 7, ENEURO.0319-19.2020. [Google Scholar] [CrossRef] [PubMed]
- Castel, P.; Ellis, H.; Bago, R.; Toska, E.; Razavi, P.; Carmona, F.J.; Kannan, S.; Verma, C.S.; Dickler, M.; Chandarlapaty, S.; et al. PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kalpha Inhibition. Cancer Cell 2016, 30, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Shimobayashi, E.; Wagner, W.; Kapfhammer, J.P. Carbonic Anhydrase 8 Expression in Purkinje Cells Is Controlled by PKCgamma Activity and Regulates Purkinje Cell Dendritic Growth. Mol. Neurobiol. 2016, 53, 5149–5160. [Google Scholar] [CrossRef]
- Trzesniewski JAltmann, S.; Jager, L.; Kapfhammer, J.P. Reduced Purkinje cell size is compatible with near normal morphology and function of the cerebellar cortex in a mouse model of spinocerebellar ataxia. Exp. Neurol. 2019, 311, 205–212. [Google Scholar] [CrossRef]
- Winkler, S.C.; Shimobayashi, E.; Kapfhammer, J.P. PKCgamma-Mediated Phosphorylation of CRMP2 Regulates Dendritic Outgrowth in Cerebellar Purkinje Cells. Mol. Neurobiol. 2020, 57, 5150–5166. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Shirai, Y.; Matsubara, T.; Sanse, K.; Kuriyama, M.; Oshiro, N.; Yoshino, K.; Yonezawa, K.; Ono, Y.; Saito, N. Phosphorylation and up-regulation of diacylglycerol kinase gamma via its interaction with protein kinase C gamma. J. Biol. Chem. 2006, 281, 31627–31637. [Google Scholar] [CrossRef]
- Niewiadomska-Cimicka, A.; Doussau, F.; Perot, J.B.; Roux, M.J.; Keime, C.; Hache, A.; Piguet, F.; Novati, A.; Weber, C.; Yalcin, B.; et al. SCA7 Mouse Cerebellar Pathology Reveals Preferential Downregulation of Key Purkinje Cell-Identity Genes and Shared Disease Signature with SCA1 and SCA2. J. Neurosci. 2021, 41, 4910–4936. [Google Scholar] [CrossRef]
- Dulneva, A.; Lee, S.; Oliver, P.L.; Di Gleria, K.; Kessler, B.M.; Davies, K.E.; Becker, E.B. The mutant Moonwalker TRPC3 channel links calcium signaling to lipid metabolism in the developing cerebellum. Hum. Mol. Genet. 2015, 24, 4114–4125. [Google Scholar] [CrossRef]
- Becker, E.B. The Moonwalker mouse: New insights into TRPC3 function, cerebellar development, and ataxia. Cerebellum 2014, 13, 628–636. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rajamanickam, J.; Palmada, M.; Lang, F.; Boehmer, C. EAAT4 phosphorylation at the SGK1 consensus site is required for transport modulation by the kinase. J. Neurochem. 2007, 102, 858–866. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, J.M.; Alessi, D.R. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 2008, 416, 375–385. [Google Scholar] [CrossRef]
- Rabenstein, M.; Peter, F.; Rolfs, A.; Frech, M.J. Impact of Reduced Cerebellar EAAT Expression on Purkinje Cell Firing Pattern of NPC1-deficient Mice. Sci. Rep. 2018, 8, 3318. [Google Scholar] [CrossRef] [PubMed]
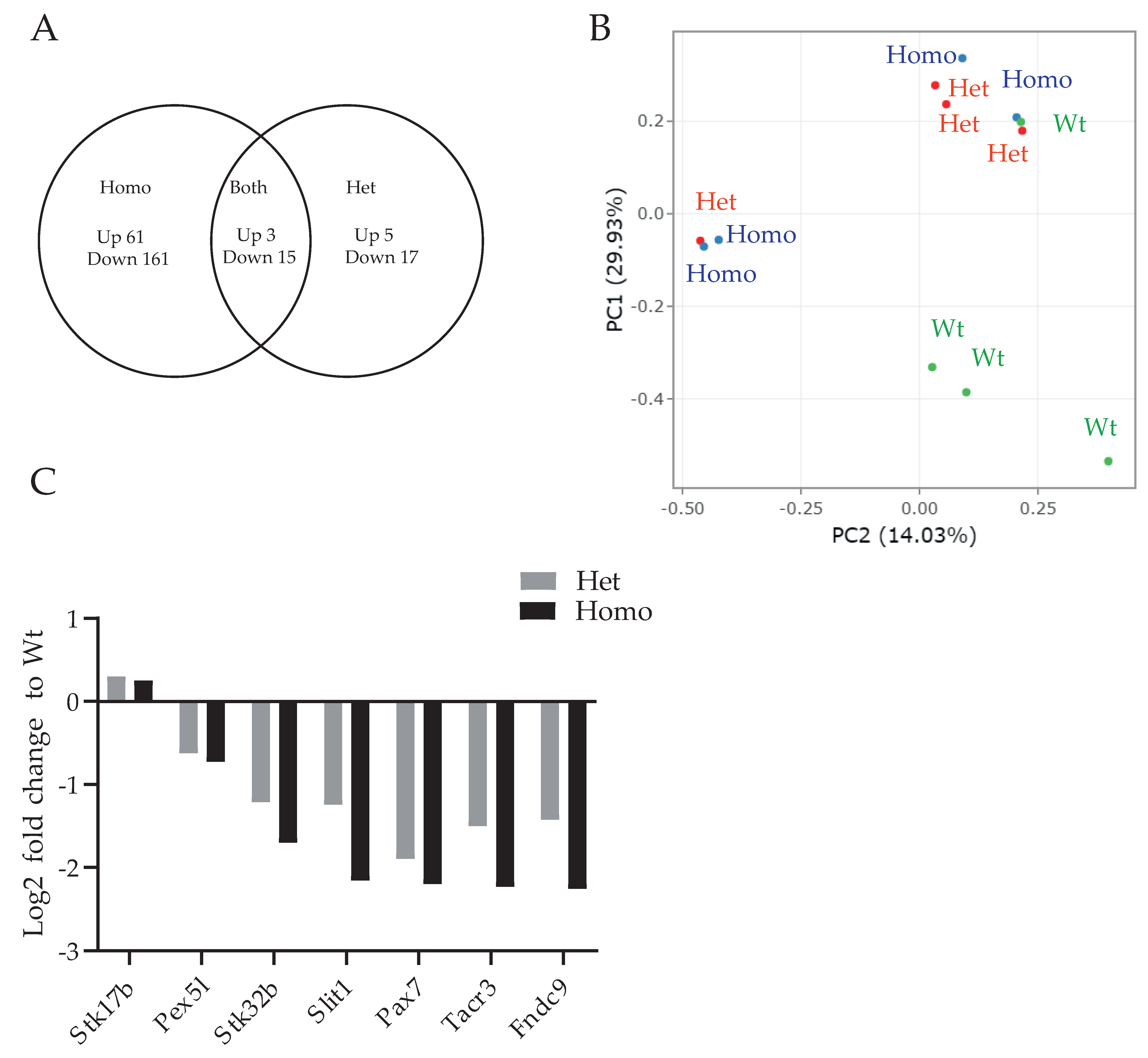

| Homo vs. WT | Hetero vs. WT | Homo vs. Hetero | |
|---|---|---|---|
| Number of genes altogether | 20,510 | 20,510 | 20,510 |
| No significant change | 20,285 | 20,488 | 20,510 |
| Upregulated genes | 61 | 5 | 0 |
| Downregulated genes | 164 | 17 | 0 |
| Genes | Log2FC | p-Value | FDR p-Value |
|---|---|---|---|
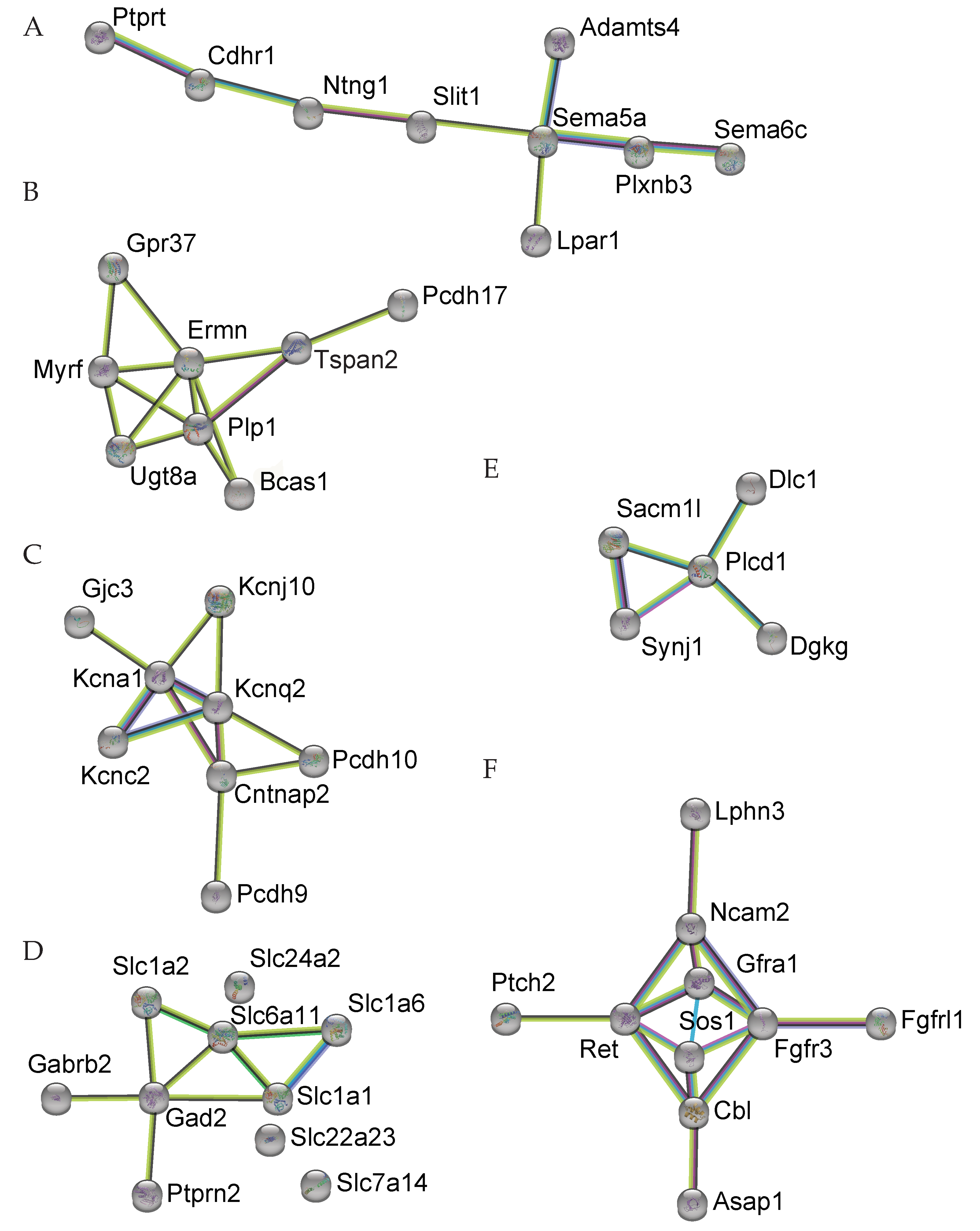
| Cbl | −0.219 | 0.0000233 | 0.0236 |
| Fgfr3 | −0.216 | 0.000263 | 0.0464 |
| Itgb8 | −0.36 | 0.000115 | 0.0382 |
| Prex2 | −0.276 | 0.0000319 | 0.0271 |
| Sos1 | −0.175 | 0.000346 | 0.0465 |
| Synj1 | −0.16 | 0.000403 | 0.0471 |
| Genes | Log2FC | p-Value | FDR p-Value |
|---|---|---|---|
| Adgra1 | −0.535 | 0.000494 | 0.0485 |
| Adgrl3 | −0.195 | 0.000158 | 0.0399 |
| Casr | −0.904 | 0.0000363 | 0.0286 |
| Cckbr | −1.398 | 0.000236 | 0.0461 |
| Gdpd1 | 0.167 | 0.000519 | 0.0493 |
| Gpr37 | −0.56 | 0.000165 | 0.0399 |
| Kcnq2 | −0.23 | 0.0003 | 0.0464 |
| Lpar1 | −0.297 | 0.000438 | 0.0474 |
| Nfatc4 | 0.481 | 0.000159 | 0.0399 |
| Pde11a | −1.625 | 0.000314 | 0.0464 |
| Pde9a | 0.285 | 0.000242 | 0.0464 |
| Prex2 | −0.276 | 0.0000319 | 0.0271 |
| Sos1 | −0.175 | 0.000346 | 0.0465 |
| Tacr3 | −2.234 | 0.000526 | 0.0494 |
| Genes | Log2FC | p-Value | FDR p-Value |
|---|---|---|---|
| Adamts4 | −0.43 | 0.000202 | 0.0426 |
| Itgb8 | −0.36 | 0.000115 | 0.0382 |
| Nfatc4 | 0.481 | 0.000159 | 0.0399 |
| Ntng1 | −0.499 | 0.00000993 | 0.0236 |
| Plcd1 | 0.369 | 0.000128 | 0.0392 |
| Plxnb3 | −0.345 | 0.000445 | 0.0474 |
| Ptch2 | 0.658 | 0.0000237 | 0.0236 |
| Sema5a | −0.389 | 0.00000821 | 0.0236 |
| Sema6c | 0.273 | 0.000275 | 0.0464 |
| Slit1 | −2.16 | 0.000303 | 0.0464 |
| Sos1 | −0.175 | 0.000346 | 0.0465 |
| Genes | Log2FC | p-Value | FDR p-Value |
|---|---|---|---|
| Gabrb2 | −0.324 | 0.000314 | 0.0464 |
| Gad2 | −0.301 | 0.000313 | 0.0464 |
| Gpr37 | −0.56 | 0.000165 | 0.0399 |
| Kcnq2 | −0.23 | 0.0003 | 0.0464 |
| Slc6a11 | −1.096 | 0.000134 | 0.0393 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mezey, S.E.; Kapfhammer, J.P.; Shimobayashi, E. Transcriptome Profile of a New Mouse Model of Spinocerebellar Ataxia Type 14 Implies Changes in Cerebellar Development. Genes 2022, 13, 1417. https://doi.org/10.3390/genes13081417
Mezey SE, Kapfhammer JP, Shimobayashi E. Transcriptome Profile of a New Mouse Model of Spinocerebellar Ataxia Type 14 Implies Changes in Cerebellar Development. Genes. 2022; 13(8):1417. https://doi.org/10.3390/genes13081417
Chicago/Turabian StyleMezey, Szilvia E., Josef P. Kapfhammer, and Etsuko Shimobayashi. 2022. "Transcriptome Profile of a New Mouse Model of Spinocerebellar Ataxia Type 14 Implies Changes in Cerebellar Development" Genes 13, no. 8: 1417. https://doi.org/10.3390/genes13081417
APA StyleMezey, S. E., Kapfhammer, J. P., & Shimobayashi, E. (2022). Transcriptome Profile of a New Mouse Model of Spinocerebellar Ataxia Type 14 Implies Changes in Cerebellar Development. Genes, 13(8), 1417. https://doi.org/10.3390/genes13081417






