Phenotypic Variation in Two Siblings Affected with Shwachman-Diamond Syndrome: The Use of Expert Variant Interpreter (eVai) Suggests Clinical Relevance of a Variant in the KMT2A Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Presentation
2.1.1. UPN42
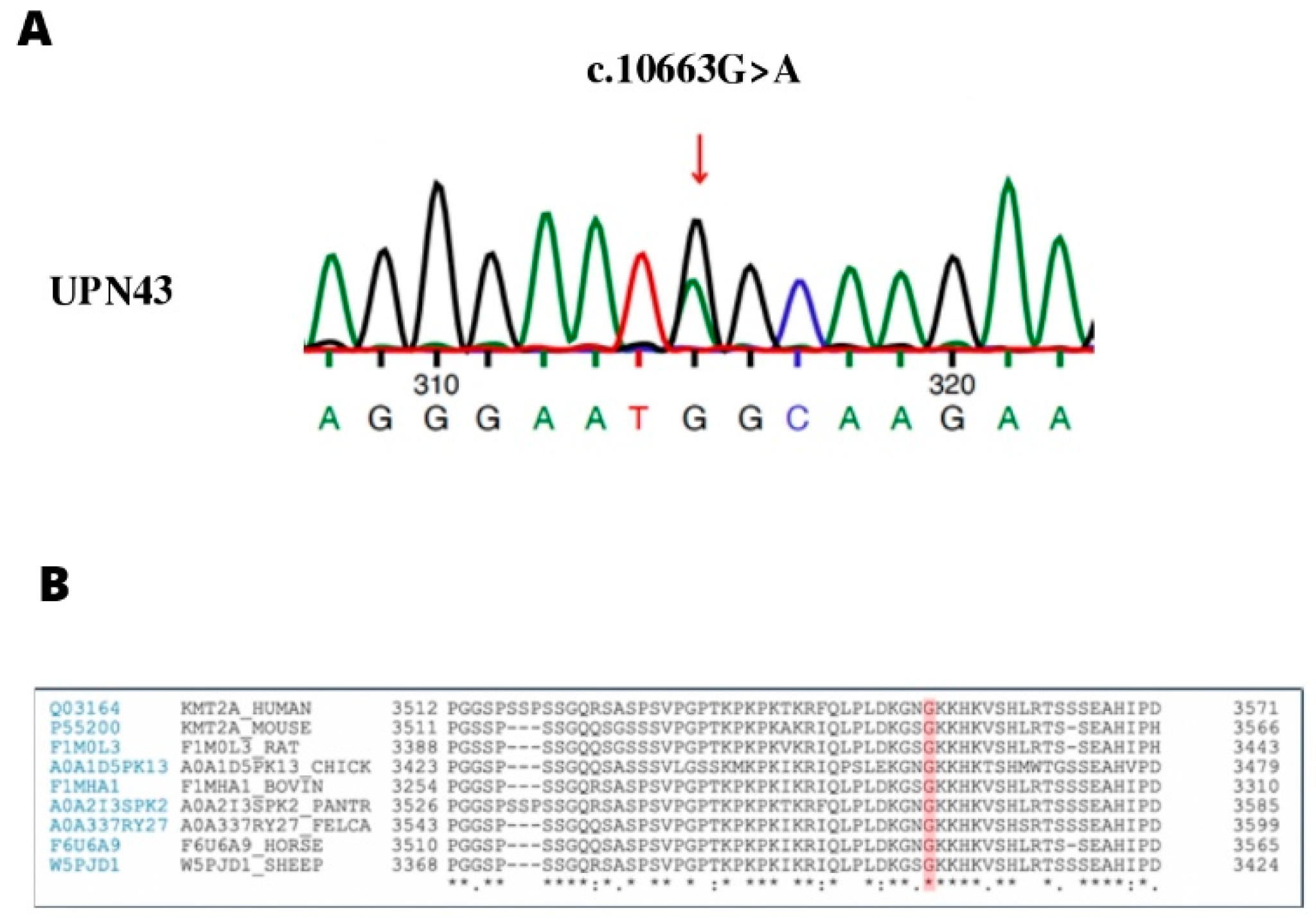
2.1.2. UPN43
2.2. Genetic Analysis
2.3. Bioinformatics Analysis
2.3.1. The Human Phenotype Ontology (HPO) Terms
2.3.2. Filtering and Variant Prioritization
2.3.3. Variant Interpretation
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dror, Y.; Donadieu, J.; Koglmeier, J.; Dodge, J.; Toiviainen-Salo, S.; Makitie, O.; Kerr, E.; Zeidler, C.; Shimamura, A.; Shah, N.; et al. Draft consensus guidelines for diagnosis and treatment of Shwachman-Diamond syndrome: Dror et al. Ann. N. Y. Acad. Sci. 2011, 1242, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Dror, Y. Shwachman-Diamond syndrome. Pediatr. Blood Cancer 2005, 45, 892–901. [Google Scholar] [CrossRef]
- Kerr, E.N.; Ellis, L.; Dupuis, A.; Rommens, J.M.; Durie, P.R. The Behavioral Phenotype of School-Age Children with Shwachman Diamond Syndrome Indicates Neurocognitive Dysfunction with Loss of Shwachman-Bodian-Diamond Syndrome Gene Function. J. Pediatrics 2010, 156, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.S.; Myers, K.C. Diagnosis, Treatment, and Molecular Pathology of Shwachman-Diamond Syndrome. Hematol. Oncol. Clin. N. Am. 2018, 32, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Bogusz-Wójcik, A.; Kołodziejczyk, H.; Moszczyńska, E.; Klaudel-Dreszler, M.; Oracz, G.; Pawłowska, J.; Szalecki, M. Endocrine dysfunction in children with Shwachman-Diamond syndrome. Endokrynol. Pol. 2021, 72, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Boocock, G.R.B.; Morrison, J.A.; Popovic, M.; Richards, N.; Ellis, L.; Durie, P.R.; Rommens, J.M. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 2003, 33, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Carapito, R.; Konantz, M.; Paillard, C.; Miao, Z.; Pichot, A.; Leduc, M.S.; Yang, Y.; Bergstrom, K.L.; Mahoney, D.H.; Shardy, D.L.; et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J. Clin. Invest 2017, 127, 4090–4103. [Google Scholar] [CrossRef]
- Dhanraj, S.; Matveev, A.; Li, H.; Lauhasurayotin, S.; Jardine, L.; Cada, M.; Zlateska, B.; Tailor, C.S.; Zhou, J.; Mendoza-Londono, R.; et al. Biallelic mutations in DNAJC21 cause Shwachman-Diamond syndrome. Blood 2017, 129, 1557–1562. [Google Scholar] [CrossRef]
- Stepensky, P.; Chacón-Flores, M.; Kim, K.H.; Abuzaitoun, O.; Bautista-Santos, A.; Simanovsky, N.; Siliqi, D.; Altamura, D.; Méndez-Godoy, A.; Gijsbers, A.; et al. Mutations in EFL1, an SBDS partner, are associated with infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies in aShwachman-Diamond like syndrome. J. Med. Genet. 2017, 54, 558–566. [Google Scholar] [CrossRef]
- Tan, S.; Kermasson, L.; Hoslin, A.; Jaako, P.; Faille, A.; Acevedo-Arozena, A.; Lengline, E.; Ranta, D.; Poirée, M.; Fenneteau, O.; et al. EFL1 mutations impair eIF6 release to cause Shwachman-Diamond syndrome. Blood 2019, 134, 277–290. [Google Scholar] [CrossRef]
- Alsavaf, M.B.; Verboon, J.M.; Dogan, M.E.; Azizoglu, Z.B.; Okus, F.Z.; Ozcan, A.; Dundar, M.; Eken, A.; Donmez-Altuntas, H.; Sankaran, V.G.; et al. A novel missense mutation outside the DNAJ domain of DNAJC21 is associated with Shwachman-Diamond syndrome. Br. J. Haematol. 2022, 43, S61–S62. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, P.; Cotter, M.; Smith, O.P. Autosomal dominant Shwachman–Diamond syndrome with a novel heterozygous missense variant in the SRP54 gene causing severe phenotypic features. Br. J. Haematol. 2022, 196, e39–e42. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, L.; Woolfrey, A.; Shimamura, A. Shwachman-Diamond Syndrome: A Review of the Clinical Presentation, Molecular Pathogenesis, Diagnosis, and Treatment. Hematol. Oncol. Clin. N. Am. 2009, 23, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Ginzberg, H.; Shin, J.; Ellis, L.; Morrison, J.; Ip, W.; Dror, Y.; Freedman, M.; Heitlinger, L.A.; Belt, M.A.; Corey, M.; et al. Shwachman syndrome: Phenotypic manifestations of sibling sets and isolated cases in a large patient cohort are similar. J. Pediatr. 1999, 135, 81–88. [Google Scholar] [CrossRef]
- Donadieu, J.; Fenneteau, O.; Beaupain, B.; Beaufils, S.; Bellanger, F.; Mahlaoui, N.; Lambilliotte, A.; Aladjidi, N.; Bertrand, Y.; Mialou, V.; et al. Classification of and risk factors for hematologic complications in a French national cohort of 102 patients with Shwachman-Diamond syndrome. Haematologica 2012, 97, 1312–1319. [Google Scholar] [CrossRef]
- Furutani, E.; Liu, S.; Galvin, A.; Steltz, S.; Malsch, M.M.; Loveless, S.K.; Mount, L.; Larson, J.H.; Queenan, K.; Bertuch, A.A.; et al. Hematologic complications with age in Shwachman-Diamond syndrome. Blood Adv. 2022, 6, 297–306. [Google Scholar] [CrossRef]
- Thompson, A.S.; Giri, N.; Gianferante, D.M.; Jones, K.; Savage, S.A.; Alter, B.P.; McReynolds, L.J. Shwachman Diamond syndrome: Narrow genotypic spectrum and variable clinical features. Pediatr. Res. 2022, 1–10. [Google Scholar] [CrossRef]
- Kent, A.; Murphy, G.H.; Milla, P. Psychological characteristics of children with Shwachman syndrome. Arch. Dis. Child. 1990, 65, 1349–1352. [Google Scholar] [CrossRef]
- Nelson, A.; Myers, K. Shwachman-Diamond Syndrome. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Mirzaa, G., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Kawakami, T.; Mitsui, T.; Kanai, M.; Shirahata, E.; Sendo, D.; Kanno, M.; Noro, M.; Endoh, M.; Hama, A.; Tono, C.; et al. Genetic analysis of Shwachman-Diamond syndrome: Phenotypic heterogeneity in patients carrying identical SBDS mutations. Tohoku J. Exp. Med. 2005, 206, 253–259. [Google Scholar] [CrossRef][Green Version]
- Minelli, A.; Maserati, E.; Nicolis, E.; Zecca, M.; Sainati, L.; Longoni, D.; Lo Curto, F.; Menna, G.; Poli, F.; De Paoli, E.; et al. The isochromosome i(7)(q10) carrying c.258+2t>c mutation of the SBDS gene does not promote development of myeloid malignancies in patients with Shwachman syndrome. Leukemia 2009, 23, 708–711. [Google Scholar] [CrossRef]
- Pressato, B.; Valli, R.; Marletta, C.; Mare, L.; Montalbano, G.; Curto, F.L.; Pasquali, F.; Maserati, E. Deletion of chromosome 20 in bone marrow of patients with Shwachman-Diamond syndrome, loss of the EIF6 gene and benign prognosis. Br. J. Haematol. 2012, 157, 503–505. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.W.; Kennedy, A.; Furutani, E.; Myers, K.; Frattini, A.; Acquati, F.; Roccia, P.; Micheloni, G.; Minelli, A.; Porta, G.; et al. The frequent and clinically benign anomalies of chromosomes 7 and 20 in Shwachman-diamond syndrome may be subject to further clonal variations. Mol. Cytogenet. 2021, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Jamuar, S.S.; Tan, E.-C. Clinical application of next-generation sequencing for Mendelian diseases. Hum. Genom. 2015, 9, 10. [Google Scholar] [CrossRef]
- Qin, D. Next-generation sequencing and its clinical application. Cancer Biol. Med. 2019, 16, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wei, X.; Kong, X.; Guo, X.; Sun, Y.; Man, J.; Du, L.; Zhu, H.; Qu, Z.; Tian, P.; et al. Targeted Next-Generation Sequencing for Clinical Diagnosis of 561 Mendelian Diseases. PLoS ONE 2015, 10, e0133636. [Google Scholar] [CrossRef]
- Hartman, P.; Beckman, K.; Silverstein, K.; Yohe, S.; Schomaker, M.; Henzler, C.; Onsongo, G.; Lam, H.C.; Munro, S.; Daniel, J.; et al. Next generation sequencing for clinical diagnostics: Five year experience of an academic laboratory. Mol. Genet. Metab. Rep. 2019, 19, 100464. [Google Scholar] [CrossRef] [PubMed]
- Boycott, K.M.; Vanstone, M.R.; Bulman, D.E.; MacKenzie, A.E. Rare-disease genetics in the era of next-generation sequencing: Discovery to translation. Nat. Rev. Genet. 2013, 14, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.R.; Eng, C.M. Next-Generation Sequencing to Diagnose Suspected Genetic Disorders. N. Engl. J. Med. 2018, 379, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Morini, J.; Nacci, L.; Babini, G.; Cesaro, S.; Valli, R.; Ottolenghi, A.; Nicolis, E.; Pintani, E.; Maserati, E.; Cipolli, M.; et al. Whole exome sequencing discloses heterozygous variants in the DNAJC21 and EFL1 genes but not in SRP54 in 6 out of 16 patients with Shwachman-Diamond Syndrome carrying biallelic SBDS mutations. Br. J. Haematol. 2019, 185, 627–630. [Google Scholar] [CrossRef]
- Nicora, G.; Zucca, S.; Limongelli, I.; Bellazzi, R.; Magni, P. A machine learning approach based on ACMG/AMP guidelines for genomic variant classification and prioritization. Sci. Rep. 2022, 12, 2517. [Google Scholar] [CrossRef]
- Raczy, C.; Petrovski, R.; Saunders, C.T.; Chorny, I.; Kruglyak, S.; Margulies, E.H.; Chuang, H.-Y.; Källberg, M.; Kumar, S.A.; Liao, A.; et al. Isaac: Ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2013, 29, 2041–2043. [Google Scholar] [CrossRef] [PubMed]
- Nicora, G.; Limongelli, I.; Gambelli, P.; Memmi, M.; Malovini, A.; Mazzanti, A.; Napolitano, C.; Priori, S.; Bellazzi, R. CardioVAI: An automatic implementation of ACMG-AMP variant interpretation guidelines in the diagnosis of cardiovascular diseases. Hum. Mutat. 2018, 39, 1835–1846. [Google Scholar] [CrossRef] [PubMed]
- De Paoli, F.; Limongelli, I.; Zucca, S.; Baccalini, F.; Serpieri, V.; d’Abrusco, F.; Zarantonello, M.; Fabio, G.; Carrabba, M.; Valente, E.M. DIVAs: A phenotype-based machine-learning model to assess the pathogenicity of digenic variant combinations. In European Journal of Human Genetics; 4 CRINAN ST; N1 9XW; Springer Nature Campus: London, UK, 2022; Volume 30, p. 486. [Google Scholar]
- Ewans, L.J.; Schofield, D.; Shrestha, R.; Zhu, Y.; Gayevskiy, V.; Ying, K.; Walsh, C.; Lee, E.; Kirk, E.P.; Colley, A.; et al. Whole-exome sequencing reanalysis at 12 months boosts diagnosis and is cost-effective when applied early in Mendelian disorders. Genet. Med. 2018, 20, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Biesecker, L.G.; Green, R.C. Diagnostic clinical genome and exome sequencing. N. Engl. J. Med. 2014, 371, 1170. [Google Scholar] [CrossRef] [PubMed]
- Mourad, S.; Bilodeau, M.; Roussy, M.; Laramée, L.; Boulianne, L.; Rouette, A.; Jouan, L.; Gendron, P.; Duval, M.; Teira, P.; et al. IDH1 as a Cooperating Mutation in AML Arising in the Context of Shwachman-Diamond Syndrome. Front Oncol. 2019. Available online: https://www.frontiersin.org/article/10.3389/fonc.2019.00772 (accessed on 18 May 2022).
- Kennedy, A.L.; Myers, K.C.; Bowman, J.; Gibson, C.J.; Camarda, N.D.; Furutani, E.; Muscato, G.M.; Klein, R.H.; Ballotti, K.; Liu, S.; et al. Distinct genetic pathways define pre-malignant versus compensatory clonal hematopoiesis in Shwachman-Diamond syndrome. Nat. Commun. 2021, 12, 1334. [Google Scholar] [CrossRef] [PubMed]
- Weniger, M.A.; Küppers, R. Molecular biology of Hodgkin lymphoma. Leukemia 2021, 35, 968–981. [Google Scholar] [CrossRef]
- Baer, S.; Afenjar, A.; Smol, T.; Piton, A.; Gérard, B.; Alembik, Y.; Bienvenu, T.; Boursier, G.; Boute, O.; Colson, C.; et al. Wiedemann-Steiner syndrome as a major cause of syndromic intellectual disability: A study of 33 French cases. Clin. Genet. 2018, 94, 141–152. [Google Scholar] [CrossRef]
- Bochyńska, A.; Lüscher-Firzlaff, J.; Lüscher, B. Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin. Cells 2018, 7, 17. [Google Scholar] [CrossRef]
- Jakovcevski, M.; Ruan, H.; Shen, E.Y.; Dincer, A.; Javidfar, B.; Ma, Q.; Peter, C.J.; Cheung, I.; Mitchell, A.C.; Jiang, Y.; et al. Neuronal Kmt2a/Mll1 Histone Methyltransferase Is Essential for Prefrontal Synaptic Plasticity and Working Memory. J. Neurosci. 2015, 35, 5097–5108. [Google Scholar] [CrossRef]
- Kim, S.Y.; Levenson, J.M.; Korsmeyer, S.; Sweatt, J.D.; Schumacher, A. Developmental Regulation of Eed Complex Composition Governs a Switch in Global Histone Modification in Brain*. J. Biol. Chem. 2007, 282, 9962–9972. [Google Scholar] [CrossRef]
- Foroutan, A.; Haghshenas, S.; Bhai, P.; Levy, M.A.; Kerkhof, J.; McConkey, H.; Niceta, M.; Ciolfi, A.; Pedace, L.; Miele, E.; et al. Clinical Utility of a Unique Genome-Wide DNA Methylation Signature for KMT2A-Related Syndrome. Int. J. Mol. Sci. 2022, 23, 1815. [Google Scholar] [CrossRef]
- Fontana, P.; Passaretti, F.F.; Maioli, M.; Cantalupo, G.; Scarano, F.; Lonardo, F. Clinical and molecular spectrum of Wiedemann-Steiner syndrome, an emerging member of the chromatinopathy family. World J. Med. Genet. 2020, 9, 1–11. [Google Scholar] [CrossRef]
- Sheppard, S.E.; Campbell, I.M.; Harr, M.H.; Gold, N.; Li, D.; Bjornsson, H.T.; Cohen, J.S.; Fahrner, J.A.; Fatemi, A.; Harris, J.R.; et al. Expanding the genotypic and phenotypic spectrum in a diverse cohort of 104 individuals with Wiedemann-Steiner syndrome. Am. J. Med. Genet. Part A 2021, 185, 1649–1665. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.D.; Dafou, D.; McEntagart, M.; Woollard, W.J.; Elmslie, F.V.; Holder-Espinasse, M.; Irving, M.; Saggar, A.K.; Smithson, S.; Trembath, R.C.; et al. De Novo Mutations in MLL Cause Wiedemann-Steiner Syndrome. Am. J. Hum. Genet. 2012, 91, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Stellacci, E.; Onesimo, R.; Bruselles, A.; Pizzi, S.; Battaglia, D.; Leoni, C.; Zampino, G.; Tartaglia, M. Congenital immunodeficiency in an individual with Wiedemann–Steiner syndrome due to a novel missense mutation in KMT2A. Am. J. Med. Genet. Part A 2016, 170, 2389–2393. [Google Scholar] [CrossRef]
- Grangeia, A.; Leão, M.; Moura, C.P. Wiedemann-Steiner syndrome in two patients from Portugal. Am. J. Med. Genet. Part A 2020, 182, 25–28. [Google Scholar] [CrossRef]
- Giangiobbe, S.; Caraffi, S.G.; Ivanovski, I.; Maini, I.; Pollazzon, M.; Rosato, S.; Trimarchi, G.; Lauriello, A.; Marinelli, M.; Nicoli, D.; et al. Expanding the phenotype of Wiedemann-Steiner syndrome: Craniovertebral junction anomalies. Am. J. Med. Genet. Part A 2020, 182, 2877–2886. [Google Scholar] [CrossRef]
- Squeo, G.M.; Augello, B.; Massa, V.; Milani, D.; Colombo, E.A.; Mazza, T.; Castellana, S.; Piccione, M.; Maitz, S.; Petracca, A.; et al. Customised next-generation sequencing multigene panel to screen a large cohort of individuals with chromatin-related disorder. J. Med. Genet. 2020, 57, 760–768. [Google Scholar] [CrossRef]
- Min Ko, J.; Cho, J.S.; Yoo, Y.; Seo, J.; Choi, M.; Chae, J.-H.; Lee, H.-R.; Cho, T.-J. Wiedemann-Steiner Syndrome With 2 Novel KMT2A Mutations: Variable Severity in Psychomotor Development and Musculoskeletal Manifestation. J. Child. Neurol. 2017, 32, 237–242. [Google Scholar] [CrossRef]
- Strom, S.P.; Lozano, R.; Lee, H.; Dorrani, N.; Mann, J.; O’Lague, P.F.; Mans, N.; Deignan, J.L.; Vilain, E.; Nelson, S.F.; et al. De Novo variants in the KMT2A (MLL) gene causing atypical Wiedemann-Steiner syndrome in two unrelated individuals identified by clinical exome sequencing. BMC Med. Genet. 2014, 15, 49. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, G.; Liu, H.; Zhang, X.; Huang, Z.; Yan, H.; Wang, L.; Fan, Y.; Gu, X.; Yu, Y. Further delineation of the phenotype of truncating KMT2A mutations: The extended Wiedemann–Steiner syndrome. Am. J. Med. Genet. Part A 2017, 173, 510–514. [Google Scholar] [CrossRef]
- Bogaert, D.J.; Dullaers, M.; Kuehn, H.S.; Leroy, B.P.; Niemela, J.E.; De Wilde, H.; De Schryver, S.; De Bruyne, M.; Coppieters, F.; Lambrecht, B.N.; et al. Early-onset primary antibody deficiency resembling common variable immunodeficiency challenges the diagnosis of Wiedeman-Steiner and Roifman syndromes. Sci. Rep. 2017, 7, 3702. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; Yang, Y.; Wang, P.; Huang, H.; Xiong, S.; Sun, L.; Cheng, M.; Song, C.; Cheng, X.; et al. Description of the molecular and phenotypic spectrum of Wiedemann-Steiner syndrome in Chinese patients. Orphanet J. Rare Dis. 2018, 13, 178. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, N.; Giurgea, I.; Goldenberg, A.; Dieux, A.; Afenjar, A.; Ghoumid, J.; Diebold, B.; Mietton, L.; Briand-Suleau, A.; Billuart, P.; et al. Molecular and cellular issues of KMT2A variants involved in Wiedemann-Steiner syndrome. Eur. J. Hum. Genet. 2018, 26, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Perobelli, S.; Nicolis, E.; Assael, B.M.; Cipolli, M. Further characterization of Shwachman–Diamond syndrome: Psychological functioning and quality of life in adult and young patients. Am. J. Med. Genet. Part A 2012, 158A, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Rodriguez-Buritica, D.F.; Northrup, H. Wiedemann-Steiner syndrome: Novel pathogenic variant and review of literature. Eur. J. Med. Genet. 2017, 60, 285–288. [Google Scholar] [CrossRef]
- Castiglioni, S.; Di Fede, E.; Bernardelli, C.; Lettieri, A.; Parodi, C.; Grazioli, P.; Colombo, E.A.; Ancona, S.; Milani, D.; Ottaviano, E.; et al. KMT2A: Umbrella Gene for Multiple Diseases. Genes 2022, 13, 514. [Google Scholar] [CrossRef]
- Nardello, R.; Mangano, G.D.; Fontana, A.; Gagliardo, C.; Midiri, F.; Borgia, P.; Brighina, F.; Raieli, V.; Mangano, S.; Salpietro, V. Broad neurodevelopmental features and cortical anomalies associated with a novel de novo KMT2A variant in Wiedemann−Steiner syndrome. Eur. J. Med. Genet. 2021, 64, 104133. [Google Scholar] [CrossRef]
- Demir, S.; Gürkan, H.; Öz, V.; Yalçıntepe, S.; Atlı, E.İ.; Atlı, E. Wiedemann-Steiner Syndrome as a Differential Diagnosis of Cornelia de Lange Syndrome Using Targeted Next-Generation Sequencing: A Case Report. MSY 2021, 12, 46–51. [Google Scholar] [CrossRef]
- Suleiman, J.; Riedhammer, K.M.; Jicinsky, T.; Mundt, M.; Werner, L.; Gusic, M.; Burgemeister, A.L.; Alsaif, H.S.; Abdulrahim, M.; Moghrabi, N.N.; et al. Homozygous loss-of-function variants of TASP1, a gene encoding an activator of the histone methyltransferases KMT2A and KMT2D, cause a syndrome of developmental delay, happy demeanor, distinctive facial features, and congenital anomalies. Hum. Mutat. 2019, 40, 1985–1992. [Google Scholar] [CrossRef]
- Peterson, J.F.; Baughn, L.B.; Pearce, K.E.; Williamson, C.M.; Benevides Demasi, J.C.; Olson, R.M.; Goble, T.A.; Meyer, R.G.; Greipp, P.T.; Ketterling, R.P. KMT2A (MLL) rearrangements observed in pediatric/young adult T-lymphoblastic leukemia/lymphoma: A 10-year review from a single cytogenetic laboratory. Genes Chromosomes Cancer 2018, 57, 541–546. [Google Scholar] [CrossRef] [PubMed]
| UPN42 | UPN43 |
|---|---|
| HP:0001875 Neutropenia | HP:0000085 Horseshoe kidney |
| HP:0005528 Bone marrow hypocellularity | HP:0001875 Neutropenia |
| HP:0001738 Exocrine pancreatic insufficiency | HP:0005528 Bone marrow hypocellularity |
| HP:0001873 Thrombocytopenia | HP:0001738 Exocrine pancreatic insufficiency |
| HP:0040088 Abnormal lymphocyte count | HP:0003025 Metaphyseal irregularity |
| HP:0012189 Hodgkin lymphoma | HP:0012758 Neurodevelopmental delay |
| HP:0001873 Thrombocytopenia | |
| HP:0000028 Cryptorchidism | |
| HP:0002719 Recurrent infection | |
| HP:0000431 Wide nasal bridge | |
| HP:0000316 Hypertelorism | |
| HP:0002474 Expressive-language delay |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taha, I.; De Paoli, F.; Foroni, S.; Zucca, S.; Limongelli, I.; Cipolli, M.; Danesino, C.; Ramenghi, U.; Minelli, A. Phenotypic Variation in Two Siblings Affected with Shwachman-Diamond Syndrome: The Use of Expert Variant Interpreter (eVai) Suggests Clinical Relevance of a Variant in the KMT2A Gene. Genes 2022, 13, 1314. https://doi.org/10.3390/genes13081314
Taha I, De Paoli F, Foroni S, Zucca S, Limongelli I, Cipolli M, Danesino C, Ramenghi U, Minelli A. Phenotypic Variation in Two Siblings Affected with Shwachman-Diamond Syndrome: The Use of Expert Variant Interpreter (eVai) Suggests Clinical Relevance of a Variant in the KMT2A Gene. Genes. 2022; 13(8):1314. https://doi.org/10.3390/genes13081314
Chicago/Turabian StyleTaha, Ibrahim, Federica De Paoli, Selena Foroni, Susanna Zucca, Ivan Limongelli, Marco Cipolli, Cesare Danesino, Ugo Ramenghi, and Antonella Minelli. 2022. "Phenotypic Variation in Two Siblings Affected with Shwachman-Diamond Syndrome: The Use of Expert Variant Interpreter (eVai) Suggests Clinical Relevance of a Variant in the KMT2A Gene" Genes 13, no. 8: 1314. https://doi.org/10.3390/genes13081314
APA StyleTaha, I., De Paoli, F., Foroni, S., Zucca, S., Limongelli, I., Cipolli, M., Danesino, C., Ramenghi, U., & Minelli, A. (2022). Phenotypic Variation in Two Siblings Affected with Shwachman-Diamond Syndrome: The Use of Expert Variant Interpreter (eVai) Suggests Clinical Relevance of a Variant in the KMT2A Gene. Genes, 13(8), 1314. https://doi.org/10.3390/genes13081314






