The Identification by Exome Sequencing of Candidate Genes in BRCA-Negative Tunisian Patients at a High Risk of Hereditary Breast/Ovarian Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. DNA Extraction and Exome Sequencing
2.3. Functional Annotation and Variants’ Prioritization
2.4. Survival Analysis
3. Results
3.1. Clinical Characteristics of BRCAness Patients
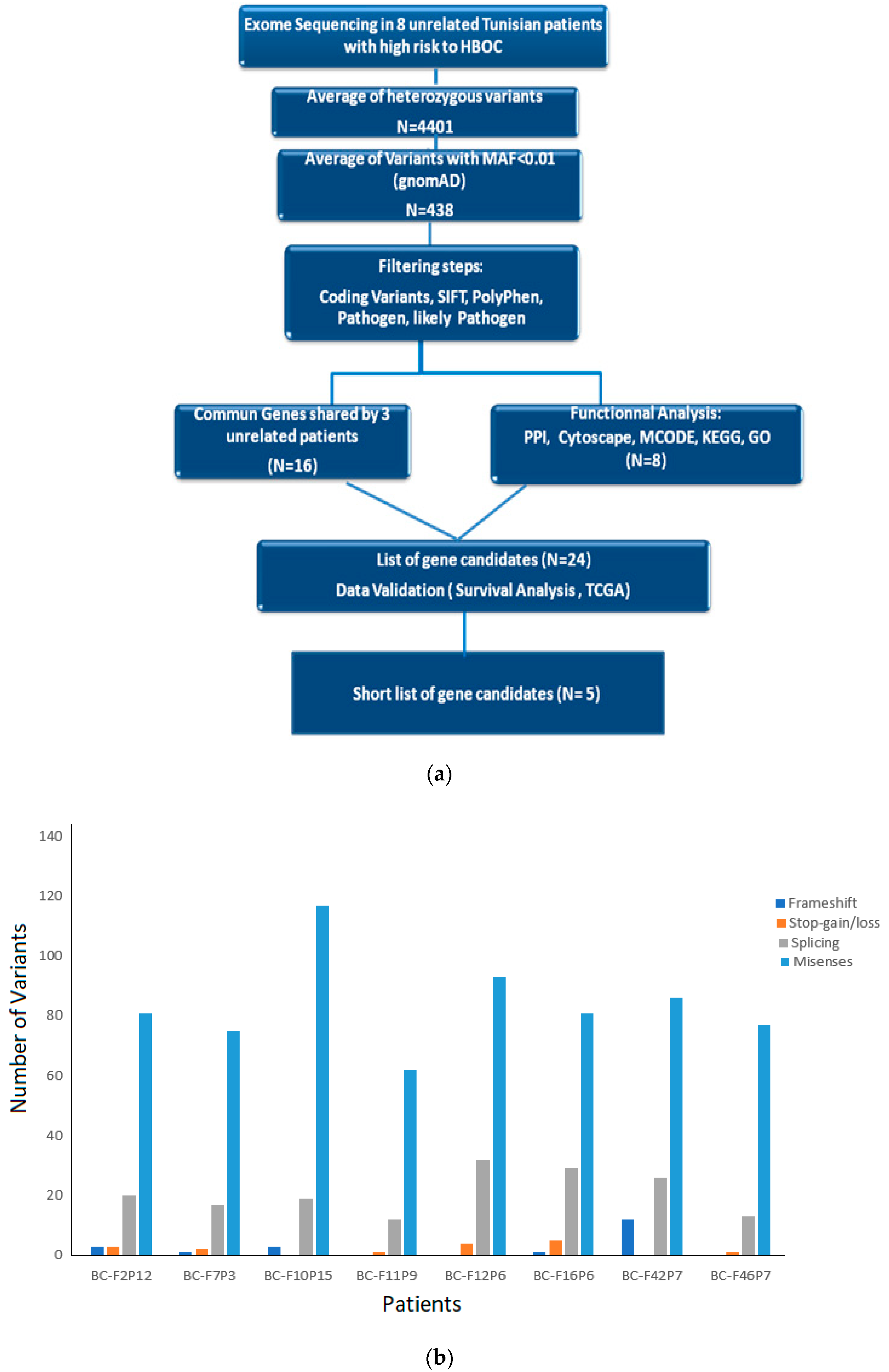
3.2. Exome Sequencing and Data Analysis
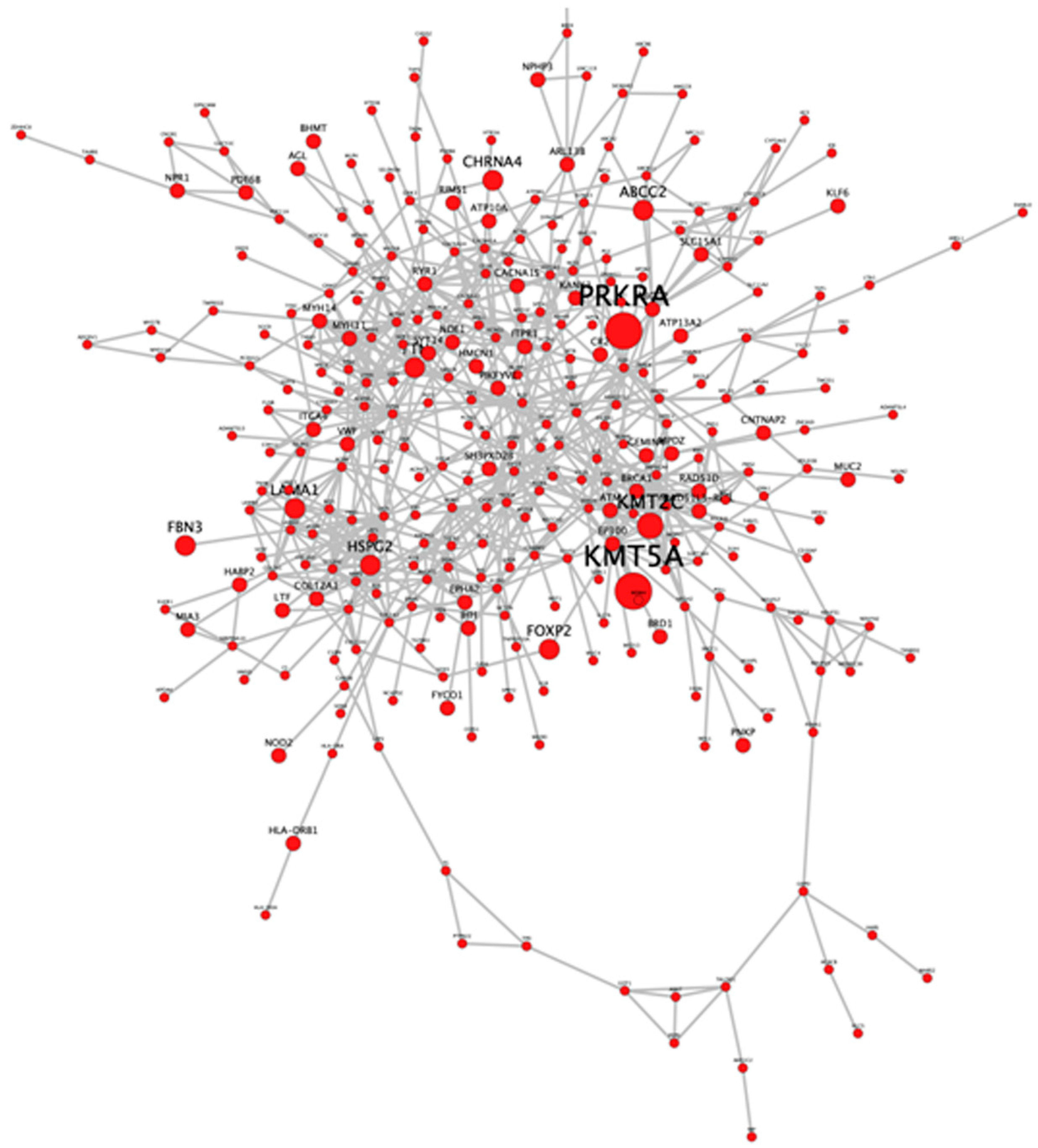
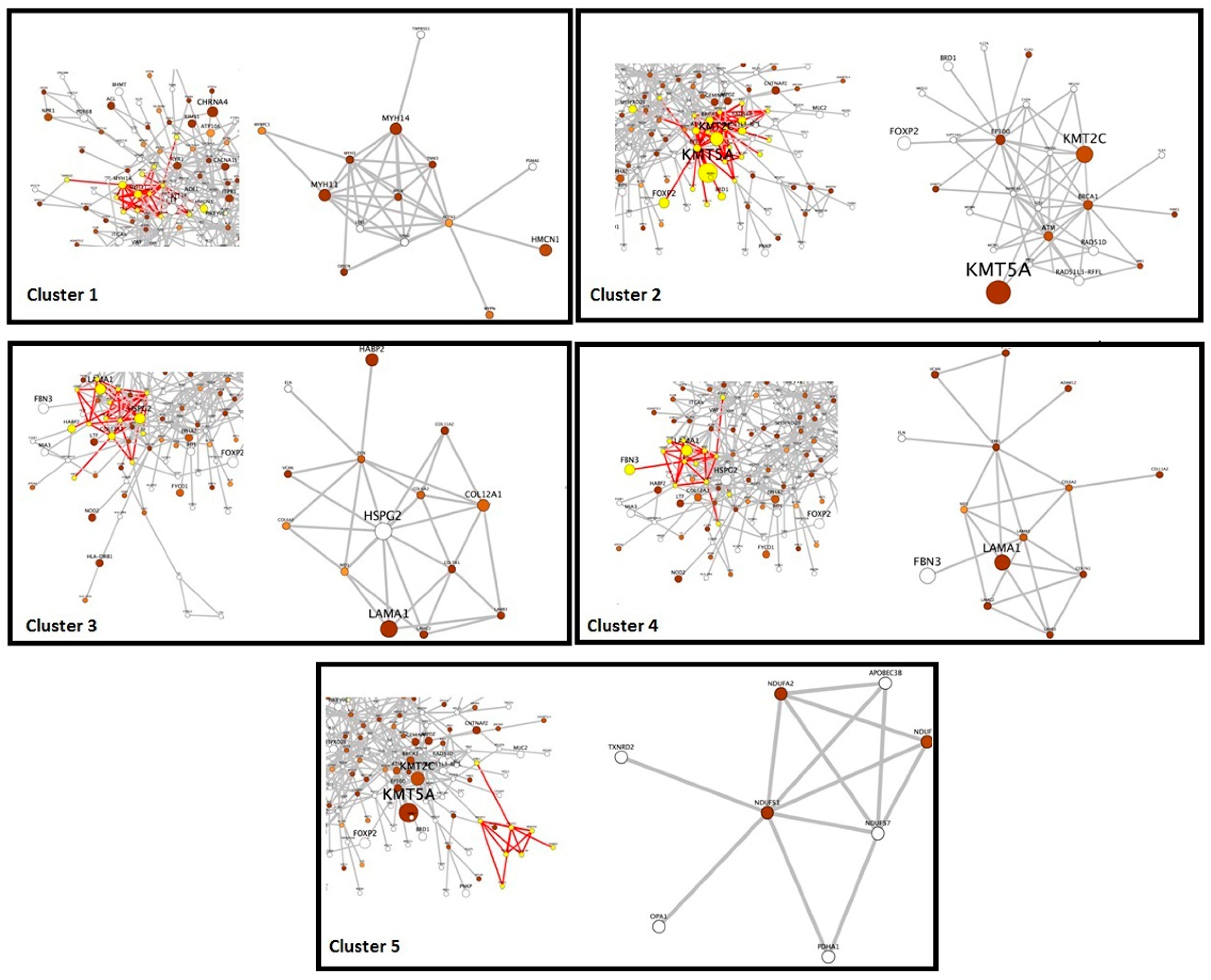
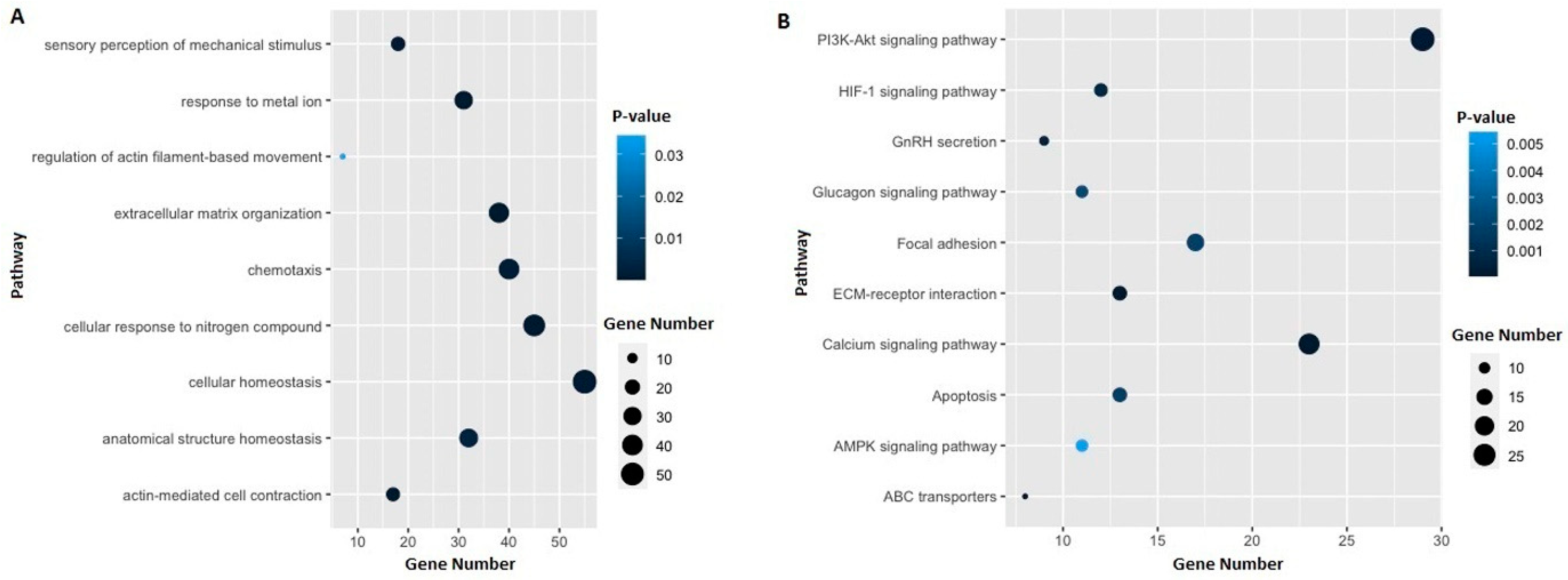
3.3. Identification of Potential Gene Candidates Predisposed to HBOC
3.4. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACMG | American College of Medical Genetics and Genomics |
| B | Benign |
| BC | Breast Cancer |
| BWA | Burrows- Wheeler Aligner |
| DFS | Disease Free Survival |
| ER | Eostrogen Receptor |
| ES | Exome Sequencing |
| GO | Gene Ontology |
| HBOC | Hereditary Breast Ovarian/Cancer |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LB | Likely Benign |
| LOF | Loss of Function |
| LP | Likely Pathogen |
| MAF | Minor Allele Frequency |
| MCODE | Molecular Complex Detection |
| NA | No Available |
| NGS | Next Generation Sequencing |
| NR | No Reported |
| OC | Ovarian Cancer |
| OS | Overall Survival |
| P | Pathogen |
| PF | Pass Filter |
| PPI | Protein-Protein Interactions |
| PR | Progesteron Receptor |
| PVs | Pathogenic Variants |
| SBR grade | SBR Scarff-Bloom-Richardson (grade) |
| TCGA | The Cancer Genome Atlas |
| TNBC | Triple Negative Breast Cancer |
| VUS | Variant Unknown Significance |
| WES | Whole Exome Sequencing |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.P.; Quaresma, M.; Berrino, F.; Lutz, J.; de Angelis, R.; Capocaccia, R.; Garber, J.E.; Boyd, J.; Lubin, M.B.; DeShano, M.L.; et al. Cancer survival in five continents: A worldwide population-based study (CONCORD). Lancet. Oncol. 2008, 9, 730–756. [Google Scholar] [CrossRef]
- Maalej, M.; Hentati, D.; Messai, T.; Kochbati, L.; El May, A.; Mrad, K.; Romdhane, K.B.; Abdallah, M.B.; Zouari, B. Breast cancer in Tunisia in 2004: A comparative clinical and epidemiological study. Bul. Cancer 2008, 95, 10005–10009. [Google Scholar]
- Missaoui, N.; Jaidene, L.; Abdelkrim, S.B.; Ben Abdelkader, A.; Beizig, N.; Ben Yaacoub, L.; Yaacoubi, M.T.; Hmissa, S. Breast cancer in Tunisia: Clinical and pathological findings. Asian Pac. J. Cancer Prev. 2011, 121, 69–72. [Google Scholar]
- Younes, N.; Zayed, H. Genetic epidemiology of ovarian cancer in the 22 Arab countries: A systematic review. Gene 2019, 684, 154–164. [Google Scholar] [CrossRef]
- Claus, E.B.; Schildkraut, J.M.; Thompson, W.D.; Risch, N.J. The genetic attributable risk of breast and ovarian cancer. Cancer 1996, 77, 2318–2324. [Google Scholar] [CrossRef]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.L.; Bennett, M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef]
- Wooster, R.; Bignell, G.; Lancaster, J.; Swift, S.; Seal, S.; Mangion, J.; Collins, N.; Gregory, S.; Gumbs, C.; Micklem, G. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995, 378, 789–792. [Google Scholar] [CrossRef]
- Antoniou, A.; Pharoah, P.D.; Narod, S.; Risch, H.A.; Eyfjord, J.E.; Hopper, J.L.; Loman, N.; Olsson, H.; Johannsson, O.; Borg, A.; et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: A combined analysis of 22 studies. Am. J. Hum. Genet. 2003, 7, 1117–1130. [Google Scholar] [CrossRef]
- Levy-Lahad, E.; Friedman, E. Cancer risks among BRCA1 and BRCA2 mutation carriers. Br. J. Cancer 2007, 96, 11–15. [Google Scholar] [CrossRef]
- Fackenthal, J.D.; Olopade, O.I. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat. Rev. Cancer 2007, 7, 937. [Google Scholar] [CrossRef] [PubMed]
- Abdulrashid, K.; AlHussaini, N.; Ahmed, W.; Thalib, L. Prevalence of BRCA mutations among hereditary breast and/or ovarian cancer patients in Arab countries: Systematic review and meta-analysis. BMC Cancer 2019, 19, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Troudi, W.; Uhrhammer, N.; Sibille, C.; Dahan, C.; Mahfoudh, W.; Bouchlaka Souissi, C.; Jalabert, T.; Chouchane, L.; Bignon, Y.J.; Ben Ayedet, F.; et al. Contribution of the BRCA1 and BRCA2 mutations to breast cancer in Tunisia. J. Hum. Genet. 2007, 52, 915–920. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fourati, A.; Louchez, M.M.; Fournier, J.; Amor, G.; Rahal, K.; El May, M.V.; El May, A.; Revillion, F.; Peyrat, J.P. Screening for common mutations in BRCA1 and BRCA2 genes: Interest in genetic testing of Tunisian families with breast and/or ovarian cancer. Bull. Cancer 2014, 101, 6–40. [Google Scholar] [CrossRef] [PubMed]
- Riahi, A.; Kharrat, M.; Ghourabi, M.E.; Khomsi, F.; Gamoudi, A.; Lariani, I.; May, A.E.; Rahal, K.; Chaabouni-Bouhamed, H. Mutation spectrum and prevalence of BRCA1 and BRCA2 genes in patients with familial and early-onset breast/ovarian cancer from Tunisia. Clin. Genet. 2015, 87, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Riahi, A.; Radmanesh, H.; Schürmann, P.; Bogdanova, N.; Geffers, R.; Meddeb, R.; Kharrat, M.; Dörk, T. Exome sequencing and case-control analyses identify RCC1 as a candidate breast cancer susceptibility gene. Int. J. Cancer 2018, 142, 2512–2517. [Google Scholar] [CrossRef]
- Ben Ayed-Guerfali, D.; Ben Kridis-Rejab, W.; Ammous-Boukhris, N.; Ayadi, W.; Charfi, S.; Khanfir, A.; Sellami-Boudawara, T.; Frikha, M.; Daoud, J.; Mokdad-Gargouri, R. Novel and recurrent BRCA1/BRCA2 germline mutations in patients with breast/ovarian cancer: A series from the south of Tunisia. J. Transl. Med. 2021, 19, 108. [Google Scholar] [CrossRef]
- Bakkach, J.; Mansouri, M.; Derkaoui, T.; Loudiyi, A.; El Fahime, E.M.; Barakat, A.; Ghailani Nourouti, N.; De Villarrea, J.M.; Bringas, C.C.; Mechita, M.B. Contribution of BRCA1 and BRCA2 germline mutations to early onset breast cancer: A series from north of Morocco. BMC Cancer 2020, 20, 859. [Google Scholar] [CrossRef]
- Mehemmai, C.; Cherbal, F.; Hamdi, Y.; Guedioura, A.; Benbrahim, W.; Bakour, R.; Abdelhak, S. BRCA1 and BRCA2 Germline Mutation Analysis in Hereditary Breast/Ovarian Cancer Families from the Aures Region (Eastern Algeria): First Report. Pathol. Oncol. Res. 2020, 26, 715–726. [Google Scholar] [CrossRef]
- Saied, M.H.; Elkaffash, D.; Fadl, R.; Abdel Haleem, R.; Refeat, A.; Ibrahim, I.; Tahoun, M.; Elkayal, A.; Tayae, E. Preliminary results of targeted sequencing of BRCA1 and BRCA2 in a cohort of breast cancer families: New insight into pathogenic variants in patients and at-risk relatives. Mol. Med. Rep. 2021, 24, 678. [Google Scholar] [CrossRef]
- Mighri, N.; Hamdi, Y.; Boujemaa, M.; Othman, H.; Ben Nasr, S.; El Benna, H.; Mejri, N.; Labidi, S.; Ayari, J.; Jaidene, O.; et al. Identification of Novel BRCA1 and RAD50 Mutations Associated with Breast Cancer Predisposition in Tunisian Patients. Front. Genet. 2020, 11, 552971. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, K.; Lindström, S.; Beesley, D.J.; Hui, S.; Kar, S.; Lemaçon, A.; Penny, S.; Glubb, D.; Rostamianfar, A.; Bolla, M.K.; et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017, 551, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Peock, S.; Frost, D.; Ellis, S.; Platte, R.; Fineberg, E.; Gareth Evans, D.; Izatt, L.; AEeles, R.; Adlard, J.; et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from prospective analysis of EMBRACE. J. Natl. Cancer Inst. 2013, 105, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Sokolenko, A.P.; Suspitsin, E.N.; Kuligina, E.S.; Bizin, I.V.; Frishman, D.; Imyanitov, E.N. Identification of novel hereditary cancer genes by whole exome sequencing. Cancer Lett. 2015, 369, 274–288. [Google Scholar] [CrossRef]
- Zelli, V.; Compagnoni, C.; Cannita, K.; Capelli, R.; Capalbo, C.; Di Vito Nolfi, M.; Alesse, E.; Zazzeroni, F.; Tessitore, A. Applications of Next Generation Sequencing to the Analysis of Familial Breast/Ovarian Cancer. High Throughput 2020, 9, 1. [Google Scholar] [CrossRef]
- Hamdi, Y.; Boujemaa, M.; Ben Rekaya, M.; Ben Hamda, C.; Mighri, N.; El Benna, H.; Mejri, N.; Labidi, S.; Daoud, N.; Naouali, C.; et al. Family specific genetic predisposition to breast cancer: Results from Tunisian whole exome sequenced breast cancer cases. J. Trans. Med. 2018, 16, 158. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galonet, J.; et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.R.; Bilgili, E.P.; Merner, N.D. A review of whole-exome sequencing efforts toward hereditary breast cancer susceptibility gene discovery. Hum. Mutat. 2016, 37, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cai, Q.; Shu, X.O.; Gao, Y.T.; Li, C.; Zheng, W.; Long, J. Whole-Exome Sequencing Identifies Novel Somatic Mutations in Chinese Breast Cancer Patients. Mol. Genet. Med. 2015, 9, 183. [Google Scholar] [CrossRef] [PubMed]
- Jalkh, N.; Chouery, E.; Haidar, Z.; Khater, C.; Atallah, D.; Ali, H. Next-generation sequencing in familial breast cancer patients from Lebanon. BMC Med. Genom. 2017, 10, 8. [Google Scholar] [CrossRef]
- Kuligina, E.S.; Sokolenko, A.P.; Bizin, I.V.; Haidar, Z.; Khater, C.; Atallah, D.; Ali, H.; Marafie MJAl-Mulla, M.R.; Al-Mulla, F.; Andre, M. Exome sequencing study of Russian breast cancer patients suggests a predisposing role for USP39. Breast Cancer Res. Treat. 2020, 179, 731–742. [Google Scholar] [CrossRef]
- Doddato, G.; Valentino, F.; Giliberti, A.; Papa, F.T.; Tita, R.; Bruno, L.P.; Resciniti, S.; Fallerini, C.; Benetti, E.; Palmieri, M.; et al. Exome sequencing in BRCA1-2 candidate familias: The contribution of other cancer susceptibility genes. Front. Oncol. 2021, 11, 649435. [Google Scholar]
- Felicio, P.S.; Grasel, R.S.; Campacci, N.P.; Galvão, A.E.; Torrezan, H.C.R.; Sabato, C.S.; Fernandes, G.C.; Souza, C.P.; Michelli, R.D. Whole-exome sequencing of non-BRCA1/BRCA2 mutation carrier cases at high-risk for hereditary breast/ovarian cancer. Hum. Mutat. 2021, 42, 290–299. [Google Scholar] [CrossRef]
- Easton, D.F.; Pharoah, P.D.; Antoniou, A.C.; Tischkowitz, M.; Tavtigian, S.V.; Nathanson, K.L.; Devilee, P.; Meindl, A.; Couch, F.J.; Southey, M.; et al. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 2015, 372, 2243–2257. [Google Scholar] [CrossRef]
- Southey, M.C.; Goldgar, D.E.; Winqvist, R.; Pylkäs, K.; Couch, F.; Tischkowitz, M.; Foulkes, W.D.; Dennis, J.; Michailidou, K.; Rensburget, E.J.; et al. PALB2, CHEK2 and ATM rare variants and cancer risk: Data from COGS. J. Med. Genet. 2016, 53, 800–811. [Google Scholar] [CrossRef]
- Seals, D.F.; Azucena, E.F.; Pass, I.; Tesfay, L.; Gordon, R.; Woodrow, M.; Resau, J.H.; Courtneidge, S.A. The adaptor protein Tks5/Fish isrequired for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell 2005, 7, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Carman, C.V.; Sage, P.T.; Sciuto, T.E.; de la Fuente, M.A.; Geha, R.S.; Ochs, H.D.; Dvorak, H.F.; Dvorak, A.M.; Springer, T.A. Transcellular diapedesis is initiated by invasive podosomes. Immunity 2007, 26, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gammon, M.D.; Zeisel, S.H.; Lee, Y.L.; Wetmur, J.G.; Teitelbaum, S.L.; Bradshaw, P.T.; Neugut, A.I.; Santella, R.M.; Choline, J.C. Metabolism and risk of breast cancer in a population-based study. FASEB J. 2008, 22, 2045–2052. [Google Scholar] [CrossRef]
- Glunde, K.; Bhujwalla, Z.M.; Ronen, S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer 2011, 11, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Swanton, C.; McGranahan, N.; Starrett, G.J.; Harris, R.S. APOBEC enzymes: Mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015, 5, 704–712. [Google Scholar] [CrossRef]
- Asaoka, M.; Santosh, K.P.; Ishikawa, T.; Takabe, K. Different members of the APOBEC3 family of DNA mutators have opposing associations with the landscape of breast cancer. Am. J. Cancer Res. 2021, 11, 5111–5125. [Google Scholar]
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366–370. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Willis, S.; Burns, M.B.; Look, M.P.; Meijer-Van Gelder, M.E.; Schlicker, A.; Heideman, M.R.; Jacobs, H.; Wessels, L.; Leyland-Jones, B. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm. Cancer 2014, 5, 405–413. [Google Scholar] [CrossRef]
- Harris, R.S. Molecular mechanism and clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer. Breast Cancer Res. 2015, 17, 8. [Google Scholar] [CrossRef]
- Høye, A.M.; Erler, J.T. Structural ECM components in the premetastatic and metastatic niche. Am. J. Physiol. Cell 2016, 310, 955–967. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, X.; Zheng, Y.; Chen, Y.; Fei, W.; Wang, F.; Zheng, C. Extracellular Matrix: Emerging Roles and Potential Therapeutic Targets for Breast Cancer. Front. Oncol. 2021, 11, 650453. [Google Scholar] [CrossRef] [PubMed]
- Gala, K.; Li, Q.; Sinha, A.; Razavi, P.; Dorso, M.; Sanchez-Vega, F.; Chung, Y.R.; Hendrickson, R.; Hsieh, J.J. KMT2C mediates the estrogen dependence of breast cancer through regulation of ERα enhancer function. Oncogene 2018, 37, 4692–4710. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Qiao, P.; Yin, G.; Sun, Y.; Yu, X.; Sun, X.; Chu, Y.; Wang, Y. RHPN2 Promotes Malignant Cell Behaviours in Ovarian Cancer by Activating STAT3 Signalling. Onco. Targets Ther. 2020, 10, 11517–11527. [Google Scholar] [CrossRef] [PubMed]
- Myatt, S.S.; Lam, E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 2007, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- López de Silanes, I.; Paz Quesada, M.; Esteller, M. Aberrant regulation of messenger RNA 3’-untranslated region in human cancer. Cell Oncol. 2007, 29, 1–17. [Google Scholar] [CrossRef]
- Vislovukh, A.; Rivera Vargas, T.; Polesskaya, A.; Groisman, I. Role of 3′-untranslated region translational control in cancer development, diagnostics and treatment. World J. Biol. Chem. 2014, 26, 40–57. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Tzanakakis, G.N.; Karamanos, N.K. Proteoglycans in health and disease: Novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J. 2010, 277, 3904–3923. [Google Scholar] [CrossRef]
- Kalscheuer, S.; Khanna, V.; Kim, H.; Li, S.; Sachdev, D.; DeCarlo, A.; Yang, D.; Panyam, J. Discovery of HSPG2 (Perlecan) as a Therapeutic Target in Triple Negative Breast Cancer. Sci. Rep. 2019, 9, 12492. [Google Scholar] [CrossRef]
- Johrer, K.; Zelle-Rieser, C.; Perathoner, A.; Moser, P.; Hager, M.; Ramoner, R.; Gander, H.; Höltl, L.; Bartsch, G.; Greil, R. Up-regulation of functional chemokine receptor CCR3 in human renal cell carcinoma. Clin. Cancer Res. 2005, 11, 2459–2465. [Google Scholar] [CrossRef]
- Miyagaki, T.; Sugaya, M.; Fujita, H.; Ohmatsu, H.; Kakinuma, T.; Kadono, T.; Tamaki, K.; Sato, S. Eotxins and CCR3 interaction regulates the Th2 environment of cutaneous T-cell lymphoma. J. Investig. Dermatol. 2010, 130, 2304–2311. [Google Scholar] [CrossRef]
- Miyagaki, T.; Sugaya, M.; Murakami, T.; Asano, Y.; Tada, Y.; Kadono, T.; Okochi, H.; Tamaki, K.; Sato, S. CCL11-CCR3 interactions promote survival of anaplastic large cell lymphoma cells via ERK1/2 activation. Cancer Res. 2011, 71, 2056–2065. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.H.; Fan, L.; Chen, H.Y.; Ding, K.F.; Yu, K.D. Intratumoral expression of CCR3 in breast cancer is associated with improved relapse-free survival in luminal-like disease. Oncotarget 2016, 7, 28570–28578. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yamaguchi, M.; Takagi, K.; Narita, K.; Miki, Y.; Onodera, Y.; Miyashita, M.; Sasano, H.; Suzuki, T. Stromal CCL5 Promotes Breast Cancer Progression by Interacting with CCR3 in Tumor Cells. Int. J. Mol. Sci. 2021, 22, 1918. [Google Scholar] [CrossRef] [PubMed]
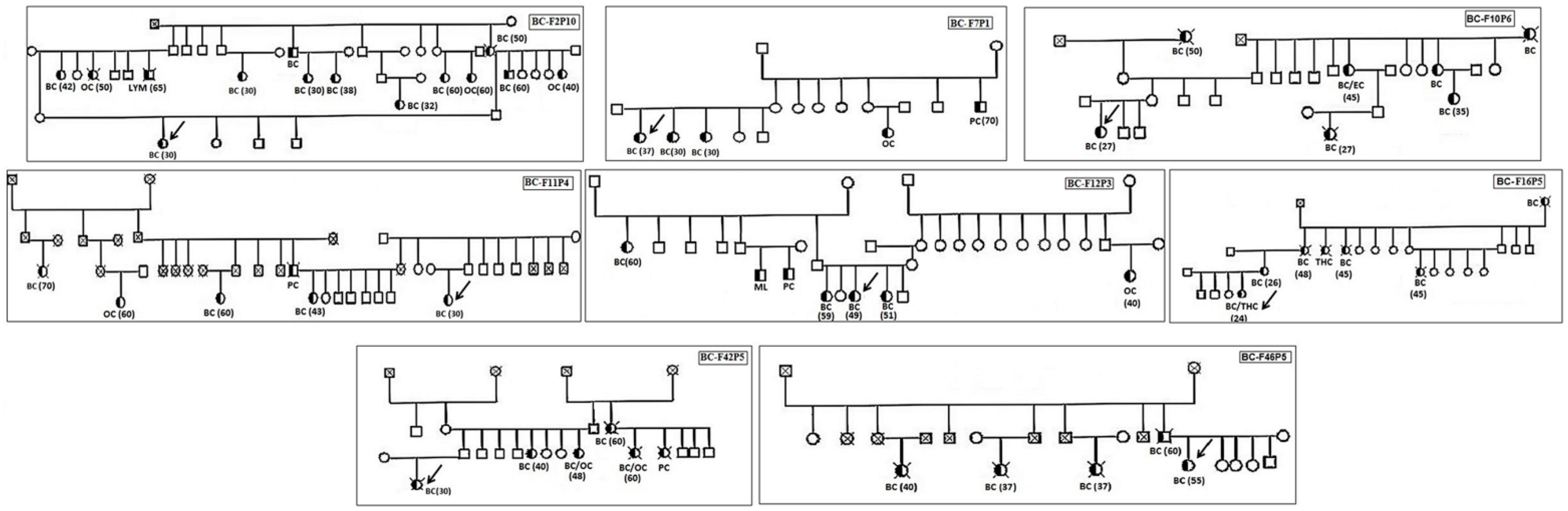


| Patients | Age at Diagnosis | Family History of BC/OC | Family History of Other Cancers | Histological Type | SBR Grade | Tumor Size | TNM | ER | PR | Her-2 | Therapy | Follow-Up |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BC-F2P10 | 30 | 9BC, 2MBC, 3OC | - | IDC | II | 2 | I | - | - | - | TAM, FEC, TXT, RT | NA |
| BC-F7P1 | 37 | 3BC, 1OC | Prostate | IDC | II | 3 | IIB | + | + | - | MCA, TAM, FEC, TXT, RT | Metastasis after 3 y of BC. Died with the disease progression |
| BC-F10P6 | 27 | 7BC | Endometrial | IDC | III | 4.5 | IIA | + | + | - | MCA, TAM, FEC, TXT, RT | Remission with OS of 1 y |
| BC-F11P4 | 30 | 4BC, 1OC | Prostate | IDC | III | 5.3 | IIB | - | - | - | FEC, RT | Remission with OS of 16 y |
| BC-F12P3 | 49 | 4BC, 1OC | Prostate Melanoma | IDC | II | 4.5 | IIB | + | - | - | MCA, TAM, RT | Remission with OS of 6.5 y |
| BC-F16P5 | 24 | 5BC, 1BC/Thy | Thyroid | IDC | II | 2.5 | IIA | + | + | - | TAM, FEC, TXT, RT | Remission with OS of 4.5 y |
| BC-F42P5 | 30 | 3BC, 2BC/OC | Prostate | IDC | II | 4 | IIB | + | + | - | MCA, TAM, FEC, C75, RT | NA |
| BC-F46P5 | 55 | 4BC, 1MBC | - | IDC | II | 2 | IA | + | + | - | TAM, FEC, TXT, RT | Remission with OS of 1 y |
| Patients | Gene | rs | Variation | MAF | Consequence | ClinVar |
|---|---|---|---|---|---|---|
| BC-F2P10 | PMS1 | 778185859 | c.278G>A; p.Arg93His | 0.000062 | VUS | NR |
| RAD54B | 35973866 | c.247A>G; p.(Ser83Gly) | 0.0408 | VUS | NR | |
| ATM | 587781352 | c.2492A>G; p.(Asp831Gly) | 0.000016 | VUS | R | |
| RAD51D | c.413A>G; p.(Asn138Ser) | 0.000756 | VUS | R | ||
| EP300 | 1268191227 | c.3976G>A; p.(Val1326Ile) | P | R | ||
| MUTYH | 140118273 | c.1544C>T; p.(Ser515Phe) | 0.0234 | B | R | |
| BC-F7P1 | ATM | 2227922 | c.1810C>T; p.(Pro604Ser) | 0.0317 | LB | R |
| RAD52 | 867412462 | c.1048G>A; p.(Asp350Asn)) | 0.000018 | VUS | NR | |
| RAD54B | 116312454 | c.2639A>G; p.(Asp880Gly) | 0.0043 | LB | R | |
| BC-F10P6 | FANCE | 768911543 | c.298T>A; p.(Ser100Thr) | 0.00072 | VUS | R |
| PMS2 | c.706-4dup | VUS | NR | |||
| EXO1 | 4150001 | c.2276G>A; p.Gly759Glu | 0.0338 | LB | R | |
| RAD52 | 4987206 | c.661C>G; p.Gln221Glu | 0.0325 | B | R | |
| BC-F11P4 | RAD51D | NR | c.*1056C>T | NA | LB | NR |
| BC-F12P3 | FANCL | 770368316 | c.288G>T; p.Lys96Asn | 0.000416 | VUS | R |
| POLL | 61757734 | c.169C>T; p.Arg57Trp | 0.0026 | VUS | NR | |
| POLH | 35675573 | c.986C>T; p.Thr329Ile | 0.017 | LB | R | |
| BC-F16P5 | ATM | 864622251 | c.6115G>A; p.Glu2039Lys | 0.000033 | VUS | R |
| DCC | 138724679 | c.527A>G; p.Asn176Ser | 0.0005 | VUS | R | |
| BARD1 | 61754118 | c.2212A>G; p.Ile738Val | 0.027 | LB | R | |
| XRCC1 | 143917286 | c.818C>T; p.Pro273Leu | 0.00311 | LB | R | |
| RAD52 | 4987206 | c.661C>G; p.Gln221Glu | 0.0325 | B | R | |
| RAD51D | NR | c.*106G>A | NA | LB | NR | |
| BC-F42P5 | BLM | 141503266 | c.254G>C; p.Arg85Thr | 0.01 | LB | R |
| PMS2 | 63750055 | c.1711C>A; p.Leu571Ile | 0.02 | LB | NR | |
| BC-F46P5 | EP300 | c.227C>G; p.Ser76Cys | VUS | |||
| RAD54B | 2919661 | cc.289G>C; p.Asp97His | 0.018 | VUS | NR |
| Patient | Gene | rs | Variation | MAF | Consequence | ClinVar |
|---|---|---|---|---|---|---|
| BC-F2P10 | PCK2 | 753706965 | c.577c>T; p.Arg193* | 0.000415 | P | R |
| PDE6B | NR | c.125_126insTGCGA; p.Asp43Alafs*109 | NA | LP | NR | |
| PDE6B | NR | c.120_121insGAGGA; p.Pro41Glufs*111 | NA | LP | NR | |
| IL31RA | 144337484 | c.700C>T; p.Arg234* | 0.000163 | LP | NR | |
| TTC37 | 768215813 | c.1708C>T; p.Arg570* | 0.000054 | LP | NR | |
| SH3PXD2B | 551498843 | c.2626_2629del; p.Phe876Lysfs*16 | 0.005594 | P | R | |
| BC-F7P1 | ZnHIT6 | NR | c.1114G>T; p.Glu372* | NA | LP | NR |
| BHMT | 763726268 | c.1200del; p.Lys400Asnfs*15 | 0.000746 | LP | R | |
| MEGF10 | NR | c.122C>A; p.Ser41* | NA | LP | NR | |
| BC-F10P6 | BHMT | 763726268 | c.1200del; p.Lys400AsnfsTer15 | 0.000746 | R | |
| Cyp3A5 | 28383469 | c.92dup; p.Leu32Thrfs*3 | 0.011928 | LP | NR | |
| ALOX15 | 781725832 | c.316del; p.Leu106* | 0.000229 | LP | NR | |
| SMUG1 | 2233919 | c.7C>T; p.Gln3* | 0.054463 | LB | NR | |
| BC-F11P4 | MAN1B1 | NR | c.383T>G; p.Leu128* | NA | LP | NR |
| BC-F12P3 | PIKFYVE | NR | c.914C>A; p.Ser305* | NA | LP | NR |
| IL3 | 373251020 | c.337C>T; p.Arg113* | 0.000056 | LP | NR | |
| TSSK4 | 200353859 | c.895A>T; p.Lys299* | 0.002882 | LP | NR | |
| APOBEC3B | 199817842 | c.166C>T; p.Arg56* | 0.002176 | LP | NR | |
| BC-F16P5 | CERKL | 121909398 | c.847C>T; p.Arg283* | 0.000538 | P | R |
| PIKFYVE | NR | c.573C>A; p.Cys191* | NA | LP | NR | |
| TM4SF19 | NR | c.273T>A; p.Cys91* | NA | LP | NR | |
| ALG1 | NR | c.297_298del; p.Val100Phefs*37 | NA | LP | NR | |
| BC-F42P5 | SH3PXD2P | 551498843 | c.2626_2629del; p.Phe876Lysfs*16 | 0.00559 | R | |
| HMSD | 559021231 | c.105_120del; p.Asp35Glufs*49 | 0.013616 | LP | NR | |
| BC-F46P5 | SPG11 | NR | c.3235G>T; p.Gly1079* | NA | LP | NR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
BenAyed-Guerfali, D.; Kifagi, C.; BenKridis-Rejeb, W.; Ammous-Boukhris, N.; Ayedi, W.; Khanfir, A.; Daoud, J.; Mokdad-Gargouri, R. The Identification by Exome Sequencing of Candidate Genes in BRCA-Negative Tunisian Patients at a High Risk of Hereditary Breast/Ovarian Cancer. Genes 2022, 13, 1296. https://doi.org/10.3390/genes13081296
BenAyed-Guerfali D, Kifagi C, BenKridis-Rejeb W, Ammous-Boukhris N, Ayedi W, Khanfir A, Daoud J, Mokdad-Gargouri R. The Identification by Exome Sequencing of Candidate Genes in BRCA-Negative Tunisian Patients at a High Risk of Hereditary Breast/Ovarian Cancer. Genes. 2022; 13(8):1296. https://doi.org/10.3390/genes13081296
Chicago/Turabian StyleBenAyed-Guerfali, Dorra, Chamseddine Kifagi, Wala BenKridis-Rejeb, Nihel Ammous-Boukhris, Wajdi Ayedi, Afef Khanfir, Jamel Daoud, and Raja Mokdad-Gargouri. 2022. "The Identification by Exome Sequencing of Candidate Genes in BRCA-Negative Tunisian Patients at a High Risk of Hereditary Breast/Ovarian Cancer" Genes 13, no. 8: 1296. https://doi.org/10.3390/genes13081296
APA StyleBenAyed-Guerfali, D., Kifagi, C., BenKridis-Rejeb, W., Ammous-Boukhris, N., Ayedi, W., Khanfir, A., Daoud, J., & Mokdad-Gargouri, R. (2022). The Identification by Exome Sequencing of Candidate Genes in BRCA-Negative Tunisian Patients at a High Risk of Hereditary Breast/Ovarian Cancer. Genes, 13(8), 1296. https://doi.org/10.3390/genes13081296






