Abstract
Autism spectrum disorder (ASD) affects more than 1% of children, and there is no viable pharmacotherapeutic agent to treat the core symptoms of ASD. Studies have shown that children with ASD show changes in their levels of immune response molecules. Our previous studies have shown that ASD is more common in children with folate receptor autoantibodies. We also found that children with ASD have abnormal gut immune function, which was characterized by a significant increase in the content of immunoglobulin A and an increase in gut-microbiota-associated epitope diversity. These studies suggest that the immune mechanism plays an important role in the occurrence of ASD. The present study aims to systematically assess gene mutations in immune mediators in patients with ASD. We collected genetic samples from 72 children with ASD (2–12 years old) and 107 healthy controls without ASD (20–78 years old). We used our previously-designed immune gene panel, which can capture cytokine and receptor genes, the coding regions of MHC genes, and genes of innate immunity. Target region sequencing (500×) and bioinformatics analytical methods were used to identify variants in immune response genes associated with patients with ASD. A total of 4 rare variants were found to be associated with ASD, including HLA-B: p.A93G, HLA-DQB1: p.S229N, LILRB2: p.R322H, and LILRB2: c.956-4C>T. These variants were present in 44.44% (32/72) of the ASD patients and were detected in 3.74% (4/107) of the healthy controls. We expect these genetic variants will serve as new targets for the clinical genetic assessment of ASD, and our findings suggest that immune abnormalities in children with ASD may have a genetic basis.
1. Introduction
Autism spectrum disorders (ASD) are a group of neurodevelopmental disorders which affect around 1% of school-age children and which impose severe economic and social burdens on ordinary families and society [1,2,3,4]. Currently, there is no pharmacotherapeutic agent to treat the core symptoms of ASD [5]. Genetic factors play an important role in autism, and we have previously applied whole-genome sequencing to autism genetic studies and found autism-related mutations [6,7,8]; However, like the clinical heterogeneity of autism, autism also has a complex genetic structure, and large-scale ASD genomic studies have identified a large number of clinically relevant ASD variants; these variants involve common and rare variants in more than 100 risk genes, from point mutations to copy number variations, either inherited or de novo [9,10,11,12,13]. Recent studies have suggested an association of MHC regional variation with ASD and neuropsychiatric diseases [14,15]. Our previous study showed that 77.5% (31/40) of children with ASD were found to be positive for folate receptor autoantibodies (FRAA) [16]; this is consistent with the results of Frye et al. [17,18]. Furthermore, Frye et al. found that exposure to FRAA in pregnant rats was associated with behavioral deficits in offspring [19]; they also found that folinic acid was associated with improvements in core and related symptoms of ASD [20,21]. A number of maternal autoantibodies that can serve as biomarkers for ASD have been identified [22,23,24,25]; meanwhile, animal studies have confirmed the correlation between continuous exposure to ASD-specific maternal autoantibodies throughout gestation in mice and ASD-like behavior in offspring [26,27]. In short, these suggest a possible correlation between autoimmune problems and the development of ASD, and it also suggests the need for systematic research on the MHC region of ASD patients. Considering the importance of the MHC region in mediating host immune responses and its inherent high mutagenicity, high-throughput sequencing of the MHC region is necessary. Cytokines have an important role in neurodevelopment [28,29], and studies have demonstrated that peripheral cytokine profiles at birth are associated with ASD in later childhood [30,31,32,33]. Maternal immune activation (MIA) is an environmental risk factor for ASD, and MIA-mouse offspring often exhibit ASD-like behavior. These mice are widely used as a model of ASD [34]. Choi et al. [35] found that MIA-induced, ASD-like behavior is dependent on maternal IL-17A and that this behavioral abnormality can be improved by blocking IL-17A. There is increasing evidence that serum cytokine abnormalities are present in patients with ASD [36]. Tsilioni et al. found that elevated serum IL-6 and TNF can define the ASD subgroup, a group that benefits from treatment with the natural flavonoid luteolin [37]. A cross-sectional study by Al-Ayadhi of 45 children, aged 6–12 years, with ASD showed that the serum levels of IL-17A were positively correlated with the severity of autism, and nearly 50% of children with autism had elevated serum IL-17A levels. Of these, 67.9% had severe ASD, and 17% had mild-to-moderate ASD [38]. The imbalance of the immune system in patients with ASD was also characterized by an abnormal lymphocyte count, the presence of serum brain-specific autoantibodies, and intestinal immune dysfunction [39]. Imbalanced immune responses in children with ASD may be affected by changes in gut microbiota [40,41], and a large number of studies on ASD gut microbiota have been published [42,43,44,45,46,47]; our previous studies have shown that abnormal intestinal or oral IgA is a potential biomarker for ASD [48,49,50], and that abnormal gut microbiota structure, especially an epitope-associated gut microbiota structure, is associated with increased IgA [51]. In short, these studies suggest an abnormality in immune function in ASD. The aim of this study was to systematically assess gene mutations in immune mediators of children with ASD to determine if there are immune-related genetic defects in children with ASD.
2. Method
2.1. Study Participants
A total of 72 patients with ASD, 54 males (2–8 years old) and eighteen females (2–11 years old) were recruited from the Department of Psychology at Shenzhen Children’s Hospital. The specific process was as described in our previous study [50]. In short, inclusion criteria were as follows: the patient must be age <14 years, must meet the diagnosis of ASD, and must be classified as “requiring very substantial support” according to the 5th edition of the Diagnostic and Statistical Manual of Mental Disorders. Furthermore, inclusion was not limited by the sex of the patient. The exclusion criteria were as follow: The patient must not (1) be suffering from other mental illnesses (such as obsessive–compulsive disorder or ADHD); (2) be suffering from other neurodevelopmental disorders; (3) be suffering from genetic metabolic diseases; (4) be suffering from serious neurological diseases or have a history of brain injury or other major physical diseases; (5) be acutely physically ill. Healthy controls totaled 107 individuals, 57 males (20–78 years old) and 50 females (24–73 years old), from a previously published hepatitis B virus (HBV) cohort study. All healthy controls were non-ASD individuals—adults who were not diagnosed with ASD in childhood as determined by medical history examination, and the inclusion and exclusion criteria are detailed in previously published articles [52,53]. All enrolled individuals were informed of the study, and the enrolled individuals signed informed consent forms. The study was approved by the Ethics Committee of Shenzhen Children’s Hospital.
2.2. DNA Extraction and Quality Control
A volume of 3 to 5 mL of whole blood was drawn and stored at −80 °C. Then, genomic DNA was extracted from these whole-blood samples using a PureLink™ Genomic DNA Mini Kit (Thermo Fisher, Foster City, CA, USA). The genomic DNA samples were quantified and controlled for quality using NanoDrop ND2000 (Thermo Fisher). The total amount of DNA was required to be above 1 µg; the purity ratio of A260/280 was required to be in the range of 1.8–2.0; the main DNA band had to be clearly visible; and the fragment size had to appear around 23 K Da, as measured by 0.3% agarose gel electrophoresis.
2.3. Library Construction
A DNA sample of 1 µg was fragmented into 150–250 bp-sized fragments using a Bioruptor interrupter (Diagenode, Seraing, Belgium); then, the fragmented DNA was subjected to end-filling, 5′ segment phosphate group repair, and 3′ segment plus A (Enzymatics Inc., Beverly, MA, USA) using an ABI 2720 type of PCR instrument (Thermo Fisher, Hudson, NH, USA). Finally, a synthetic paired-end adapter (Thermo Fisher) suitable for the Illumina HiSeq sequencer (Illumina, San Diego, CA, USA) was ligated using an ABI 2720 PCR machine (Thermo Fisher), and the ligation product was purified using MagPure A3 XP beads (Magen, Guangzhou, China). For the purified ligation product, PCR pre-amplification (KAPA Biosystems, USA) was performed using an ABI 2720 PCR machine (Thermo Fisher), and a synthetic index sequence (Thermo Fisher) capable of distinguishing individual samples was introduced. The parameters were as follows: 95 °C for 4 min, 98 °C for 20 s, 65 °C for 30 s, 5 cycles, 72 °C for 30 s, 72 °C for 5 min, and 12 °C hold. A small-fragment sequencing library was obtained, and 1 µL of the small-fragment library was subjected to quantification using a Qubit dsDNA HS Assay Kit (Thermo Fisher). The concentration of the capture library was determined, and the standard of the qualified library was measured to be greater than 3 ng/µL.
2.4. Targeted Region Sequencing
The panel, which targeted the coding region of 404 immune-response molecule genes, included genes related to cytokines, receptors, innate immunity, and adaptive immunity and was designed in our previous study [52]; target region sequencing of the 404 immune-response genes was performed according to our published studies [54,55]. In brief, the small-fragment library was mixed with a hybridization block (iGeneTech, Beijing, China). Then, commercially acquired TargetSeq hybridization buffer (iGeneTech) was thawed at room temperature, mixed, and placed in a 65 °C water bath to preheat until the solution was completely dissolved. For each sample, 20 µL of the hybridization buffer was placed in a PCR tube and incubated in a water bath at 65 °C. Before hybridization, 5 µL of RNase block (Thermo Fisher) was also prepared and mixed with a single-stranded RNA probe to prevent probe degradation. Hybridization captures were performed on an ABI 2720 PCR machine (Thermo Fisher). The hybridization mixture was covered and incubated overnight (8–16 h) at 65 °C. After the hybridization was complete, the resulting DNA–RNA hybrids were bound to the magnetic beads (Dynabeads MyOne Streptavidin T1, Thermo Fisher). To remove non-specific binding, the magnetic bead–DNA–RNA complex was washed with TargetSeq washing solution (iGeneTech). Finally, PCR enrichment was performed on an ABI 2720 PCR machine (Thermo Fisher) for the target region obtained by hybridization, with parameters as follow: 95 °C for 4 min, 98 °C for 20 s, 65 °C for 30 s, 16 cycles, 72 °C for 30 s, 72 °C for 5 min, and 12 °C hold. The PCR amplification reagent was commercially purchased from KAPA Biosystems. The Nextflex primer was synthesized by Invitrogen, China. The amplified target-region capture library was found to be greater than 3 ng/µL, as determined by a Qubit dsDNA HS Assay Kit (Thermo Fisher). The constructed sequencing library was sequenced on a HiSeq X Ten sequencer (Illumina, San Diego, CA, USA) PE150.
2.5. Bioinformatics Analysis
Previously published studies [7,8,16] provide complete details; a brief introduction is as follows: The raw data were first filtered using Trimmomatic software [56] to filter the linker sequences (GATCGGAAGAGCACACGTCT and AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT) and low-quality sequences. Low-quality sequences were defined as having a base quality value of less than or equal to 20, corresponding to an accuracy of 99%. Bases that did not satisfy the conditions in the sequence were removed, and sequences having a length of less than 40 bp after base filtration were removed. Finally, FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 June 2022), version 0.11.9) was used to perform a quality assessment and ensure clean reads of greater than 95%, with a quality value greater than 30. The clean reads were aligned to the human reference genome (hg19, downloaded from UCSC) using BWA-MEM software [57] to generate an aligned BAM file. To improve the accuracy of the final results the effects of PCR, repetitions in the experiments were removed using Samtools [58] and Picard software (http://broadinstitute.github.io/picard/ (accessed on 15 June 2022), version 2.26.5). Then, a GATK (Genome Analysis Toolkit) [59] was used to detect mutations, such as SNP and InDel, in the results of the alignment. Finally, the results of the detected mutations were annotated with ANNOVAR software [60], and the sequencing depth and coverage were also evaluated.
2.6. Association Analysis
The correlation analysis of point mutations and gene levels was completed with reference to a previously published study [61]. In short, We first calculated the groups carrying rare variants, including heterozygous or homozygous mutations, referring to the gnomAD database [62] containing large-scale, whole exomes (123,136 samples, including 8624 people of East Asian ethnicity) and whole-genome sequencing samples. Variants with an East Asian (EAS) allele frequency of less than 0.1 are considered rare. Then, Fisher’s exact test was used to calculate the p value of each group; the variants with significant differences were defined as characterized by: (1) both the discovery and validation stages were significant (p value of less than 0.05), (2) the combined two-stage data were also significant (FDR value less than 0.05 using Fisher’s exact test); (3) the gene-level association was significant as performed using the SKAT-O method [63] (p value of less than 0.001).
2.7. Sanger Sequencing
To confirm the four rare variants, PCR was performed using the ABI9700 PCR system (Thermo Fisher), and the PCR product was then sequenced in an ABI 3730 xl sequencer (Thermo Fisher).
2.8. Other Analysis
The lm.fit() function in R software (version 4.0.4) was used to perform a linear fitting analysis on the age and the total number of immune-gene variants in each sample. The violinplot() function in the Python (version 3.7.5) package of Seaborn (version 0.11.1) was used to draw the violinplot; the add_stat_annotation() function in the Python package statannot (version 0.2.3) was used for the statistical significance test, and the set parameters were: test = ‘t-test_ind’, text_format = ‘star’, loc = ‘inside’, verbose = 2.
3. Results
As shown in Figure 1, the study was conducted in two stages: the discovery stage and the validation stage. In the discovery stage, 37 patients with ASD and 55 healthy controls were recruited. targeted region sequencing was performed using our previously designed panel of 404 immune-response-molecule genes. The target region size was approximately 500 Kbp, the average coverage depth was >1000×, and the 10× coverage rate was >99%. A total of 7526 point mutations were detected, 285 of which differed significantly between the ASD and the healthy control groups (p value less than 0.05, Fisher’s exact test, Supplementary Table S1). During the validation stage, 35 patients with ASD and 52 healthy controls were enrolled. Statistical analysis showed that 231 mutations were significantly different between the ASD and healthy control groups (p value less than 0.05, Fisher’s exact test, Supplementary Table S2).
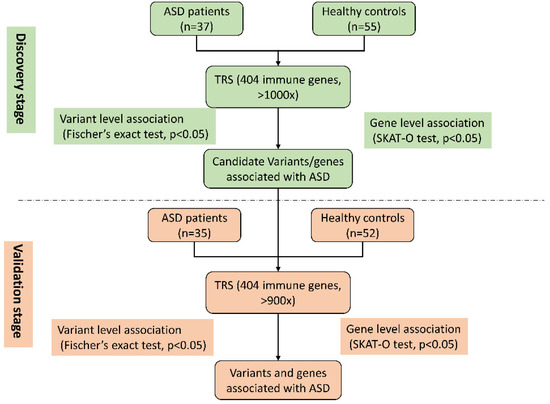
Figure 1.
Flowchart of study design.
Combining these two stages, a total of 61 ASD-related point mutations were found (Supplementary Table S3), 4 of which were rare, functional mutations. These included the missense mutation p.R322H and the splicing mutation c.956-4C>T of the LILRB2 gene, the missense mutation p.S229N of the HLA-DQB1 gene, and the missense mutation p.A93G of the HLA-B gene (Table 1). These 4 mutations were verified by Sanger sequencing with the primers listed in Supplementary Table S4.

Table 1.
Rare functional mutations in immune-response-molecule genes associated with ASD.
The SKAT-O method, which considers both common and rare variants as risk factors for disease, was used to evaluate the association between the 404 immune-response genes and ASD. The results showed that the correlation p value of HLA-B and LILRB2 reached a level of less than 1 × 10−5.
Considering that age is an important confounding factor, this controlled study included adults without ASD as controls for individuals with ASD. To assess the potential impact of age on immune-gene-variant counts, we used the lm.fit() function in R software to perform a linear fitting analysis on the age and the total number of immune-gene variants in each sample (Supplementary Table S5). The calculated p value was 0.4556, and R-squared was 0.0032 (Supplementary Figure S1), indicating that age effect on the number of immune-gene variants is limited. In ASD individuals, the ratio of male:female was extremely unbalanced, usually 3~4:1, or even higher. In this study, the number of male ASD individuals was three times that of female ASD individuals, which is also typical. Considering that gender is a potential confounding factor for ASD, we used gender as a covariate to evaluate its effect on the four candidate ASD-related immune-gene variants, showing that gender had no effect on these variants (Supplementary Figure S2).
4. Discussion
The imbalance of the immune response is considered to play a very important role in ASD. Early studies focused on the discovery of ASD-specific cytokine profiles, lymphocyte subsets, etc. [30,64]; in recent years, researchers have begun to focus on the discovery of maternal autoantibodies [18,21,22,23,25,26] and understanding the role of gut microbiota in it [65,66,67]. Genetic mechanisms are considered to play an important role in ASD, and although a large number of studies have focused on the discovery of ASD-related clinical variants [9,10,11,13,68,69], there is still a lack of systematic evaluation of ASD-related immune-response-gene mutations. In the present study, we systematically evaluated mutations in immune-response molecular genes, including HLA region genes and cytokine and receptor genes, in patients with ASD. Maternal autoantibodies are potential biomarkers for ASD [21,24]; the exposure to maternal autoantibodies during pregnancy is closely related to the ASD-like behavior of the offspring [26,27]. Targeted intervention for ASD patients with positive maternal autoantibodies is a potential treatment for ASD [20]. The HLA system plays an important role in the recognition of autoantibodies; recent studies have found that the HLA system is involved in the regulation of neural development and neuroplasticity, especially through microglia regulation and synaptic pruning [15]. HLA Gene variants are associated with neurodevelopmental diseases such as ASD [70,71]. We found that the missense mutation p.A93G of the HLA-B gene from HLA Class I is a risk factor for ASD, which supports the previously published findings that HLA-B*07 alleles are more common in patients with ASD [72] and that HLA-B gene polymorphism is related to ASD [73]. We also found that the frequency of the missense mutation p.S229N of HLA-DQB1 gene in ASD patients (15/72 = 20.83%) was significantly higher than that in healthy controls (4/107 = 3.73%); it is well known that HLA-DQB1 is a susceptibility gene for celiac disease [74]. Celiac disease is an immune enteropathy, mainly manifested by gluten intolerance, and the symptoms of celiac disease are similar to the gastrointestinal dysfunction prevalent in ASD [75]. Although there is no scientific evidence, the method of intervening in celiac disease using a glutamate-free/casein-free diet is widely used in ASD, and 20–29% of parents report significant improvement in ASD symptoms [76]. We found that 20.83% of patients with ASD had a mutation in a genetic sequence associated with celiac disease risk, which may provide theoretical support for gluten-free/casein-free (GFCF) dietary interventions in patients with ASD; meanwhile, given the high prevalence of gastrointestinal problems in ASD [77], the identified associated variants in HLA-DQB1 may provide a more prevalent gastrointestinal phenotype than specifically gluten sensitivity in the face of individuals with ASD and gastrointestinal problems. Furthermore, we found that the 2 closely linked mutations, p.R322H and c.956-4C>T, from the LILRB2 gene had a mutation frequency of 20.83% (15/72) in the ASD patient group, and that these 2 mutations were rare in the control group (2/107 = 1.87%). The LILRB2 gene is an inhibitory receptor gene of HLA Class I, and it was found that the LILRB2 gene encodes a neuronal cell-surface receptor that is associated with occurrences of Alzheimer disease as a receptor for β-amyloid [78]. The inhibition of the binding of β-amyloid to LilrB2 has become a potential pathway for the treatment of Alzheimer disease [79]. The results of this study suggest that the LILRB2 gene may be a potential target for ASD therapy.
The 4 immune-response-gene mutations found in this study were present in 44.44% (32/72) of the ASD patient group and are expected to serve as new targets for the clinical genetic evaluation of ASD. The clinical genetic evaluation of ASD is usually performed using a chromosome microarray combined with whole exome/genome sequencing, which is costly, difficult for data interpretation, and has limited use for clinical genetic evaluation in patients with ASD. The combined mutation frequency of the 4 mutations in ASD patients was 44.44%, with a single mutation frequency of 20.83%. Whole-exome sequencing, whole-genome sequencing, and robust bioinformatic analysis methods have successfully identified high-confidence ASD candidate genes from a large number of de novo and inherited variants [7,8,9,10,11,13,80]. However, despite great efforts, the genetic structure interpretation of ASD using these candidate genes is still incomplete; high-throughput sequencing of immune response genes is expected to complement the ASD genetic machinery.
To our knowledge, this is the first study to screen mutations in immune-response-molecule genes in patients with ASD. This approach can prove helpful in answering whether ASD immune dysfunction has a certain genetic basis. However, this study has certain limitations. First, the genes of the immune-response molecules that we were concerned with may not be comprehensive. This current study only covered cytokines and receptors, HLA regions, and other genes. Second, although we have completely covered the HLA gene-coding region, it does not cover the entire HLA region, and the haplotype of the HLA region could not be obtained. Future studies are planned to evaluate more genes of the immune-response molecule and to perform HLA haplotype detection. Finally, as an exploratory ASD genetic research project, we did not collect detailed psychological or behavioral assessment parameters when we included ASD individuals; the main reason was that we included ASD individuals from several different institutions, and the criteria used for detailed psychological and behavioral assessment of ASD vary from institution to institution. In the future, we will further evaluate the association of immune-gene mutations with psychological and behavioral assessment parameters.
In conclusion, our results suggest that immune dysfunction in patients with ASD may have a genetic basis. The identification of these genetic mutations may also facilitate the development of new genetic, clinical ASD assessment tools.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13061098/s1: Supplementary Table S1. Significantly different point mutations identified in the discovery stage; Supplementary Table S2. Significantly different point mutations identified in the validation stage; Supplementary Table S3. Significantly different point mutations identified in the discovery and validation stages; Supplementary Table S4. Primers for four rare variants; and Supplementary Table S5. Metadata and immune-gene variant counts. Supplementary Figure S1. The effect of age on immune gene variant counts; Supplementary Figure S2. The effect of sex on ASD-related immune gene variants.
Author Contributions
Conceptualization, M.W. and J.Z.; methodology, A.L.; software, S.Z.; formal analysis, M.W., C.C. and Z.Y.; investigation, C.C. and Z.Y.; resources, H.W. and S.L.; data curation, A.L. and M.W.; writing—original draft preparation, M.W., C.C., Z.Y. and A.L.; writing—review and editing, M.W., Z.Y., A.L., J.Z., S.L. and C.C.; visualization, C.C.; supervision, M.W. and J.Z.; project administration, Z.W. and H.Q.; funding acquisition, M.W., Z.Y., S.L. and C.C.; All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by Supported by the National Natural Science Foundation of China (Program Nos. 82071733, 81960290 and 81971603), the Shenzhen Science Technology and Innovation Commission (JCYJ20190807152403624), the Shanghai Talent Development Funding (No. 2020115), the Baoan Science Technology and Innovation Commission of Shenzhen (2019JD486), the Longgang Science Technology and Innovation Commission of Shenzhen (LGKCYLWS2021000006), the High Level Project of Medicine of Longhua (HLPM201907020103), the Beijing Municipal Science & Technology Commission (Z181100001518005), and the Capital’s Funds for Health Improvement and Research (CFH2018-2Z-5012).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Shenzhen Children’s Hospital (protocol code 2017031 and date of approval: 15 December 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Considering the control of human genetic resources, all of the original sequencing data have not been uploaded to the public database. Readers can ask the corresponding author for the variant summary table of the samples.
Acknowledgments
We are sincerely grateful to the children and their parents who participated in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shaw, K.A.; Maenner, M.J.; Baio, J.; Washington, A.; Christensen, D.L.; Wiggins, L.D.; Pettygrove, S.; Andrews, J.G.; White, T.; Rosenberg, C.R.; et al. Early Identification of Autism Spectrum Disorder Among Children Aged 4 Years—Early Autism and Developmental Disabilities Monitoring Network, Six Sites, United States, 2016. MMWR Surveill. Summ. 2020, 69, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Hirota, T.; Sakamoto, Y.; Adachi, M.; Takahashi, M.; Osato-Kaneda, A.; Kim, Y.S.; Leventhal, B.; Shui, A.; Kato, S.; et al. Prevalence and cumulative incidence of autism spectrum disorders and the patterns of co-occurring neurodevelopmental disorders in a total population sample of 5-year-old children. Mol. Autism 2020, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xu, X.; Yan, W.; Zou, X.; Wu, L.; Luo, X.; Li, T.; Huang, Y.; Guan, H.; Chen, X.; et al. Prevalence of Autism Spectrum Disorder in China: A Nationwide Multi-center Population-based Study Among Children Aged 6 to 12 Years. Neurosci. Bull. 2020, 36, 961–971. [Google Scholar] [CrossRef]
- Zablotsky, B.; Black, L.I.; Maenner, M.J.; Schieve, L.A.; Blumberg, S.J. Estimated Prevalence of Autism and Other Developmental Disabilities Following Questionnaire Changes in the 2014 National Health Interview Survey. Natl. Health Stat. Rep. 2015, 87, 1–20. [Google Scholar]
- Muhle, R.A.; Reed, H.E.; Stratigos, K.A.; Veenstra-VanderWeele, J. The Emerging Clinical Neuroscience of Autism Spectrum Disorder: A Review. JAMA Psychiatry 2018, 75, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Yuen, R.K.; Jin, X.; Wang, M.; Chen, N.; Wu, X.; Ju, J.; Mei, J.; Shi, Y.; He, M.; et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet. 2013, 93, 249–263. [Google Scholar] [CrossRef] [Green Version]
- Yuen, R.K.; Merico, D.; Cao, H.; Pellecchia, G.; Alipanahi, B.; Thiruvahindrapuram, B.; Tong, X.; Sun, Y.; Cao, D.; Zhang, T.; et al. Genome-wide characteristics of de novo mutations in autism. NPJ Genom. Med. 2016, 1, 160271–1602710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Yu, P.; Jin, X.; Xu, X.; Li, J.; Li, Z.; Wang, M.; Wang, T.; Wu, X.; Jiang, Y.; et al. Genomic landscapes of Chinese sporadic autism spectrum disorders revealed by whole-genome sequencing. J. Genet. Genom. 2018, 45, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Choi, L.; An, J.Y. Genetic architecture of autism spectrum disorder: Lessons from large-scale genomic studies. Neurosci. Biobehav. Rev. 2021, 128, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Havdahl, A.; Niarchou, M.; Starnawska, A.; Uddin, M.; van der Merwe, C.; Warrier, V. Genetic contributions to autism spectrum disorder. Psychol. Med. 2021, 51, 2260–2273. [Google Scholar] [CrossRef] [PubMed]
- Wilfert, A.B.; Turner, T.N.; Murali, S.C.; Hsieh, P.; Sulovari, A.; Wang, T.; Coe, B.P.; Guo, H.; Hoekzema, K.; Bakken, T.E.; et al. Recent ultra-rare inherited variants implicate new autism candidate risk genes. Nat. Genet. 2021, 53, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Antaki, D.; Guevara, J.; Maihofer, A.X.; Klein, M.; Gujral, M.; Grove, J.; Carey, C.E.; Hong, O.; Arranz, M.J.; Hervas, A.; et al. A phenotypic spectrum of autis.sm is attributable to the combined effects of rare variants, polygenic risk and sex. Nat. Genet. 2022. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhao, P.A.; Eichler, E.E. Rare variants and the oligogenic architecture of autism. Trends Genet. 2022. [Google Scholar] [CrossRef] [PubMed]
- Guerini, F.R.; Bolognesi, E.; Sotgiu, S.; Carta, A.; Clerici, C.; Chiappedi, M.; Ghezzo, A.; Zanette, M.; Mensi, M.M.; Canevini, M.P.; et al. HLA-G allelic distribution in Sardinian children with Autism spectrum disorders: A replication study. Brain Behav. Immun. 2019, 79, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Tamouza, R.; Krishnamoorthy, R.; Leboyer, M. Understanding the genetic contribution of the human leukocyte antigen system to common major psychiatric disorders in a world pandemic context. Brain Behav. Immun. 2021, 91, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, A.; He, F.; Jin, Y.; Zhou, S.; Xu, R.; Guo, H.; Zhou, W.; Wei, Q.; Wang, M. High prevalence of serum folate receptor autoantibodies in children with autism spectrum disorders. Biomarkers 2018, 23, 622–624. [Google Scholar] [CrossRef]
- Frye, R.E.; Delhey, L.; Slattery, J.; Tippett, M.; Wynne, R.; Rose, S.; Kahler, S.G.; Bennuri, S.C.; Melnyk, S.; Sequeira, J.M.; et al. Blocking and Binding Folate Receptor Alpha Autoantibodies Identify Novel Autism Spectrum Disorder Subgroups. Front. Neurosci. 2016, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Frye, R.E.; Wynne, R.; Rose, S.; Slattery, J.; Delhey, L.; Tippett, M.; Kahler, S.G.; Bennuri, S.C.; Melnyk, S.; Sequeira, J.M.; et al. Thyroid dysfunction in children with autism spectrum disorder is associated with folate receptor alpha autoimmune disorder. J. Neuroendocrinol. 2017, 29. [Google Scholar] [CrossRef]
- Desai, A.; Sequeira, J.M.; Quadros, E.V. Prevention of behavioral deficits in rats exposed to folate receptor antibodies: Implication in autism. Mol. Psychiatry 2017, 22, 1291–1297. [Google Scholar] [CrossRef]
- Frye, R.E.; Slattery, J.; Delhey, L.; Furgerson, B.; Strickland, T.; Tippett, M.; Sailey, A.; Wynne, R.; Rose, S.; Melnyk, S.; et al. Folinic acid improves verbal communication in children with autism and language impairment: A randomized double-blind placebo-controlled trial. Mol. Psychiatry 2018, 23, 247–256. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, D.A.; Frye, R.E. Cerebral Folate Deficiency, Folate Receptor Alpha Autoantibodies and Leucovorin (Folinic Acid) Treatment in Autism Spectrum Disorders: A Systematic Review and Meta-Analysis. J. Pers Med. 2021, 11, 1141. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.L.; Van de Water, J. Maternal autoantibody related autism: Mechanisms and pathways. Mol. Psychiatry 2019, 24, 252–265. [Google Scholar] [CrossRef]
- Ramirez-Celis, A.; Edmiston, E.; Schauer, J.; Vu, T.; Van de Water, J. Peptides of neuron specific enolase as potential ASD biomarkers: From discovery to epitope mapping. Brain Behav. Immun. 2020, 84, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Celis, A.; Becker, M.; Nuno, M.; Schauer, J.; Aghaeepour, N.; Van de Water, J. Risk assessment analysis for maternal autoantibody-related autism (MAR-ASD): A subtype of autism. Mol. Psychiatry 2021, 26, 1551–1560. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Celis, A.; Croen, L.A.; Yoshida, C.K.; Alexeeff, S.E.; Schauer, J.; Yolken, R.H.; Ashwood, P.; Van de Water, J. Maternal autoantibody profiles as biomarkers for ASD and ASD with co-occurring intellectual disability. Mol. Psychiatry 2022. [Google Scholar] [CrossRef]
- Jones, K.L.; Pride, M.C.; Edmiston, E.; Yang, M.; Silverman, J.L.; Crawley, J.N.; Van de Water, J. Autism-specific maternal autoantibodies produce behavioral abnormalities in an endogenous antigen-driven mouse model of autism. Mol. Psychiatry 2020, 25, 2994–3009. [Google Scholar] [CrossRef] [PubMed]
- Bruce, M.R.; Jones, K.L.; Vernon, A.C.; Silverman, J.L.; Crawley, J.N.; Ellegood, J.; Lerch, J.P.; Van de Water, J. Sexually dimorphic neuroanatomical differences relate to ASD-relevant behavioral outcomes in a maternal autoantibody mouse model. Mol. Psychiatry 2021, 26, 7530–7537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, B.; Li, H.; Wang, Y.; Zhou, Y.; Wang, W.; Song, T.; Du, N.; Gu, X.; Luo, Y.; et al. SOCS3 Attenuates GM-CSF/IFN-gamma-Mediated Inflammation During Spontaneous Spinal Cord Regeneration. Neurosci. Bull. 2020, 36, 778–792. [Google Scholar] [CrossRef]
- Kim, J.; Erice, C.; Rohlwink, U.K.; Tucker, E.W. Infections in the Developing Brain: The Role of the Neuro-Immune Axis. Front. Neurol. 2022, 13, 805786. [Google Scholar] [CrossRef] [PubMed]
- Krakowiak, P.; Goines, P.E.; Tancredi, D.J.; Ashwood, P.; Hansen, R.L.; Hertz-Picciotto, I.; Van de Water, J. Neonatal Cytokine Profiles Associated With Autism Spectrum Disorder. Biol. Psychiatry 2017, 81, 442–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodnar, T.S.; Raineki, C.; Wertelecki, W.; Yevtushok, L.; Plotka, L.; Zymak-Zakutnya, N.; Honerkamp-Smith, G.; Wells, A.; Rolland, M.; Woodward, T.S.; et al. Altered maternal immune networks are associated with adverse child neurodevelopment: Impact of alcohol consumption during pregnancy. Brain Behav. Immun. 2018, 73, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, T.S.; Raineki, C.; Wertelecki, W.; Yevtushok, L.; Plotka, L.; Granovska, I.; Zymak-Zakutnya, N.; Pashtepa, A.; Wells, A.; Honerkamp-Smith, G.; et al. Immune network dysregulation associated with child neurodevelopmental delay: Modulatory role of prenatal alcohol exposure. J. Neuroinflamm. 2020, 17, 39. [Google Scholar] [CrossRef]
- Kim, D.H.; Krakowiak, P.; Meltzer, A.; Hertz-Picciotto, I.; Van de Water, J. Neonatal chemokine markers predict subsequent diagnosis of autism spectrum disorder and delayed development. Brain Behav. Immun. 2022, 100, 121–133. [Google Scholar] [CrossRef]
- Mueller, F.S.; Polesel, M.; Richetto, J.; Meyer, U.; Weber-Stadlbauer, U. Mouse models of maternal immune activation: Mind your caging system! Brain Behav. Immun. 2018, 73, 643–660. [Google Scholar] [CrossRef]
- Choi, G.B.; Yim, Y.S.; Wong, H.; Kim, S.; Kim, H.; Kim, S.V.; Hoeffer, C.A.; Littman, D.R.; Huh, J.R. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 2016, 351, 933–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eftekharian, M.M.; Ghafouri-Fard, S.; Noroozi, R.; Omrani, M.D.; Arsang-Jang, S.; Ganji, M.; Gharzi, V.; Noroozi, H.; Komaki, A.; Mazdeh, M.; et al. Cytokine profile in autistic patients. Cytokine 2018, 108, 120–126. [Google Scholar] [CrossRef]
- Tsilioni, I.; Taliou, A.; Francis, K.; Theoharides, T.C. Children with autism spectrum disorders, who improved with a luteolin-containing dietary formulation, show reduced serum levels of TNF and IL-6. Transl. Psychiatry 2015, 5, e647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Ayadhi, L.Y.; Mostafa, G.A. Elevated serum levels of interleukin-17A in children with autism. J. Neuroinflamm. 2012, 9, 158. [Google Scholar] [CrossRef] [Green Version]
- Mostafa, G.A.; Al-Ayadhi, L.Y. Increased serum levels of anti-ganglioside M1 auto-antibodies in autistic children: Relation to the disease severity. J. Neuroinflamm. 2011, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Rose, D.R.; Yang, H.; Serena, G.; Sturgeon, C.; Ma, B.; Careaga, M.; Hughes, H.K.; Angkustsiri, K.; Rose, M.; Hertz-Picciotto, I.; et al. Differential immune responses and microbiota profiles in children with autism spectrum disorders and co-morbid gastrointestinal symptoms. Brain Behav. Immun. 2018, 70, 354–368. [Google Scholar] [CrossRef]
- Leader, G.; Abberton, C.; Cunningham, S.; Gilmartin, K.; Grudzien, M.; Higgins, E.; Joshi, L.; Whelan, S.; Mannion, A. Gastrointestinal Symptoms in Autism Spectrum Disorder: A Systematic Review. Nutrients 2022, 14, 1471. [Google Scholar] [CrossRef]
- Wang, M.; Wan, J.; Rong, H.; He, F.; Wang, H.; Zhou, J.; Cai, C.; Wang, Y.; Xu, R.; Yin, Z.; et al. Alterations in Gut Glutamate Metabolism Associated with Changes in Gut Microbiota Composition in Children with Autism Spectrum Disorder. mSystems 2019, 4, e00321-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dan, Z.; Mao, X.; Liu, Q.; Guo, M.; Zhuang, Y.; Liu, Z.; Chen, K.; Chen, J.; Xu, R.; Tang, J.; et al. Altered gut microbial profile is associated with abnormal metabolism activity of Autism Spectrum Disorder. Gut Microbes 2020, 11, 1246–1267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chu, Y.; Meng, Q.; Ding, R.; Shi, X.; Wang, Z.; He, Y.; Zhang, J.; Liu, J.; Zhang, J.; et al. A quasi-paired cohort strategy reveals the impaired detoxifying function of microbes in the gut of autistic children. Sci. Adv. 2020, 6, eaba3760. [Google Scholar] [CrossRef] [PubMed]
- Hazan, S.; Spradling-Reeves, K.D.; Papoutsis, A.; Walker, S.J. Shotgun Metagenomic Sequencing Identifies Dysbiosis in Triplet Sibling with Gastrointestinal Symptoms and ASD. Children 2020, 7, 255. [Google Scholar] [CrossRef]
- Wan, Y.; Zuo, T.; Xu, Z.; Zhang, F.; Zhan, H.; Chan, D.; Leung, T.F.; Yeoh, Y.K.; Chan, F.K.L.; Chan, R.; et al. Underdevelopment of the gut microbiota and bacteria species as non-invasive markers of prediction in children with autism spectrum disorder. Gut 2022, 71, 910–918. [Google Scholar] [CrossRef]
- Yap, C.X.; Henders, A.K.; Alvares, G.A.; Wood, D.L.A.; Krause, L.; Tyson, G.W.; Restuadi, R.; Wallace, L.; McLaren, T.; Hansell, N.K.; et al. Autism-related dietary preferences mediate autism-gut microbiome associations. Cell 2021, 184, 5916–5931. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Doenyas, C.; Wan, J.; Zeng, S.; Cai, C.; Zhou, J.; Liu, Y.; Yin, Z.; Zhou, W. Virulence factor-related gut microbiota genes and immunoglobulin A levels as novel markers for machine learning-based classification of autism spectrum disorder. Comput. Struct. Biotechnol. J. 2021, 19, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Qiao, Y.; Li, B.; Zheng, X.; Xu, R.; Wang, M.; Mi, X.; Li, Y. The Alteration of Salivary Immunoglobulin A in Autism Spectrum Disorders. Front. Psychiatry 2021, 12, 669193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; He, F.; Yang, F.; Yang, Z.; Xie, Y.; Zhou, S.; Liang, J.; Xu, R.; Wang, Y.; Guo, H.; et al. Increased stool immunoglobulin A level in children with autism spectrum disorders. Res. Dev. Disabil. 2018, 82, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhou, J.; He, F.; Cai, C.; Wang, H.; Wang, Y.; Lin, Y.; Rong, H.; Cheng, G.; Xu, R.; et al. Alteration of gut microbiota-associated epitopes in children with autism spectrum disorders. Brain Behav. Immun. 2019, 75, 192–199. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, X.; Tian, S.; Geng, L.; Xu, W.; Ma, N.; Wang, M.; Jia, Y.; Liu, X.; Ma, J.; et al. Comprehensive investigation of cytokine- and immune-related gene variants in HBV-associated hepatocellular carcinoma patients. Biosci. Rep. 2017, 37, BSR20171263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Ma, N.; Zhang, X.; Tian, S.; Geng, L.; Xu, W.; Wang, M.; Jia, Y.; Liu, X.; Ma, J.; et al. Comprehensive investigating of cytokine and receptor related genes variants in patients with chronic hepatitis B virus infection. Cytokine 2018, 103, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, J.; He, F.; Huang, Y.; Lu, S.; Fan, H.; Wang, M.; Xu, R. Design of a Targeted Sequencing Assay to Detect Rare Mutations in Circulating Tumor DNA. Genet. Test. Mol. Biomark. 2019, 23, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Wen, F.; Zhou, S.; Su, Z.; Liu, G.; Wang, M.; Zhou, J.; He, F. Association of MTTP gene variants with pediatric NAFLD: A candidate-gene-based analysis of single nucleotide variations in obese children. PLoS ONE 2017, 12, e0185396. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 2014, 30, 2843–2851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Wang, M.; Zhuang, D.; Mei, M.; Ma, H.; Li, Z.; He, F.; Cheng, G.; Lin, G.; Zhou, W. Frequent mutation of hypoxia-related genes in persistent pulmonary hypertension of the newborn. Respir Res. 2020, 21, 53. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Emond, M.J.; Bamshad, M.J.; Barnes, K.C.; Rieder, M.J.; Nickerson, D.A.; Christiani, D.C.; Wurfel, M.M.; Lin, X.; NHLBI GO Exome Sequencing Project—ESP Lung Project Team. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 2012, 91, 224–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuer, L.S.; Croen, L.A.; Jones, K.L.; Yoshida, C.K.; Hansen, R.L.; Yolken, R.; Zerbo, O.; DeLorenze, G.; Kharrazi, M.; Ashwood, P.; et al. An Exploratory Examination of Neonatal Cytokines and Chemokines as Predictors of Autism Risk: The Early Markers for Autism Study. Biol. Psychiatry 2019, 86, 255–264. [Google Scholar] [CrossRef]
- Laue, H.E.; Coker, M.O.; Madan, J.C. The Developing Microbiome From Birth to 3 Years: The Gut-Brain Axis and Neurodevelopmental Outcomes. Front. Pediatr. 2022, 10, 815885. [Google Scholar] [CrossRef] [PubMed]
- Prosperi, M.; Santocchi, E.; Guiducci, L.; Frinzi, J.; Morales, M.A.; Tancredi, R.; Muratori, F.; Calderoni, S. Interventions on Microbiota: Where Do We Stand on a Gut-Brain Link in Autism? A Systematic Review. Nutrients 2022, 14, 462. [Google Scholar] [CrossRef]
- Puricelli, C.; Rolla, R.; Gigliotti, L.; Boggio, E.; Beltrami, E.; Dianzani, U.; Keller, R. The Gut-Brain-Immune Axis in Autism Spectrum Disorders: A State-of-Art Report. Front. Psychiatry 2021, 12, 755171. [Google Scholar] [CrossRef]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584. [Google Scholar] [CrossRef]
- Doan, R.N.; Lim, E.T.; De Rubeis, S.; Betancur, C.; Cutler, D.J.; Chiocchetti, A.G.; Overman, L.M.; Soucy, A.; Goetze, S.; Autism Sequencing, C.; et al. Recessive gene disruptions in autism spectrum disorder. Nat. Genet. 2019, 51, 1092–1098. [Google Scholar] [CrossRef]
- Rosenspire, A.; Yoo, W.; Menard, S.; Torres, A.R. Autism spectrum disorders are associated with an elevated autoantibody response to tissue transglutaminase-2. Autism Res. 2011, 4, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.R.; Suenaga, T.; Edamura, M.; Fukuda, A.; Ishida, Y.; Nakahara, D.; Murakami, G. Functional MHCI deficiency induces ADHD-like symptoms with increased dopamine D1 receptor expression. Brain Behav. Immun. 2021, 97, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Al-Hakbany, M.; Awadallah, S.; Al-Ayadhi, L. The Relationship of HLA Class I and II Alleles and Haplotypes with Autism: A Case Control Study. Autism Res. Treat. 2014, 2014, 242048. [Google Scholar] [CrossRef] [PubMed]
- Puangpetch, A.; Suwannarat, P.; Chamnanphol, M.; Koomdee, N.; Ngamsamut, N.; Limsila, P.; Sukasem, C. Significant Association of HLA-B Alleles and Genotypes in Thai Children with Autism Spectrum Disorders: A Case-Control Study. Dis. Markers 2015, 2015, 724935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sollid, L.M.; Thorsby, E. HLA susceptibility genes in celiac disease: Genetic mapping and role in pathogenesis. Gastroenterology 1993, 105, 910–922. [Google Scholar] [CrossRef]
- Bennabi, M.; Gaman, A.; Delorme, R.; Boukouaci, W.; Manier, C.; Scheid, I.; Si Mohammed, N.; Bengoufa, D.; Charron, D.; Krishnamoorthy, R.; et al. HLA-class II haplotypes and Autism Spectrum Disorders. Sci. Rep. 2018, 8, 7639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, K.W.; Hauser, J.; Reissmann, A. Gluten-free and casein-free diets in the therapy of autism. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 572–575. [Google Scholar] [CrossRef]
- Restrepo, B.; Angkustsiri, K.; Taylor, S.L.; Rogers, S.J.; Cabral, J.; Heath, B.; Hechtman, A.; Solomon, M.; Ashwood, P.; Amaral, D.G.; et al. Developmental-behavioral profiles in children with autism spectrum disorder and co-occurring gastrointestinal symptoms. Autism Res. 2020, 13, 1778–1789. [Google Scholar] [CrossRef]
- Kim, T.; Vidal, G.S.; Djurisic, M.; William, C.M.; Birnbaum, M.E.; Garcia, K.C.; Hyman, B.T.; Shatz, C.J. Human LilrB2 is a beta-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer’s model. Science 2013, 341, 1399–1404. [Google Scholar] [CrossRef] [Green Version]
- Cao, Q.; Shin, W.S.; Chan, H.; Vuong, C.K.; Dubois, B.; Li, B.; Murray, K.A.; Sawaya, M.R.; Feigon, J.; Black, D.L.; et al. Inhibiting amyloid-beta cytotoxicity through its interaction with the cell surface receptor LilrB2 by structure-based design. Nat. Chem. 2018, 10, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Fan, X.; Wang, T.; Wu, J. High-throughput sequencing of autism spectrum disorders comes of age. Genet. Res. 2013, 95, 121–129. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).