Abstract
Beak color diversity is a broadly occurring phenomenon in birds. Here, we used ducks to identify candidate genes for yellow, black, and spotted beaks. For this, an F2 population consisting of 275 ducks was genotyped using whole genome resequencing containing 12.6 M single-nucleotide polymorphisms (SNPs) and three beak colors. Genome-wide association studies (GWAS) was used to identify the candidate and potential SNPs for three beak colors in ducks (yellow, spotted, and black). The results showed that 2753 significant SNPs were associated with black beaks, 7462 with yellow, and 17 potential SNPs with spotted beaks. Based on SNP annotation, MITF, EDNRB2, members of the POU family, and the SLC superfamily were the candidate genes regulating pigmentation. Meanwhile, isoforms MITF-M and EDNRB2 were significantly different between black and yellow beaks. MITF and EDNRB2 likely play a synergistic role in the regulation of melanin synthesis, and their mutations contribute to phenotypic differences in beak melanin deposition among individuals. This study provides new insights into genetic factors that may influence the diversity of beak color.
1. Introduction
Beak pigmentation is a common phenomenon observed in most birds. The color of feathers, coats, and skin is primarily determined by melanocytes, which are involved in the synthesis of melanin and play an important role in cosmetic change, heat regulation, camouflage, and protection against UV radiation from sun exposure [1,2,3]. In addition, melanin accumulation can lead to hyperpigmentation of the skin [4,5]. Previous studies have shown that skin color is highly heritable and one of the most variable phenotypes. This phenotype is influenced not only by genetic factors, but also by the environment [6,7]. Skin pigmentation is highly associated with latitude and fundamentally, the distribution of ultraviolet (UV) radiation [8]. Skin pigmentation is also influenced by the concerted action of different types of natural selection, including climate, lifestyle, diet, and metabolism [9,10].
With rapid development in genetics and genomics, researchers have gradually realized that human skin color diversity is due to the natural positive selection of those genes that affect human pigmentation, especially in melanosome biogenesis or melanin biosynthetic pathways [11,12,13,14]. Recently, a large number of genome-wide association studies (GWAS) of pigmentation have established that some single-nucleotide polymorphisms (SNPs) in TYR, IRF4, TYRP1, OCA2, SLC45A2, MC1R, and KITLG genes are significantly associated with human skin color [15,16,17]. Moreover, the α-MSH gene is a significant inherited factor that acts mainly as an agonist of MC1R [18]. Furthermore, SLC45A2 (also known as AIM1 or MATP) encodes a transporter that mediates melanin synthesis and is expressed in a high percentage of melanoma cell lines [19,20]. Several SLC45A2 mutations have been reported to lead to OCA4, and polymorphisms of this gene are significantly associated with human skin, hair, and eye pigmentation [21,22,23]. Selected signatures of skin pigmentation loci have been revealed by studies of modern and ancient populations, with some genes showing variation associated with light skin pigmentation also showing polygenic selection in western Eurasia [24]. However, this observation is the only recorded sign of polygenic selection for skin pigmentation based on only four loci (SLC24A5, SLC45A2, TYR, and APBA2/OCA2) [25].
Ducks, the second-largest poultry species in the world, mostly have yellow and black beaks. Occasionally, spotting occurs, which directly affects carcass sales. However, genetic factors that lead to the appearance of spot color remain unclear. Based on an F2 cross between the Cherry Valley Duck and Runzhou White Crested Duck, we performed a genome-wide association study (GWAS) with 275 birds to gain insight into the effects of genetic factors on beak pigmentation. These studies provided insight into the molecular regulatory mechanisms and genetic improvement of melanin deposition in duck beaks.
2. Materials and Methods
2.1. Ethical Approval
All experiments on ducks were performed in accordance with the Regulations on the Administration of Experimental Animals issued by the Ministry of Science and Technology (Beijing, China) in 1988 (last modified in 2001). The experimental protocols were approved by the Animal Care and Use Committee of the Yangzhou University (YZUDWSY2017-11-07). All efforts were made to minimize animal discomfort and suffering.
2.2. Samples and Sequencing
The F2 resource population, which crosses the Chinese Crested duck (CC duck) and Cherry Valley duck (CV duck), was obtained from the Laboratory of Poultry Genetic Resources Evaluation and Germplasm Utilization at Yangzhou University. The ducks were raised in stair-step cages under the recommended environmental and nutritional conditions at the conservation farm of Ecolovo Group, China. The CC duck is a Chinese indigenous breed that has a black shank and beak, while the CV duck is a commercial breed that has a yellow shank and beak and white plumage. In the F1 generation, 30 CC ducks and six CV ducks were randomly selected and divided into six families to interbreed. To generate F2 progeny, 30 males and 150 unrelated female ducks were used as hybrids. A total of 275 ducks were used in the next experiment. To identify candidate genes associated with beak color, we classified beak color as yellow, spotted, or black. The R/ggcor package was used to calculate the correlation between beak color and sex.
Blood samples were used to collect high-quality DNA at 42 days of age. Genomic DNA (gDNA) was extracted from blood samples by using the DNA extraction kit method (QIAampR DNA Blood Mini Kit), following the manufacturer’s protocol. Two paired-end sequencing libraries with insert sizes of 350 bp were constructed according to the Illumina protocol (Illumina, San Diego, CA, USA). All libraries were sequenced using the Illumina NovaSeq platform.
2.3. Genotyping
Raw reads were filtered using the NGS QC Toolkit (version 2.3) with default parameters [26]. The clean reads were mapped to the duck reference genome (the Chinese Crested duck genome was assembled by our lab (unpublished) with a Burrows–Wheeler alignment (BWA aln) using the default parameters) [27]. GATK then performed SNP calling [28]. VCF tools were used to further filter the raw data [29]. The SNPs were screened with parameters of a minor allele frequency (MAF) > 0.05, maximum allele frequency < 0.99, and maximum missing rate < 0.01. After filtering, SNPs showed a mean density of 8.5 SNPs/kb across the genome. All filtered SNPs were distributed on 37 autosomal chromosomes, ChrZ, and ChrU (unplaced scaffolds).
2.4. Population Structure
The population structure was assessed with multidimensional scaling (MDS) using PLINK 1.9 software [30]. Independent SNPs were obtained on all autosomes using the in-dep-pairwise option, with a 1000 bp window, five steps, and an r2 threshold of 0.2. Pairwise identity-by-state (IBS) distances between all individuals were calculated using these independent SNP markers, and MDS components were acquired using the mds-plot option based on the IBS matrix. A relative kinship matrix was constructed using these independent SNP markers.
2.5. Whole-Genome Association Analysis and Linkage Disequilibrium Analysis
The GWAS analysis of beak color used the linear mixed model in the Effective Mixed Model Association eXpedited (EMMAX) software [31]. EMMAX makes the simplifying and time-saving assumption that any given SNP’s effect on the trait is typically small, and therefore only estimates the model variance components once per analysis to account for population structure. EMMAX estimates variance components using the REML model.
where y is a vector of beak colors, X is the incidence matrix for a random additive effect, a is the column vector of random additive effects, Z is the genotype value of the candidate SNP, b is the regression coefficient of the candidate SNP, and e is the random residual. The phenotypic variance–covariance matrix is var(y) = Var(a) + var(e) = K σa2 + I σe2, where K is the IBS kinship matrix, I is the identity matrix, σa2 is the additive variance, and σe2 is the variance of random residuals. The regional Manhattan plot and LD heatmap were obtained using LDBlockShow software.
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses
Based on the LD attenuation distance calculated using PopLDdecay [32], annotation of related genes in a certain region upstream and downstream of the physical location of the significant SNPs were performed. The sequences of the relevant genes were extracted from the mallard genome and translated into a protein sequence, which was then analyzed using KOBAS 3.0 software [33]. Chicken was selected as the reference species, and hypergeometric tests along with Fisher’s exact test were used as the statistical methods.
3. Results
3.1. Phenotypic Description and Population Structure Analysis
To identify the candidate genes for beak color, we first focused on the correlation between beak color and sex to determine whether there was a correlation between these two traits. The results showed that there was no correlation between black, yellow, or spotted beak color and sex (Figure 1a). We also identified the population structure of all the samples used in the present study using MDS. The results showed that the three different beak-colored ducks had no obvious clustering and were evenly distributed (Figure 1b).

Figure 1.
Beak color and sex correlation analysis (a); population structure analysis (b).
3.2. Genome-Wide Association Study Identified the Candidate Variants for Beak Color
EMMAX software was used to conduct the genome-wide association analysis in the present study. The Q–Q plot illustrated that the model used for the GWAS analysis was reasonable. The lambdas (inflation factor (λ)) of the three different color beaks were 0.98 (black beak), 1.05 (spotted beak), and 0.99 (yellow beak), and the points at the upper right corner of the Q–Q plots were the significant markers associated with the traits under study (Figure 2). Thus, population stratification was adequately controlled. However, no significantly associated sites were found in the GWAS analysis of spotted beaks in the Q–Q plot, but we found a large number of potential associated sites for spotted beak through the Manhattan plot.
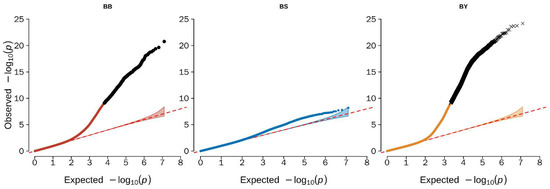
Figure 2.
Quantile–quantile (Q–Q) from GWAS for beak color trait in duck. Q–Q plot showing the late separation between observed and expected values. The red lines indicate the null hypothesis of no true association. Deviation from the expected p-value distribution is evident only in the tail area for each trait, indicating that population stratification was properly controlled. BB refers to black beak; BS refers to spotted beak; BY refers to yellow beak.
The Manhattan plot of beak color showed that a total of 2753 significant SNPs associated with black beak were identified using the threshold of significant p-value (threshold = 0.05/total number of all SNPs = 3.94885 × 10−09), and 1916 extremely significant SNPs were identified using the threshold of significant p-value (threshold = 0.01/total number of all SNPs = 7.8977 × 10−10), most of which were located on chromosome 14 (ALP 14) (2708 SNPs) and ALP 11 (45 SNP) (Figure 3, top).
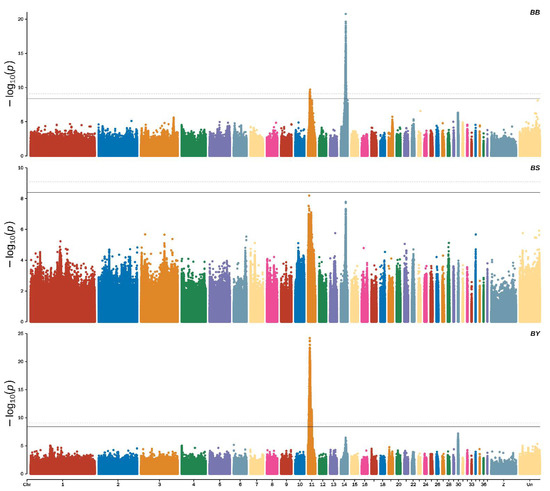
Figure 3.
Manhattan plots showing the significance of genetic effects on the beak color according to the GWAS.
For yellow beak, 7462 significant SNPs were identified using the threshold of significant p-value (threshold = 0.05/total number of all SNPs = 3.94885 × 10−09), and 5878 extremely significant SNPs were identified using the threshold of significant p-value (threshold = 0.01/total number of all SNPs = 7.8977 × 10−10), most of which were located in ALP11 (Figure 3, middle).
For spotted beaks, there were no significant SNPs associated with them (p-value ≤ 3.94885 × 10−09). However, we identified 17 potential SNPs associated with spotted beaks using the threshold of significant p-value (threshold = 1/total number of all SNPs = 7.8977 × 10−08) (Figure 3, bottom). We found 45 shared SNPs between black and yellow beaks, 11 shared SNPs between black and spotted beaks, and 5 shared SNPs between yellow and spotted beaks using a Venn analysis (Figure 4). In addition, based on the result of all SNP synonymous analysis that all exonic SNPs were synonymous and not predicted to alter protein function.
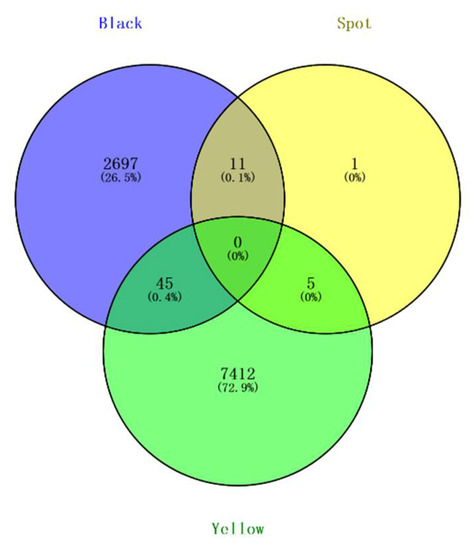
Figure 4.
Venn analysis of all beak colors showing overlap of significant SNPs.
3.3. Functional Analysis
The genes or genomic regions identified in the GWAS explained only part of the genetic variation. To overcome this limitation, the GWAS was complemented with a gene set analysis using GO and KEGG databases to detect potential functional categories underlying the beak color. Based on the SNP annotation, we found that 94 genes, including MITF, EDNRB2, SLC25A43, SLC25A5, SLC25A14, SPRY3, POU4F3, etc. (Table S1), were the most significant candidates associated with a black beak. The results of the KEGG and GO enrichment analysis showed that these candidate genes were involved in melanogenesis, necroptosis, calcium signaling, the FoxO signaling pathway, primary bile acid biosynthesis, apoptosis, the positive regulation of secretion by cells, positive regulation of secretion, and the mitochondrial part (Figure 5a). In addition, MITF, POU4F1, POU3F3B, POU1F1, POU2F1, SLC7A1, SLC46A3, SLC25A15, SLC25A30, SLC15A1, SLC10A2, SLC5A7, SLC9A2, SLC9A4, and others were most significantly associated with yellow beak (Table S2). Cytokine–cytokine receptor interaction, apoptosis, phototransduction, the thyroid hormone signaling pathway, DNA replication, axon guidance, the cell adhesion molecule pathway, cytokine receptor activity, lipid-transporting ATPase activity, gap junction channel activity, negative regulation of adherens junction organization, wide pore channel activity, axon choice point recognition, axon midline choice point recognition, negative regulation of negative chemotaxis, cellular response to vitamin K, positive regulation of small-molecule metabolic processes, microtubule cytoskeleton, and pigmentation were enriched (Figure 5b). Finally, we found MITF, TMLHE, EDNRB2, NUP98, ITPR1, CHL1, ALG8, SPRY3, and PDHB genes (Table S3), which are involved in melanogenesis, the calcium signaling pathway, the citrate cycle (TCA cycle), lysine degradation, etc.; and dolichyl pyrophosphate Glc1Man9GlcNAc2 α-1,3-glucosyltransferase activity, trimethyllysine dioxygenase activity, inositol 1,4,5-trisphosphate receptor activity involved in regulation of postsynaptic cytosolic calcium levels, pyruvate dehydrogenase (acetyl-transferring) activity, inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity, the carnitine biosynthetic process, α-1,3-mannosyltransferase activity, endothelin receptor activity, the amino acid betaine biosynthetic process, and regulation of postsynaptic cytosolic calcium ion concentration terms were the potential candidate genes for spotted beak (Figure 5c).

Figure 5.
Functional enrichment analysis of the beak color candidate genes. (a) KEGG (left) and GO (right) enrichment of black beak candidate genes; (b) KEGG (left) and GO (right) enrichment of spotted beak candidate genes; (c) KEGG (left) and GO (right) enrichment of yellow beak candidate genes.
3.4. The EDNRB2 and MITF Isoform Expression Level in Black and Yellow Beaks
The genes responsible for melanoblast migration and melanocyte development include EDN3, EDNRB, EDNRB2, MITF, KIT, and KITLG [34]. Based on the GWAS analysis, we found that MITF and EDNRB2 were candidate genes for beak color, and therefore might regulate pigmentation. Duck MITF consists of two isoforms, MITF-B and MITF-M, with isoform-specific first exons called 1B and 1M, respectively (Figure 6a). Thus, we determined the expression levels of these two isoforms in black and yellow beaks. The results showed that the MITF-M was significantly expressed in black beaks (Figure 6b). The EDNRB2 expression levels were compared between black and yellow beaks. EDNRB2 expression levels showed that EDNRB2 was significantly expressed in black beaks (Figure 6b).
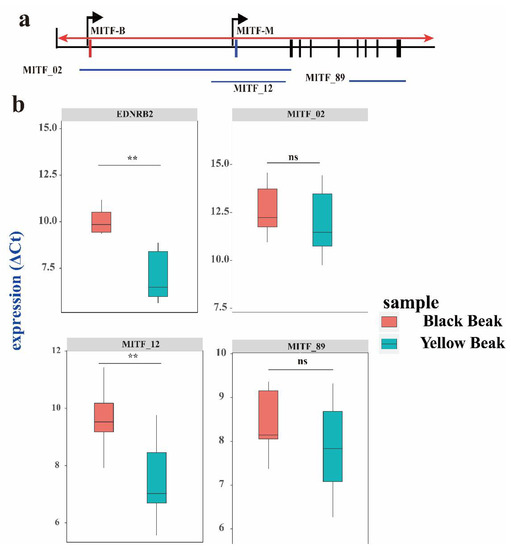
Figure 6.
Expression differences in EDNRB2 and MITF on three exon junctions between black and yellow beaks according to RT-qPCR. (a) Information on the MITF isoform. The red triangle represents the intronic insertion on chromosome 13 in Pekin ducks. Exon 1M is specific for the MITF-M transcript, while exon 1B is specific for the MITF-B transcript. (b) EDNRB2 and MITF on three exon junctions between black and yellow beaks. Each exon junction was assayed in six biological replicates with three technical replicates. The indicated p-values were based on one-way ANOVA. NS, nonsignificant; **, extremely significant.
4. Discussion
Melanin is a substance in the body that is responsible for hair, eye, and skin pigmentation [35]. Melanin is a complex polymer that originates from the amino acid tyrosine [36]. Melanin is present in human and animal skin to varying degrees, and is responsible for unique eye, hair, and skin colors [4,35,37]. The color of a bird’s beak, which is the exposed skin tissue, results from concentrations of pigments, primarily melanin and carotenoids, in the epidermal layers, including the rhamphotheca [38]. In duck beaks, melanin deposition increases with age and UV exposure [39,40]. However, under the influence of domestication and selection, many duck breeds have a significantly fixed and stable inheritance of the black beak, as seen in Chinese Crested ducks and Lianchen ducks. Most duck breeds exhibit yellow beaks. However, some duck breeds also exhibit spotted beaks. To determine the genetic basis of beak color diversity, the present study designed an F2 population cross between Chinese Crested ducks and Cherry Valley ducks. A GWAS was performed to identify candidate genes associated with different beak colors. In black beaks, we found two significantly associated signals: MITF and EDNRB2. Most genes belonging to the SLC superfamily and POU (Pit-Oct-Unc) family were significantly associated with yellow beaks. Although we did not identify a significantly associated signal in the spotted beak, we found two candidate signal loci on chromosomes 11 and 14. By annotating the candidate signals, we found that MITF and EDNRB2, two key genes responsible for melanin synthesis, were enriched.
MITF has been shown to affect pigmentation in cattle [41,42,43,44], mice [45,46,47], horses [48,49,50,51], dogs [52,53], humans [54,55], and ducks [56]. MITF belongs to the basic helix–loop–helix–leucine zipper (bHLHZip) family of proteins. Studies have shown that it regulates melanogenesis by binding to the highly conserved M-box (GTCATGTGCT) and E-box (TCATGTG) motifs upstream of the tyrosinase promoter, thereby strongly stimulating and promoting the activity of the tyrosinase promoter. Tyrosinase expression promotes melanin production [57,58,59,60]. Our results implied that melanin synthesis and metabolic pathways play crucial roles in inducing melanin deposition in beaks and genes related to melanin synthesis and metabolic pathways, such as MITF, MC1R, EDNRB2, the PMEL family, TYR, TYRP1, and TYRP2, affect melanin syntheses. Our findings showed similar results, including a significant association of MITF.
EDNRB is a gene expressed in melanocytes that are derived from the neural crest, and for this reason, EDNRB is particularly mentioned. EDNRB2 is a homolog of EDNRB, which belongs to the endothelin receptor (EDNR) gene family and has been lost in mammalian lineages. EDNRB signaling is required for melanocyte development [61,62]. Loss of function variants in EDNRB leads to white spotting phenotypes in humans, animals [63,64], and poultry [65,66]. A similar result was identified in our study, which showed that EDNRB2 regulated melanin pigmentation in ducks. In addition, our results showed that the spotted beak was mainly coregulated by MITF and EDNRB2. However, the mechanism by which MITF and EDNRB2 are coregulated requires further investigation.
Our results suggested that the POU family and SLC superfamily are significantly associated with yellow beaks. Yellow skin, beaks, and feet in most birds are caused by carotene deposits. The results showed that most members of the POU family, which share the typical POU domain structure [67], were significantly associated with yellow beaks. The POU family is a transcription factor family member that can promote the transcription of many genes related to development and metabolism, especially in Schwann and progenitor cell development [68,69]. Thirteen POU gene family members were found in the duck genome. In the present study, 44 POU transcription factors were predicted to be distributed within the promoter region of MITF [40], and POU4F1, POU3F3B, POU1F1, and POU2F1 were identified. Therefore, we speculated that the yellow beak is coregulated by the POU family and MITF, but the specific mechanism requires further experimental verification. This study provided new clues for understanding the genetic factors that may influence the diversity of beak color, but further experimental studies are needed to strengthen this hypothesis.
5. Conclusions
This study identified candidate genes closely related to duck beak color. MITF and EDNRB2 were the candidate genes associated with beak melanosis. We speculated that beak pigmentation may be coregulated by the POU family, MITF, and EDNRB2. However, the specific mechanisms require further experimental verification.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13071271/s1. Table S1. The black beak candidate associated SNP location; Table S2. The yellow beak candidate associated SNP location; Table S3. The spotted beak candidate associated SNP location.
Author Contributions
Q.G. performed the data analysis; G.C. (Guobin Chang) conceived and designed the experiments, Y.B., Y.J., H.B., Z.W., G.C. (Guohong Chen) and G.C. (Guobin Chang) contributed to the interpretation of data and to reviewing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Postgraduate Research and Innovation in Jiangsu Province (grant: KYCX21_3258), China Agriculture Research System of MOF and MARA (grant: CARS-42), and Jiangsu Agricultural Industry Technology System (grant: JATS [2021]326). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board (or Ethics Committee) of Regulations on the Administration of Experimental Animals issued by the Ministry of Science and Technology (Beijing, China) in 1988 (last modified in 2001), and Animal Care and Use Committee of the Yangzhou University (YZUDWSY2017-11-07).
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome assembly and all of the resequencing data used in this research were deposited in the Genome Sequence Archive (GSA) at National Genomics Data Center (http://bigd.big.ac.cn (accessed on: 5 February 2022)) Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA005019).
Acknowledgments
The authors thank the Zhenjiang Tiancheng Agricultural Technology Co., Ltd. for providing duck embryos.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Galván, I.; Rodríguez-Martínez, S.; Carrascal, L. Dark pigmentation limits thermal niche position in birds. Funct. Ecol. 2018, 32, 1531–1540. [Google Scholar] [CrossRef]
- Glogau, R.G. Physiologic and structural changes associated with aging skin. Dermatol. Clin. 1997, 15, 555–559. [Google Scholar] [CrossRef]
- Rittié, L.; Fisher, G.J. Natural and Sun-Induced Aging of Human Skin. Cold Spring Harb. Perspect. Med. 2015, 5, a015370. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Xu, S. Adaptation of human skin color in various populations. Hereditas 2017, 155, 1. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Hearing, V.J. Melanocytes and their diseases. Cold Spring Harb. Perspect. Med. 2014, 4, a017046. [Google Scholar] [CrossRef] [PubMed]
- Shekar, S.N.; Luciano, M.; Duffy, D.L.; Martin, N.G. Genetic and Environmental Influences on Skin Pattern Deterioration. J. Investig. Dermatol. 2005, 125, 1119–1129. [Google Scholar] [CrossRef]
- Hubbard, J.K. Environmental and Genetic Influences on Melanin-Based Plumage Coloration: Implications for Population Divergence. Ph.D. Thesis, University of Colorado at Boulder, Boulder, CO, USA, 2014. [Google Scholar]
- Lin, J.Y.; Fisher, D.E. Melanocyte biology and skin pigmentation. Nature 2007, 445, 843–850. [Google Scholar] [CrossRef]
- Huang, X.; Otecko, N.O.; Peng, M.; Weng, Z.; Li, W.; Chen, J.; Zhong, M.; Zhong, F.; Jin, S.; Geng, Z.; et al. Genome-wide genetic structure and selection signatures for color in 10 traditional Chinese yellow-feathered chicken breeds. BMC Genom. 2020, 21, 316. [Google Scholar] [CrossRef]
- Aoki, K. Sexual selection as a cause of human skin colour variation: Darwin’s hypothesis revisited. Ann. Hum. Biol. 2002, 29, 589–608. [Google Scholar] [CrossRef]
- Wang, Y. Association of pigmentation related-genes polymorphisms and geographic environmental variables in the Chinese population. Hereditas 2021, 158, 24. [Google Scholar] [CrossRef]
- Pavan, W.J.; Sturm, R.A. The Genetics of Human Skin and Hair Pigmentation. Annu. Rev. Genom. Hum. Genet. 2019, 20, 41–72. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Pacheco, N.; Flores, C.; Alonso, S.; Eng, C.; Mak, A.C.; Hunstman, S.; Hu, D.; White, M.J.; Oh, S.S.; Meade, K.; et al. Identification of a novel locus associated with skin colour in African-admixed populations. Sci. Rep. 2017, 7, 44548. [Google Scholar] [CrossRef] [PubMed]
- A Sturm, R.; Duffy, D.L. Human pigmentation genes under environmental selection. Genome Biol. 2012, 13, 248. [Google Scholar] [CrossRef]
- Gerstenblith, M.R.; Shi, J.; Landi, M.T. Genome-wide association studies of pigmentation and skin cancer: A review and meta-analysis. Pigment. Cell Melanoma Res. 2010, 23, 587–606. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.D.; Pairo-Castineira, E.; Rawlik, K.; Canela-Xandri, O.; Rees, J.; Sims, D.; Tenesa, A.; Jackson, I.J. Genome-wide study of hair colour in UK Biobank explains most of the SNP heritability. Nat. Commun. 2018, 9, 5271. [Google Scholar] [CrossRef] [PubMed]
- Zhong, K.; Verkouteren, J.A.; Jacobs, L.C.; Uitterlinden, A.G.; Hofman, A.; Liu, F.; Nijsten, T.; Kayser, M. Pigmentation-Independent Susceptibility Loci for Actinic Keratosis Highlighted by Compound Heterozygosity Analysis. J. Investig. Dermatol. 2017, 137, 77–84. [Google Scholar] [CrossRef]
- Moscowitz, A.E.; Asif, H.; Lindenmaier, L.B.; Calzadilla, A.; Zhang, C.; Mirsaeidi, M. The Importance of Melanocortin Receptors and Their Agonists in Pulmonary Disease. Front. Med. 2019, 6, 145. [Google Scholar] [CrossRef]
- Serre, C.; Busuttil, V.; Botto, J.-M. Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. Int. J. Cosmet. Sci. 2018, 40, 328–347. [Google Scholar] [CrossRef]
- Le, L.; Escobar, I.E.; Ho, T.; Lefkovith, A.J.; Latteri, E.; Haltaufderhyde, K.D.; Dennis, M.K.; Plowright, L.; Sviderskaya, E.V.; Bennett, D.C.; et al. SLC45A2 protein stability and regulation of melanosome pH determine melanocyte pigmentation. Mol. Biol. Cell 2020, 31, 2687–2702. [Google Scholar] [CrossRef]
- Tóth, L.; Fábos, B.; Farkas, K.; Sulák, A.; Tripolszki, K.; Széll, M.; Nagy, N. Identification of two novel mutations in the SLC45A2 gene in a Hungarian pedigree affected by unusual OCA type 4. BMC Med. Genet. 2017, 18, 27. [Google Scholar] [CrossRef]
- Cook, A.L.; Chen, W.; Thurber, A.E.; Smit, D.J.; Smith, A.G.; Bladen, T.G.; Brown, D.L.; Duffy, D.L.; Pastorino, L.; Bianchi-Scarra, G.; et al. Analysis of Cultured Human Melanocytes Based on Polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P Loci. J. Investig. Dermatol. 2009, 129, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Reinders, A.; Ward, J.M. Investigating polymorphisms in membrane-associated transporter protein SLC45A2, using sucrose transporters as a model. Mol. Med. Rep. 2012, 12, 1393–1398. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fang, W.; Huang, J.; Li, S. The application of genome editing technology in fish. Mar. Life Sci. Technol. 2021, 3, 326–346. [Google Scholar] [CrossRef]
- Martin, A.R.; Lin, M.; Granka, J.M.; Myrick, J.W.; Liu, X.; Sockell, A.; Atkinson, E.G.; Werely, C.J.; Möller, M.; Sandhu, M.S.; et al. An Unexpectedly Complex Architecture for Skin Pigmentation in Africans. Cell 2017, 171, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Paschon, D.E.; Lussier, S.; Wangzor, T.; Xia, D.F.; Li, P.W.; Hinkley, S.J.; Scarlott, N.A.; Lam, S.C.; Waite, A.J.; Truong, L.N.; et al. Diversifying the structure of zinc finger nucleases for high-precision genome editing. Nat. Commun. 2019, 10, 1133. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Cieslak, M.; Reissmann, M.; Hofreiter, M.; Ludwig, A. Colours of domestication. Biol. Rev. 2011, 86, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A.; Tobin, D.J.; Shibahara, S.; Wortsman, J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol. Rev. 2004, 84, 1155–1228. [Google Scholar] [CrossRef]
- Zheng, P.; Sun, H.; Liu, J.; Lin, J.; Zhang, X.; Qin, Y.; Zhang, W.; Xu, X.; Deng, X.; Yang, D.; et al. Comparative analyses of American and Asian lotus genomes reveal insights into petal color, carpel thermogenesis and domestication. Plant J. 2022, 110, 1498–1515. [Google Scholar] [CrossRef] [PubMed]
- Naik, P.P.; Farrukh, S.N. Influence of Ethnicities and Skin Color Variations in Different Populations: A Review. Ski. Pharmacol. Physiol. 2021, 35, 65–76. [Google Scholar] [CrossRef]
- Ralph, C.L. The Control of Color in Birds. Am. Zool. 1969, 9, 521–530. [Google Scholar] [CrossRef]
- Hitselberger, M.H.; Schleicher, R.L.; Beattie, C.W. Effects of Estradiol on Estrogen Receptor, Progesterone Receptor, and Tyrosinase in Hamster Melanoma Transplanted into Athymic Mice1. Cancer Res. 1988, 48, 3720–3727. [Google Scholar]
- Liu, H.; Wang, J.; Hu, J.; Wang, L.; Guo, Z.; Fan, W.; Xu, Y.; Liu, D.; Zhang, Y.; Xie, M.; et al. Genome-wide association analysis reveal the genetic reasons affect melanin spot accumulation in beak skin of ducks. BMC Genom. 2022, 23, 236. [Google Scholar] [CrossRef]
- Hofstetter, S.; Seefried, F.; Häfliger, I.; Jagannathan, V.; Leeb, T.; Drögemüller, C. A non-coding regulatory variant in the 5′-region of the MITF gene is associated with white-spotted coat in Brown Swiss cattle. Anim. Genet. 2018, 50, 27–32. [Google Scholar] [CrossRef]
- Philipp, U.; Lupp, B.; Mömke, S.; Stein, V.; Tipold, A.; Eule, J.C.; Rehage, J.; Distl, O. A MITF Mutation Associated with a Dominant White Phenotype and Bilateral Deafness in German Fleckvieh Cattle. PLoS ONE 2011, 6, e28857. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Scotti, E.; Russo, V. Haplotype variability in the bovine MITF gene and association with piebaldism in Holstein and Simmental cattle breeds. Anim. Genet. 2011, 43, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Edea, Z.; Dadi, H.; Dessie, T.; Kim, I.-H.; Kim, K.-S. Association of MITF loci with coat color spotting patterns in Ethiopian cattle. Genes Genom. 2016, 39, 285–293. [Google Scholar] [CrossRef]
- Nakayama, A.; Nguyen, M.-T.T.; Chen, C.C.; Opdecamp, K.; A Hodgkinson, C.; Arnheiter, H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech. Dev. 1998, 70, 155–166. [Google Scholar] [CrossRef]
- Cheli, Y.; Guiliano, S.; Botton, T.; Rocchi, S.; Hofman, V.; Hofman, P.; Bahadoran, P.; Bertolotto, C.; Ballotti, R. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene 2011, 30, 2307–2318. [Google Scholar] [CrossRef]
- Hou, L.; Pavan, W.J. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: Do all roads lead to Mitf? Cell Res. 2008, 18, 1163–1176. [Google Scholar] [CrossRef]
- Hauswirth, R.; Haase, B.; Blatter, M.; Brooks, S.; Burger, D.; Drögemüller, C.; Gerber, V.; Henke, D.; Janda, J.; Jude, R.; et al. Mutations in MITF and PAX3 Cause “Splashed White” and Other White Spotting Phenotypes in Horses. PLoS Genet. 2012, 8, e1002653. [Google Scholar] [CrossRef]
- Magdesian, K.G.; Tanaka, J.; Bellone, R.R. A De Novo MITF Deletion Explains a Novel Splashed White Phenotype in an American Paint Horse. J. Hered. 2020, 111, 287–293. [Google Scholar] [CrossRef]
- Henkel, J.; Lafayette, C.; Brooks, S.A.; Martin, K.; Patterson-Rosa, L.; Cook, D.; Jagannathan, V.; Leeb, T. Whole-genome sequencing reveals a large deletion in the MITF gene in horses with white spotted coat colour and increased risk of deafness. Anim. Genet. 2019, 50, 172–174. [Google Scholar] [CrossRef]
- Hauswirth, R.; Jude, R.; Haase, B.; Bellone, R.R.; Archer, S.; Holl, H.; Brooks, S.A.; Tozaki, T.; Penedo, M.C.; Rieder, S.; et al. Novel variants in the KIT and PAX3 genes in horses with white-spotted coat colour phenotypes. Anim. Genet. 2013, 44, 763–765. [Google Scholar] [CrossRef]
- Körberg, I.B.; Sundström, E.; Meadows, J.R.S.; Pielberg, G.R.; Gustafson, U.; Hedhammar, Å.; Karlsson, E.K.; Seddon, J.; Söderberg, A.; Vilà, C.; et al. A Simple Repeat Polymorphism in the MITF-M Promoter Is a Key Regulator of White Spotting in Dogs. PLoS ONE 2014, 9, e104363. [Google Scholar] [CrossRef]
- Schmutz, S.M.; Berryere, T.G.; Dreger, D.L. MITF and White Spotting in Dogs: A Population Study. J. Hered. 2009, 100, S66–S74. [Google Scholar] [CrossRef][Green Version]
- Léger, S.; Balguerie, X.; Goldenberg, A.; Drouin-Garraud, V.; Cabot, A.; Amstutz-Montadert, I.; Young, P.; Joly, P.; Bodereau, V.; Holder-Espinasse, M.; et al. Novel and recurrent non-truncating mutations of the MITF basic domain: Genotypic and phenotypic variations in Waardenburg and Tietz syndromes. Eur. J. Hum. Genet. 2012, 20, 584–587. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, K.; Yokoyama, K.; Shibata, K.; Tomita, Y.; Shibahara, S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol. Cell. Biol. 1994, 14, 8058–8070. [Google Scholar] [CrossRef]
- Zhou, Z.K.; Li, M.; Cheng, H.; Fan, W.L.; Yuan, Z.R.; Gao, Q.; Xu, Y.X.; Guo, Z.B.; Zhang, Y.S.; Hu, J.; et al. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 2018, 9, 2648. [Google Scholar] [CrossRef]
- Wang, Y.; Li, S.-M.; Huang, J.; Chen, S.-Y.; Liu, Y.-P. Mutations of TYR and MITF Genes are Associated with Plumage Colour Phenotypes in Geese. Asian-Australas. J. Anim. Sci. 2014, 27, 778–783. [Google Scholar] [CrossRef]
- Simcoe, M.; Valdes, A.; Liu, F.; Furlotte, N.A.; Evans, D.M.; Hemani, G.; Ring, S.M.; Smith, G.D.; Duffy, D.L.; Zhu, G.; et al. Genome-wide association study in almost 195,000 individuals identifies 50 previously unidentified genetic loci for eye color. Sci. Adv. 2021, 7, eabd1239. [Google Scholar] [CrossRef]
- Asgari, M.M.; Wang, W.; Ioannidis, N.M.; Itnyre, J.; Hoffmann, T.; Jorgenson, E.; Whittemore, A.S. Identification of Susceptibility Loci for Cutaneous Squamous Cell Carcinoma. J. Invest. Dermatol. 2016, 136, 930–937. [Google Scholar] [CrossRef]
- Wu, Z.; Deng, Z.; Huang, M.; Hou, Y.; Zhang, H.; Chen, H.; Ren, J. Whole-Genome Resequencing Identifies KIT New Alleles That Affect Coat Color Phenotypes in Pigs. Front. Genet. 2019, 10, 218. [Google Scholar] [CrossRef]
- Pla, P.; LaRue, L. Involvement of endothelin receptors in normal and pathological development of neural crest cells. Int. J. Dev. Biol. 2003, 47, 315–325. [Google Scholar]
- Braasch, I.; Volff, J.-N.; Schartl, M. The Endothelin System: Evolution of Vertebrate-Specific Ligand-Receptor Interactions by Three Rounds of Genome Duplication. Mol. Biol. Evol. 2009, 26, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Verheij, J.B.G.M.; Kunze, G.; Osinga, J.; van Essen, A.J.; Hofstra, R.M.W. ABCD syndrome is caused by a homozygous mutation in the EDNRB gene. Am. J. Med. Genet. 2002, 108, 223–225. [Google Scholar] [CrossRef]
- Kinoshita, K.; Akiyama, T.; Mizutani, M.; Shinomiya, A.; Ishikawa, A.; Younis, H.H.; Tsudzuki, M.; Namikawa, T.; Matsuda, Y. Endothelin receptor B2 (EDNRB2) is responsible for the tyrosinase-independent recessive white (mo(w)) and mottled (mo) plumage phenotypes in the chicken. PLoS ONE 2014, 9, e86361. [Google Scholar] [CrossRef] [PubMed]
- Vignal, A.; Boitard, S.; Thébault, N.; Dayo, G.-K.; Yapi-Gnaore, V.; Karim, I.Y.A.; Berthouly-Salazar, C.; Pálinkás-Bodzsár, N.; Guémené, D.; Thibaud-Nissen, F.; et al. A guinea fowl genome assembly provides new evidence on evolution following domestication and selection in galliformes. Mol. Ecol. Resour. 2019, 19, 997–1014. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, Q.; Huang, Q.; Ma, S.; Wang, Y.; Han, C.; Zhang, R.; Wang, J.; Liu, H.; Li, L. Genome-wide association analysis reveals that EDNRB2 causes a dose-dependent loss of pigmentation in ducks. BMC Genom. 2021, 22, 381. [Google Scholar] [CrossRef]
- Besch, R.; Berking, C. POU transcription factors in melanocytes and melanoma. Eur. J. Cell Biol. 2014, 93, 55–60. [Google Scholar] [CrossRef]
- Huang, S.; Sato, S. Progenitor cells in the adult zebrafish nervous system express a Brn-1-related POU gene, tai-ji. Mech. Dev. 1998, 71, 23–35. [Google Scholar] [CrossRef]
- Eng, S.R.; Dykes, I.M.; Lanier, J.; Fedtsova, N.; E Turner, E. POU-domain factor Brn3a regulates both distinct and common programs of gene expression in the spinal and trigeminal sensory ganglia. Neural Dev. 2007, 2, 3–17. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).