Genome-Wide Identification and Expression Analysis of Expansin Gene Family in the Storage Root Development of Diploid Wild Sweetpotato Ipomoea trifida
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Expansin Genes in I. trifida
2.2. Sequence Alignment and Phylogenetic Analysis
2.3. Chromosomal Localization and Gene Duplication Analysis
2.4. Conserved Motif, Gene Structure, and Promoter Analysis
2.5. Prediction of Protein Interaction
2.6. RNA-Sequencing (RNA-Seq) and Analysis
2.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.8. Gene Expression Analysis of Development Responsive Interacted Proteins
3. Results
3.1. Identification and Characterization of Expansin Family Members in Y22
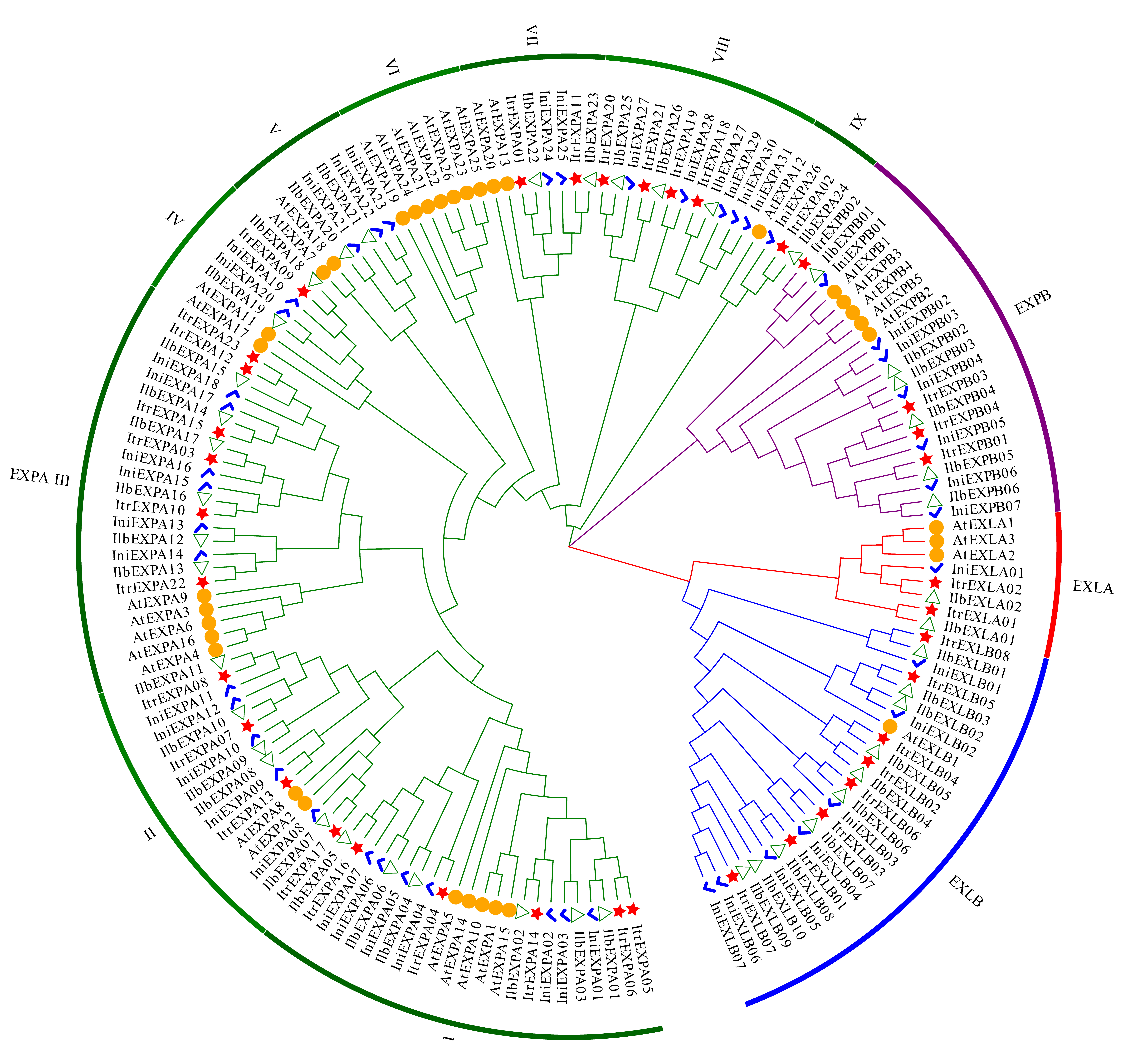
3.2. Phylogenetic Analysis of Expansins
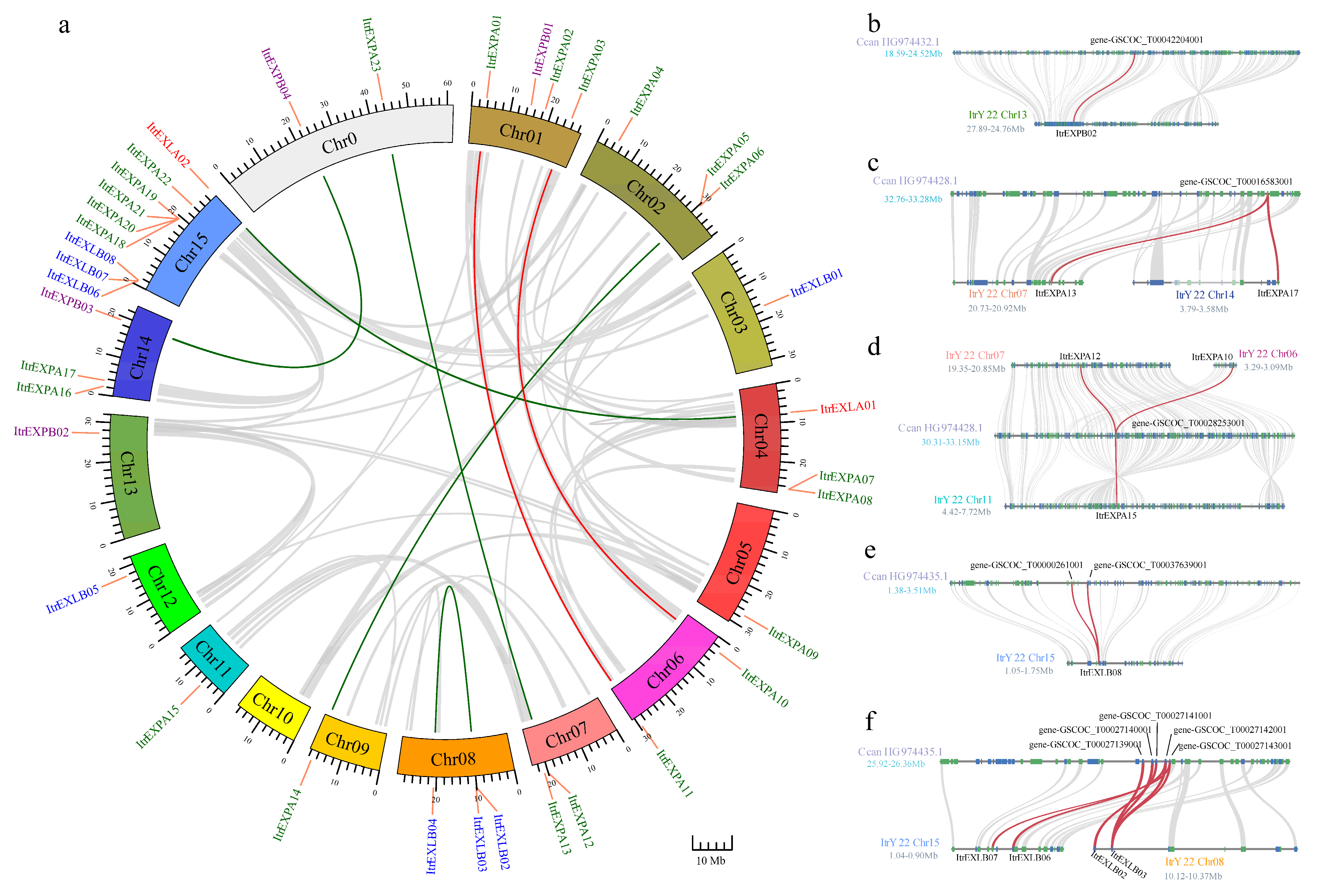
3.3. Chromosomal Location and Gene Duplication
3.4. Protein Domains and Gene Structure Analysis
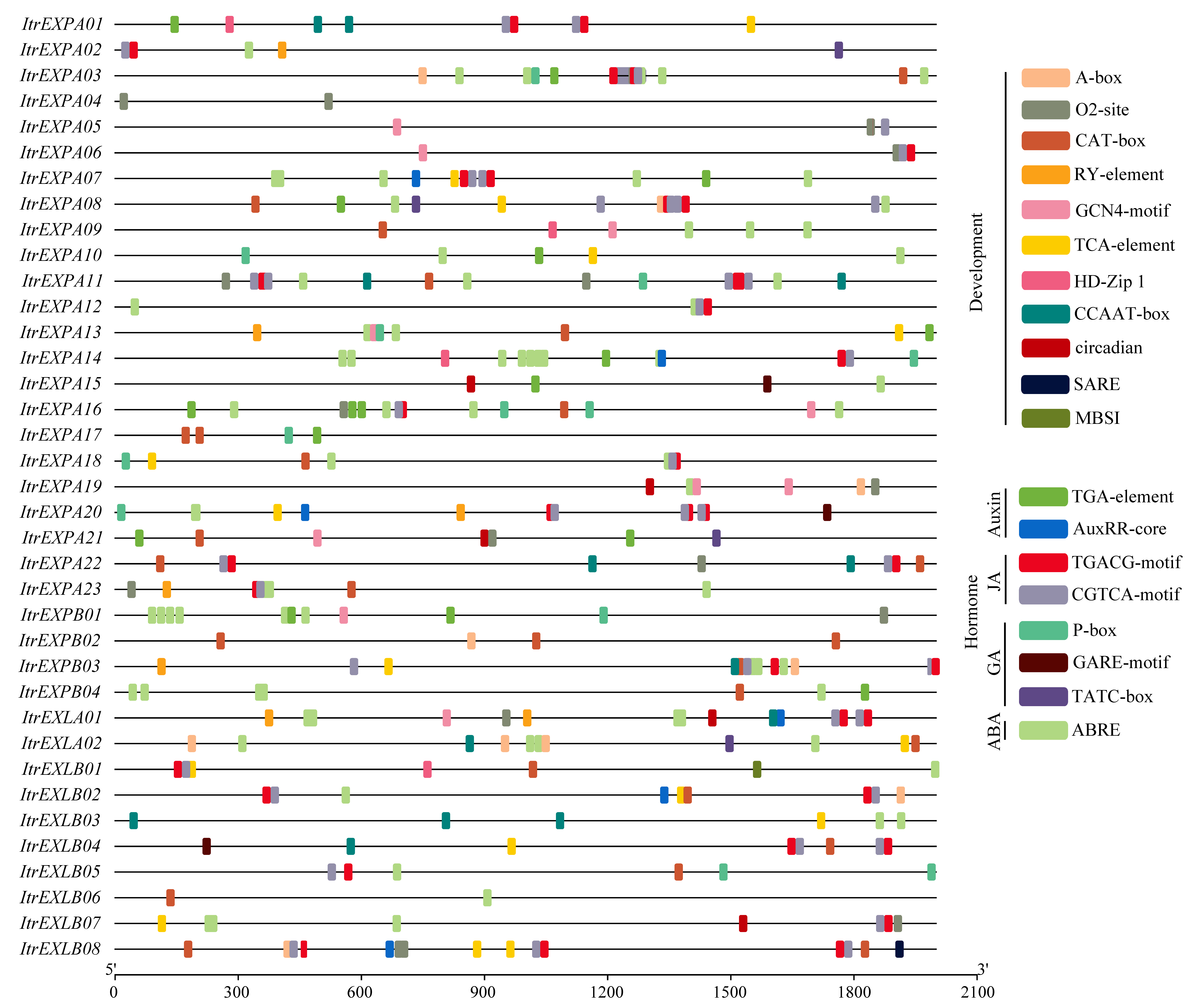
3.5. Potential Cis-Element Analysis in Gene Promoter Regions
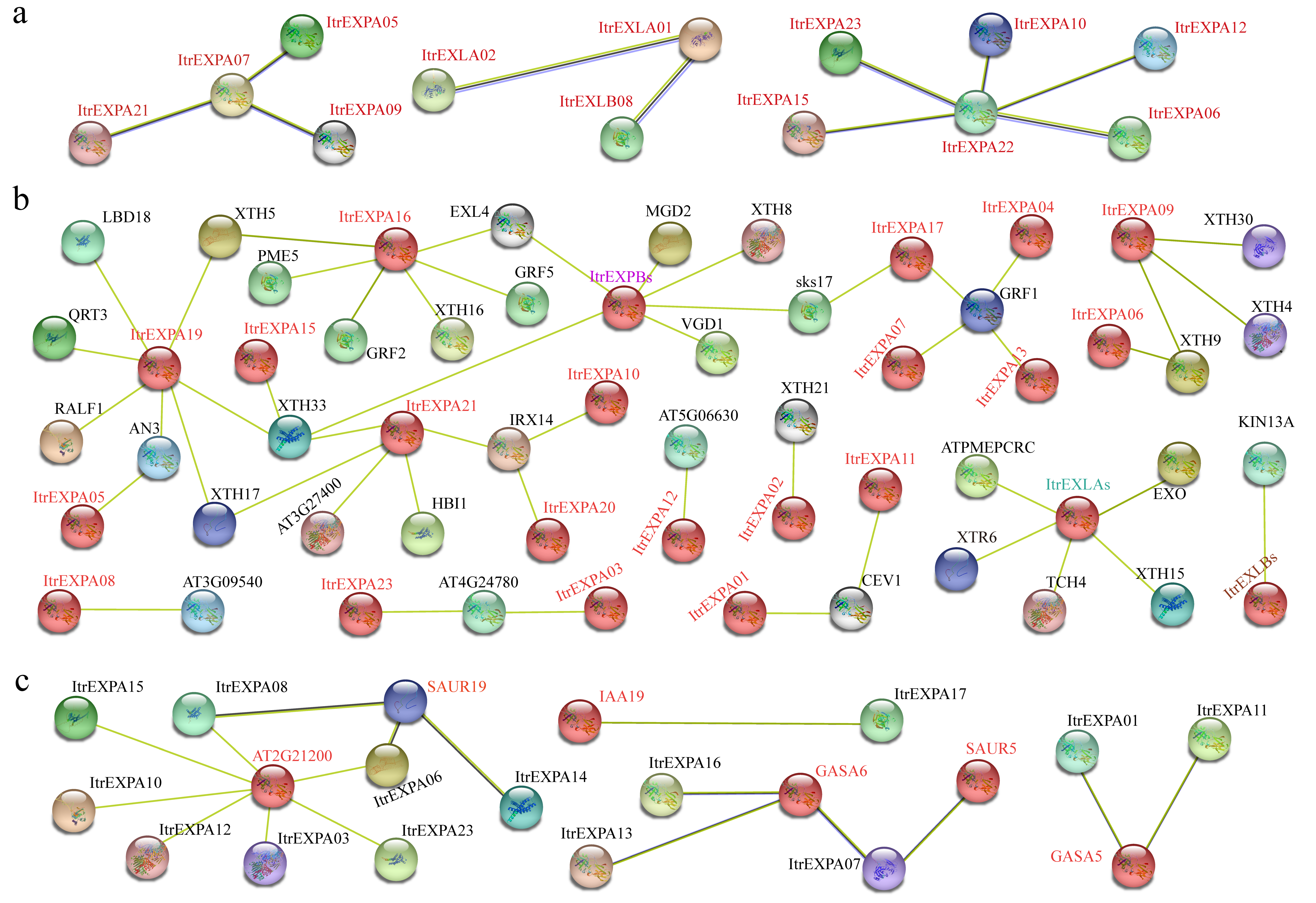
3.6. Protein Interaction Network of Expansins
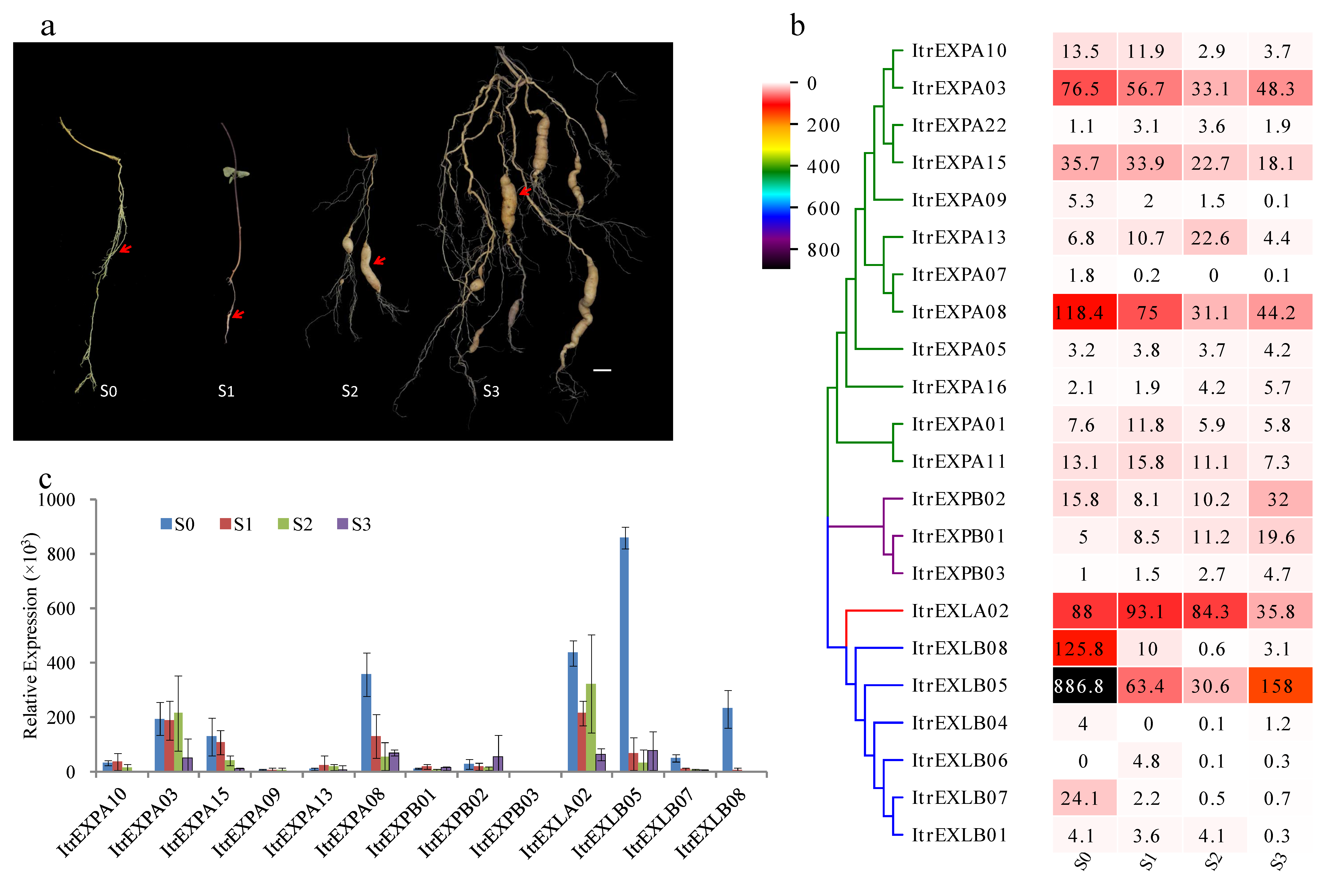
3.7. Expression Analysis of ItrEXPs in Y22 SR Development
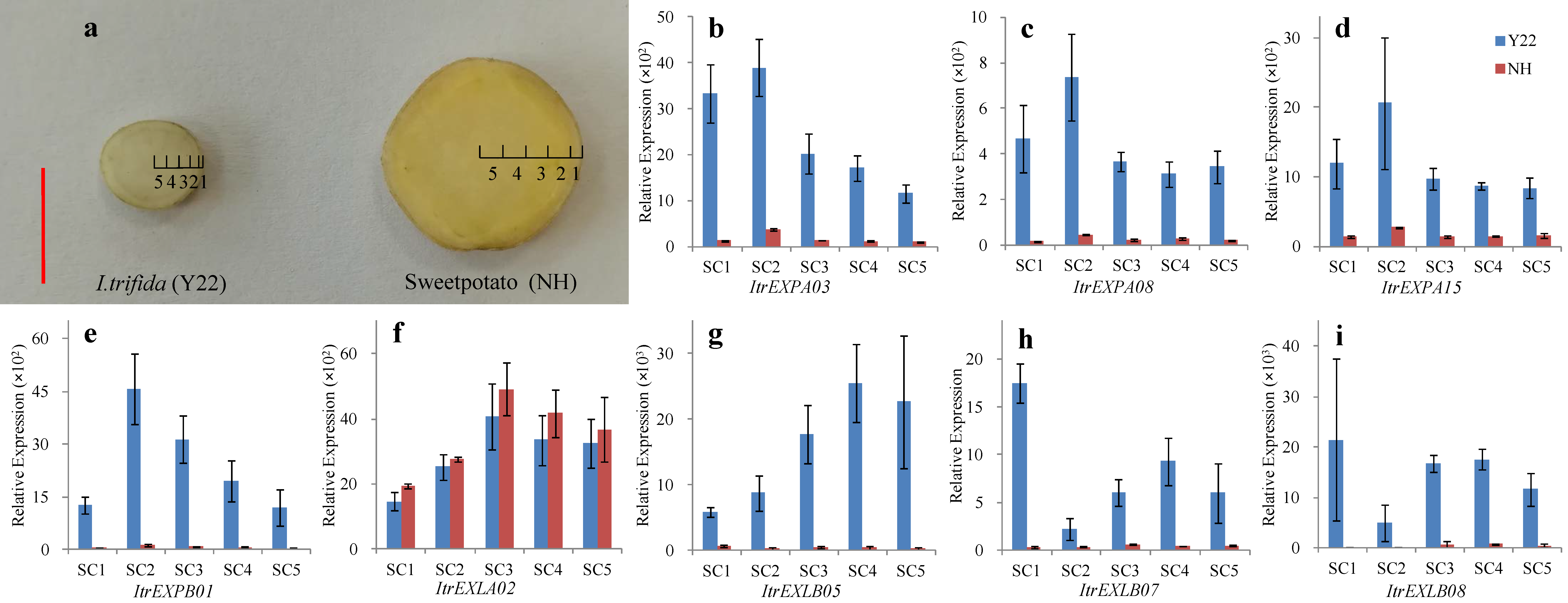
3.8. Expression Analysis of ItrEXPs in Different Tissues of SR inY22 and Sweetpotato
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef]
- Muthusamy, M.; Kim, J.Y.; Yoon, E.K.; Kim, J.A.; Lee, S.I. BrEXLB1, a Brassica rapa Expansin-Like B1 Gene is Associated with Root Development, Drought Stress Response, and Seed Germination. Genes 2020, 11, 404. [Google Scholar] [CrossRef]
- Chen, S.; Ren, H.; Luo, Y.; Feng, C.; Li, H. Genome-wide identification of wheat (Triticum aestivum L.) expansin genes and functional characterization of TaEXPB1A. Environ. Exp. Bot. 2021, 182, 104307. [Google Scholar] [CrossRef]
- Cho, H.T.; Cosgrove, D.J. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 2002, 14, 3237–3253. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013, 54, 1600–1611. [Google Scholar] [CrossRef]
- Ma, N.; Wang, Y.; Qiu, S.; Kang, Z.; Che, S.; Wang, G.; Huang, J. Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension. PLoS ONE 2013, 8, e75997. [Google Scholar] [CrossRef]
- Che, J.; Yamaji, N.; Shen, R.F.; Ma, J.F. An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J. Cell Mol. Biol. 2016, 88, 132–142. [Google Scholar] [CrossRef]
- Zou, H.; Wenwen, Y.; Kang, Z.; Wang, G.; Zhang, Z.; Zang, G.; Huang, J. OsEXPB2,a b-expansin gene, is involved in rice root system architecture. Mol. Breed. 2015, 35, 41. [Google Scholar] [CrossRef]
- Yang, J.; Moeinzadeh, M.H.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.; Zheng, J.; Sun, Z.; Fan, W.; et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef]
- Isobe, S.; Shirasawa, K.; Hirakawa, H. Challenges to genome sequence dissection in sweetpotato. Breed Sci. 2017, 67, 35–40. [Google Scholar] [CrossRef]
- Yan, M.; Nie, H.; Wang, Y.; Wang, X.; Jarret, R.; Zhao, J.; Wang, H.; Yang, J. Exploring and exploiting genetics and genomics for sweetpotato improvement: Status and perspectives. Plant Commun. 2022, 100332, in press. [Google Scholar] [CrossRef]
- Noh, S.A.; Lee, H.S.; Kim, Y.S.; Paek, K.H.; Shin, J.S.; Bae, J.M. Down-regulation of the IbEXP1 gene enhanced storage root development in sweetpotato. J. Exp. Bot. 2013, 64, 129–142. [Google Scholar] [CrossRef]
- Dong, W.; Li, L.; Cao, R.; Xu, S.; Cheng, L.; Yu, M.; Lv, Z.; Lu, G. Changes in cell wall components and polysaccharide-degrading enzymes in relation to differences in texture during sweetpotato storage root growth. J. Plant Physiol. 2020, 254, 153282. [Google Scholar] [CrossRef]
- Noh, S.A.; Park, S.H.; Huh, G.H.; Paek, K.-H.; Shin, J.S.; Bae, J.M. Growth retardation and differential regulation of expansin genes in chilling-stressed sweetpotato. Plant Biotechnol. Rep. 2008, 3, 75–85. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Zhai, H.; Gao, S.; Yang, L.; Wang, Z.; Xu, Y.; Huo, J.; Ren, Z.; Zhao, N.; et al. IbBBX24 Promotes the Jasmonic Acid Pathway and Enhances Fusarium Wilt Resistance in Sweet Potato. Plant Cell 2020, 32, 1102–1123. [Google Scholar] [CrossRef]
- Tanaka, M. Recent Progress in Molecular Studies On Storage Root Formation in Sweetpotato (Ipomoea batatas). Jpn. Agric. Res. Q. 2016, 50, 293–299. [Google Scholar] [CrossRef]
- Roullier, C.; Duputie, A.; Wennekes, P.; Benoit, L.; Fernandez Bringas, V.M.; Rossel, G.; Tay, D.; McKey, D.; Lebot, V. Disentangling the origins of cultivated sweet potato (Ipomoea batatas (L.) Lam.). PLoS ONE 2013, 8, e62707. [Google Scholar] [CrossRef]
- Ponniah, S.K.; Thimmapuram, J.; Bhide, K.; Kalavacharla, V.; Manoharan, M. Comparative analysis of the root transcriptomes of cultivated sweetpotato (Ipomoea batatas [L.] Lam) and its wild ancestor (Ipomoea trifida [Kunth] G. Don). BMC Plant Biol. 2017, 17, 9. [Google Scholar] [CrossRef]
- Munoz-Rodriguez, P.; Carruthers, T.; Wood, J.R.I.; Williams, B.R.M.; Weitemier, K.; Kronmiller, B.; Goodwin, Z.; Sumadijaya, A.; Anglin, N.L.; Filer, D.; et al. A taxonomic monograph of Ipomoea integrated across phylogenetic scales. Nat. Plants 2019, 5, 1136–1144. [Google Scholar] [CrossRef]
- Hou, W.; Ren, L.; Zhang, Y.; Sun, H.; Shi, T.; Gu, Y.; Wang, A.; Ma, D.; Li, Z.; Zhang, L. Characterization of BBX family genes and their expression profiles under various stresses in the sweet potato wild ancestor Ipomoea trifida. Sci. Hortic. 2021, 288, 110374. [Google Scholar] [CrossRef]
- Pratt, I.S.; Zhang, B. Genome-Wide Identification of ARF Transcription Factor Gene Family and Their Expression Analysis in Sweet Potato. Int. J. Mol. Sci. 2021, 22, 9391. [Google Scholar] [CrossRef]
- Wu, S.; Lau, K.H.; Cao, Q.; Hamilton, J.P.; Sun, H.; Zhou, C.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.; et al. Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef]
- Li, M.; Yang, S.; Xu, W.; Pu, Z.; Feng, J.; Wang, Z.; Zhang, C.; Peng, M.; Du, C.; Lin, F.; et al. The wild sweetpotato (Ipomoea trifida) genome provides insights into storage root development. BMC Plant Biol. 2019, 19, 119. [Google Scholar] [CrossRef]
- Bararyenya, A.; Olukolu, B.A.; Tukamuhabwa, P.; Gruneberg, W.J.; Ekaya, W.; Low, J.; Ochwo-Ssemakula, M.; Odong, T.L.; Talwana, H.; Badji, A.; et al. Genome-wide association study identified candidate genes controlling continuous storage root formation and bulking in hexaploid sweetpotato. BMC Plant Biol. 2020, 20, 3. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Jayakumar, V.; Nitasaka, E.; Toyoda, A.; Noguchi, H.; Itoh, T.; Shin, I.T.; Minakuchi, Y.; Koda, Y.; Nagano, A.J.; et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 2016, 7, 13295. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Komaki, K.; Katayama, K. Root thickness of diploid Ipomoea trifida (H. B. K.) G. Don and performance of progeny derived from the cross with sweetpotato. Breed. Sci. 1999, 49, 123–129. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Liu, X.; Dong, S.; Miao, H.; Bo, K.; Li, C.; Yang, Y.; Gu, X.; Zhang, S. Genome-Wide Analysis of Expansins and their Role in Fruit Spine Development in Cucumber (Cucumis sativus L.). Hortic. Plant J. 2021, in press. [Google Scholar] [CrossRef]
- Aleman, L.; Kitamura, J.; Abdel-mageed, H.; Lee, J.; Sun, Y.; Nakajima, M.; Ueguchi-Tanaka, M.; Matsuoka, M.; Allen, R.D. Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol. Biol 2008, 68, 1. [Google Scholar] [CrossRef]
- Kong, Y.; Wang, B.; Du, H.; Li, W.; Li, X.; Zhang, C. GmEXLB1, a Soybean Expansin-Like B Gene, Alters Root Architecture to Improve Phosphorus Acquisition in Arabidopsis. Front. Plant Sci. 2019, 10, 808. [Google Scholar] [CrossRef]
- Lu, S.; Liu, Q.; Li, W. Sweetpotato Breeding; Lu, S.-Y., Liu, Q.-C., Li, W.-J., Eds.; Chinese Agricultural Press: Beijing, China, 1998. [Google Scholar]


| Subfamily | Gene Name | Gene ID | pI | Mw/kD | Length/AA | Subcellular Localization |
|---|---|---|---|---|---|---|
| EXPA | ItrEXPA01 | Itr.Sc0000011.164 | 8.58 | 29.74 | 273 | cell wall |
| EXPA | ItrEXPA02 | Itr.xfSc0000576.12 | 10.42 | 28.18 | 254 | cell wall |
| EXPA | ItrEXPA03 | Itr.Sc0000007.290 | 9.48 | 27.92 | 257 | cell wall |
| EXPA | ItrEXPA04 | Itr.Sc0000051.50 | 9.00 | 26.88 | 251 | cell wall |
| EXPA | ItrEXPA05 | Itr.Sc0000046.34 | 9.34 | 26.05 | 248 | cell wall |
| EXPA | ItrEXPA06 | Itr.Sc0000046.36 | 9.34 | 26.05 | 248 | cell wall |
| EXPA | ItrEXPA07 | Itr.xfSc0000007.14 | 8.07 | 26.96 | 251 | cell wall |
| EXPA | ItrEXPA08 | Itr.xfSc0000007.15 | 8.07 | 26.54 | 249 | cell wall |
| EXPA | ItrEXPA09 | Itr.Sc0000078.10 | 9.38 | 28.29 | 260 | cell wall |
| EXPA | ItrEXPA10 | Itr.Sc0000001.78 | 9.60 | 27.98 | 259 | cell wall |
| EXPA | ItrEXPA11 | Itr.xfSc0000049.46 | 8.56 | 28.66 | 263 | cell wall |
| EXPA | ItrEXPA12 | Itr.Sc0000003.235 | 9.28 | 27.58 | 258 | cell wall |
| EXPA | ItrEXPA13 | Itr.Sc0000003.134 | 8.79 | 27.55 | 256 | cell wall |
| EXPA | ItrEXPA14 | Itr.Sc0000025.53 | 8.81 | 26.02 | 246 | cell wall |
| EXPA | ItrEXPA15 | Itr.Sc0000014.125 | 9.27 | 28.29 | 260 | cell wall |
| EXPA | ItrEXPA16 | Itr.Sc0000087.3 | 6.28 | 31.91 | 305 | cell wall |
| EXPA | ItrEXPA17 | Itr.Sc0000013.190 | 7.05 | 28.18 | 259 | cell wall |
| EXPA | ItrEXPA18 | Itr.xpSc0079065.36 | 6.87 | 26.43 | 244 | cell wall |
| EXPA | ItrEXPA19 | Itr.xpSc0079065.38 | 6.11 | 26.42 | 243 | cell wall |
| EXPA | ItrEXPA20 | Itr.xpSc0079065.39 | 9.35 | 26.50 | 240 | cell wall |
| EXPA | ItrEXPA21 | Itr.xpSc0079065.40 | 6.00 | 26.29 | 241 | cell wall |
| EXPA | ItrEXPA22 | Itr.xfSc0000000.33 | 9.82 | 27.89 | 257 | cell wall |
| EXPA | ItrEXPA23 | Itr.xfSc0001203.1 | 9.30 | 27.68 | 258 | cell wall |
| EXPB | ItrEXPB01 | Itr.xfSc0000185.16 | 6.29 | 28.52 | 269 | cell wall |
| EXPB | ItrEXPB02 | Itr.Sc0000024.30 | 8.74 | 28.41 | 262 | cell wall |
| EXPB | ItrEXPB03 | Itr.xfSc0000047.33 | 6.19 | 28.09 | 263 | cell wall |
| EXPB | ItrEXPB04 | Itr.xfSc0001448.1 | 6.92 | 28.04 | 263 | cell wall |
| EXLA | ItrEXLA01 | Itr.Sc0000050.70 | 5.13 | 28.57 | 266 | cell wall |
| EXLA | ItrEXLA02 | Itr.Sc0000020.14 | 6.96 | 29.41 | 268 | cell wall |
| EXLB | ItrEXLB01 | Itr.xpSc0079072.22 | 7.99 | 27.39 | 251 | cell wall |
| EXLB | ItrEXLB02 | Itr.xfSc0000061.14 | 5.71 | 25.49 | 238 | cell wall |
| EXLB | ItrEXLB03 | Itr.xfSc0000061.13 | 6.73 | 28.99 | 259 | cell wall |
| EXLB | ItrEXLB04 | Itr.xfSc0000239.16 | 8.96 | 26.56 | 248 | cell wall |
| EXLB | ItrEXLB05 | Itr.Sc0000071.6 | 6.14 | 28.24 | 253 | cell wall |
| EXLB | ItrEXLB06 | Itr.xfSc0000002.83.1 | 6.59 | 32.70 | 310 | cell wall |
| EXLB | ItrEXLB07 | Itr.xfSc0000002.86 | 8.73 | 27.01 | 246 | cell wall |
| EXLB | ItrEXLB08 | Itr.xfSc0000002.103 | 4.62 | 27.46 | 258 | cell wall |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Chen, L.; Lang, T.; Qu, H.; Zhang, C.; Feng, J.; Pu, Z.; Peng, M.; Lin, H. Genome-Wide Identification and Expression Analysis of Expansin Gene Family in the Storage Root Development of Diploid Wild Sweetpotato Ipomoea trifida. Genes 2022, 13, 1043. https://doi.org/10.3390/genes13061043
Li M, Chen L, Lang T, Qu H, Zhang C, Feng J, Pu Z, Peng M, Lin H. Genome-Wide Identification and Expression Analysis of Expansin Gene Family in the Storage Root Development of Diploid Wild Sweetpotato Ipomoea trifida. Genes. 2022; 13(6):1043. https://doi.org/10.3390/genes13061043
Chicago/Turabian StyleLi, Ming, Lianfu Chen, Tao Lang, Huijuan Qu, Cong Zhang, Junyan Feng, Zhigang Pu, Meifang Peng, and Honghui Lin. 2022. "Genome-Wide Identification and Expression Analysis of Expansin Gene Family in the Storage Root Development of Diploid Wild Sweetpotato Ipomoea trifida" Genes 13, no. 6: 1043. https://doi.org/10.3390/genes13061043
APA StyleLi, M., Chen, L., Lang, T., Qu, H., Zhang, C., Feng, J., Pu, Z., Peng, M., & Lin, H. (2022). Genome-Wide Identification and Expression Analysis of Expansin Gene Family in the Storage Root Development of Diploid Wild Sweetpotato Ipomoea trifida. Genes, 13(6), 1043. https://doi.org/10.3390/genes13061043





