Field-Evolved ΔG210-ppo2 from Palmer Amaranth Confers Pre-emergence Tolerance to PPO-Inhibitors in Rice and Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of Transgenic Rice
2.2. Transgenic Rice Response to Foliar-Applied Fomesafen
2.3. Transgenic Rice Response to Soil-Applied Fomesafen
2.4. Molecular Analysis of Transgenic Rice Plants
2.5. Agar-Based Germination Assay with Transgenic Rice Line
2.6. Generation of Transgenic A. thaliana Line
2.7. Transgenic A. thaliana Response to Soil-Applied Fomesafen and Saflufenacil
2.8. Statistical Analysis
3. Results
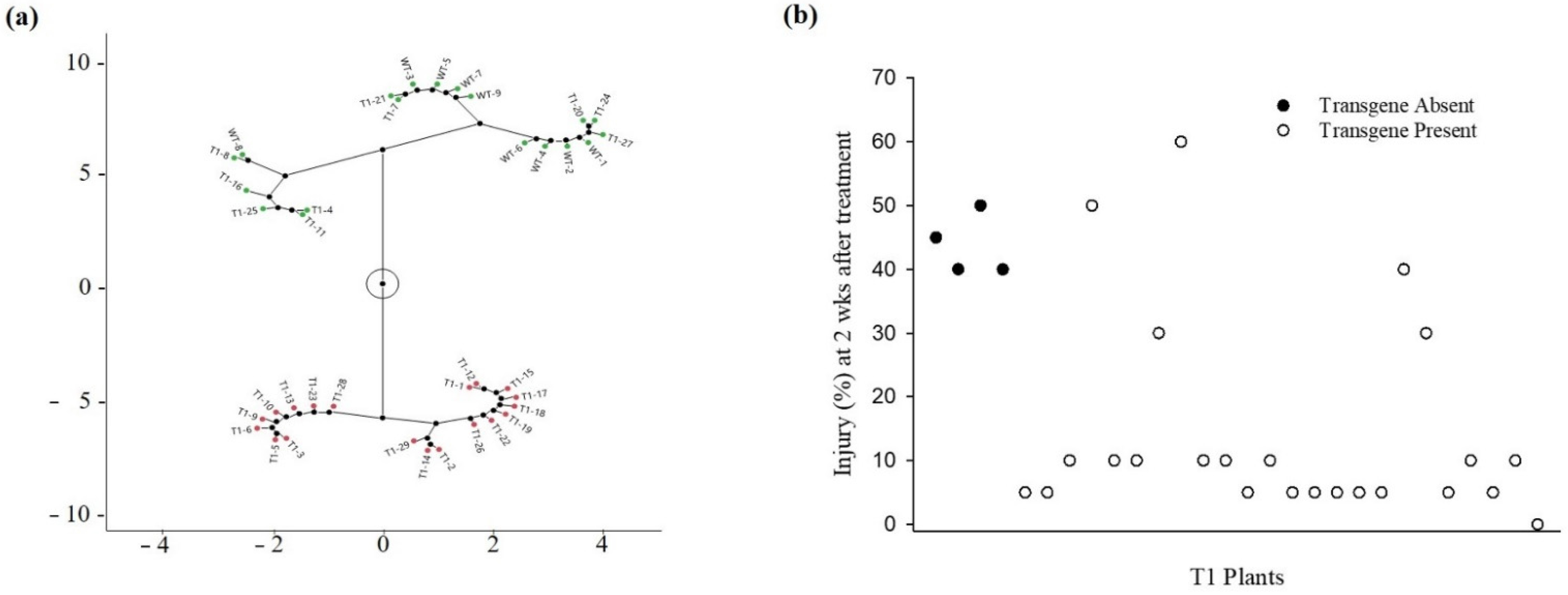
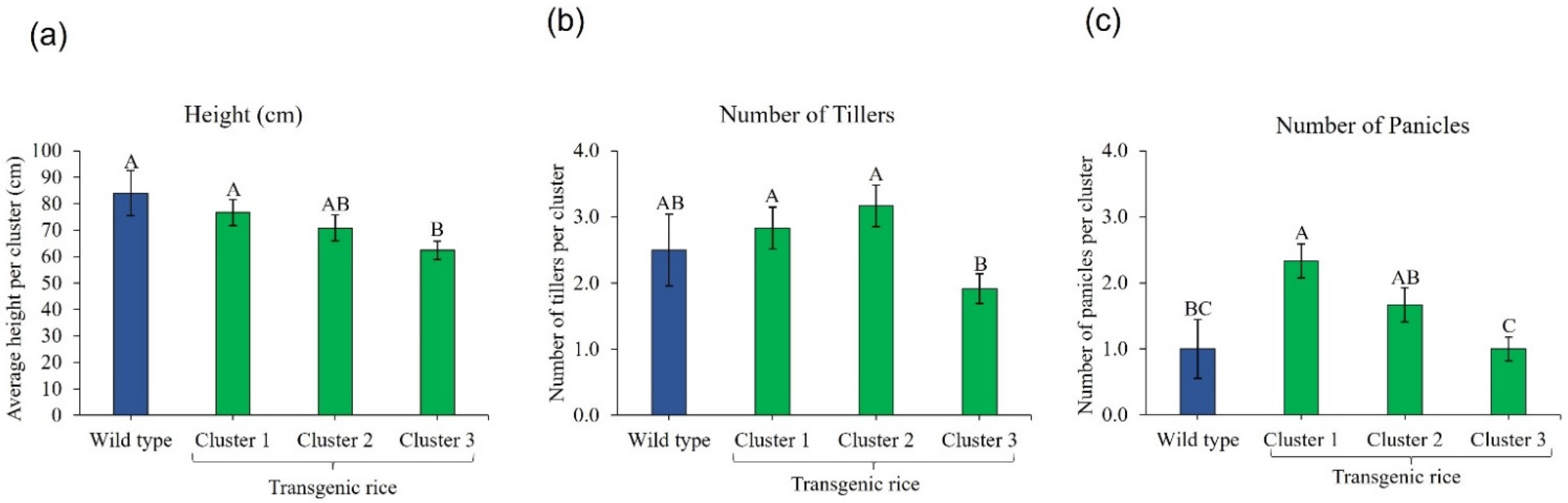
3.1. Transgenic Rice Response to Foliar-Applied Fomesafen
3.2. Transgenic Rice Response to Soil-Applied Fomesafen
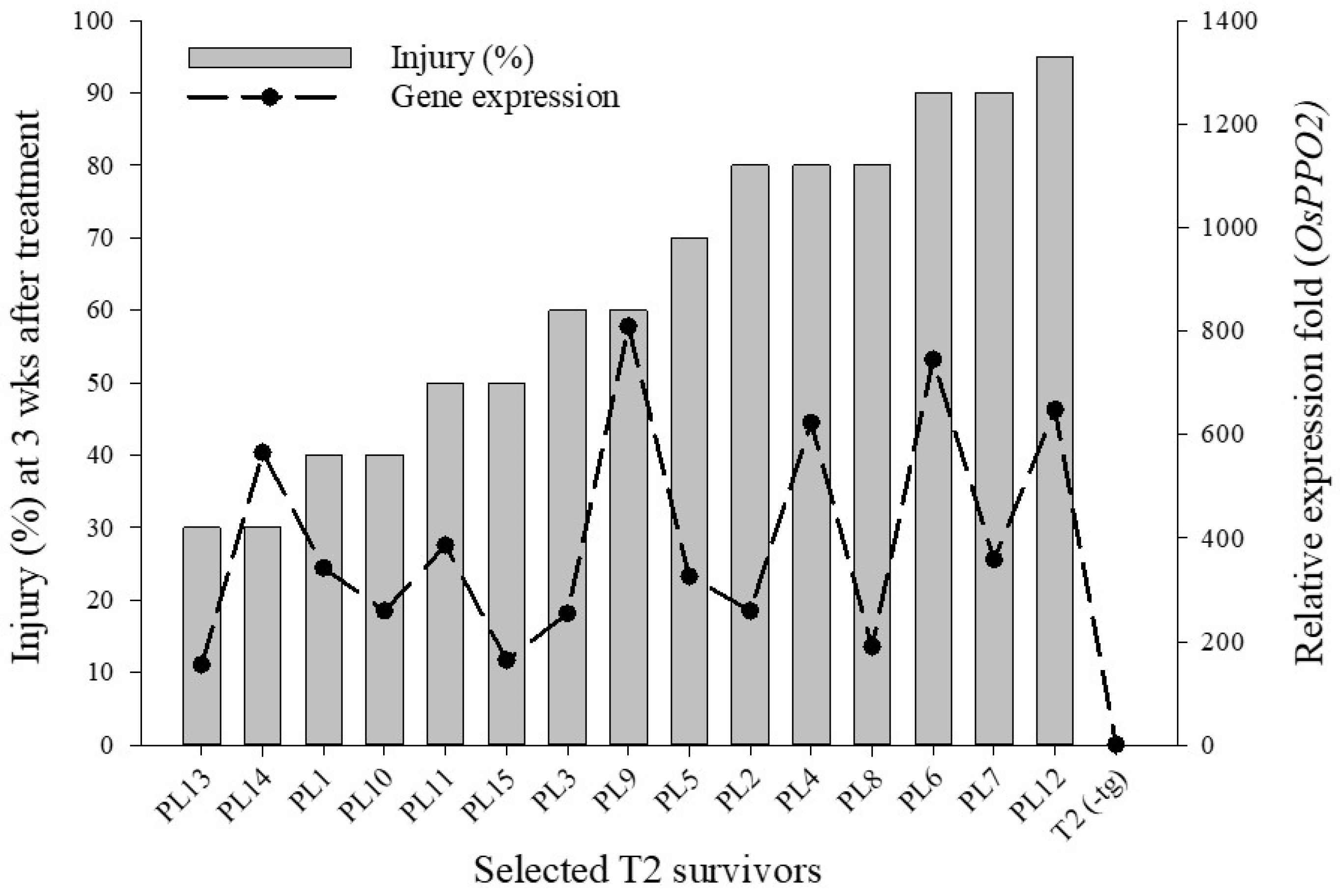
3.3. Gene Expression Analysis of Fomesafen Tolerant Rice Plants
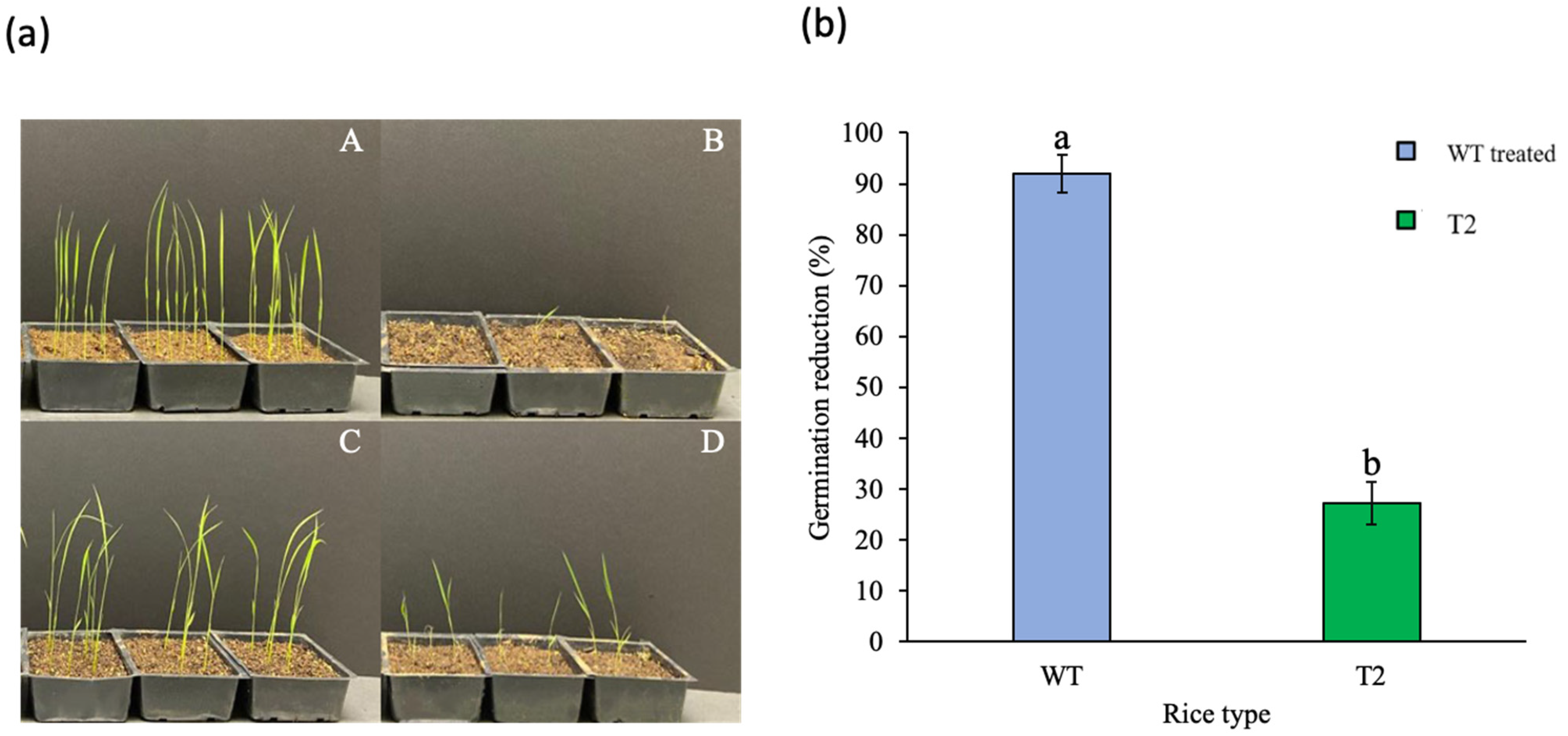
3.4. Agar-Based Germination Assay with T2 Seeds of Transgenic Rice
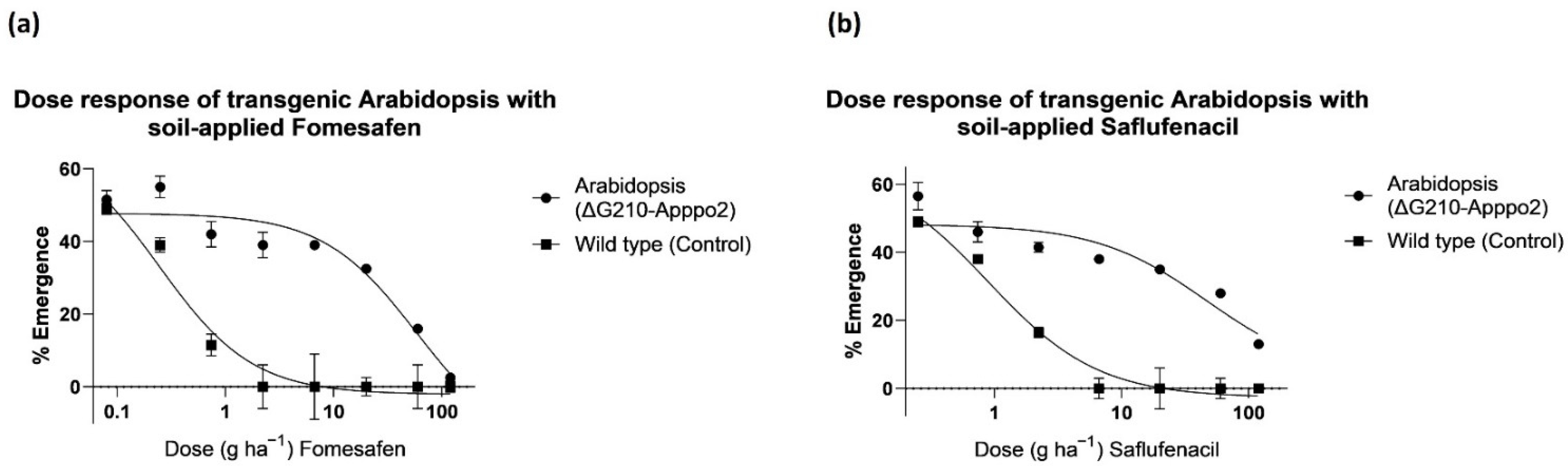
3.5. Transgenic A. thaliana Response to Soil-Applied Fomesafen and Saflufenacil
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wychen, V.L. 2019 Survey of the Most Common and Troublesome Weeds in Broadleaf Crops, Fruits & Vegetables in the United States and Canada. Weed Science Society of America National Weed Survey Dataset. Available online: https://wssanet/wp-content/uploads/2019-Weed-Survey_broadleaf-cropsxlsx (accessed on 3 March 2022).
- Heap, I. The International Survey of Herbicide Weeds. Available online: http://www.weedscience.org/ (accessed on 1 February 2022).
- Ying, Z.X.L.; Jun, X.; Fengshou, D.; Xingang, L.; Yongquan, Z. Determination of fomesafen residues in soybean and soil using QuEChERS and UPLC-MS/MS. Environ. Chem 2012, 31, 1399–1404. [Google Scholar]
- Hao, G.F.; Zuo, Y.; Yang, S.G.; Yang, G.F. Protoporphyrinogen oxidase inhibitor: An ideal target for herbicide discovery. Chimia 2011, 65, 961–969. [Google Scholar] [CrossRef]
- Poulson, R.; Polglase, W.J. The enzymic conversion of protoporphyrinogen IX to protoporphyrin IX. Protoporphyrinogen oxidase activity in mitochondrial extracts of Saccharomyces cerevisiae. J. Biol. Chem. 1975, 250, 1269–1274. [Google Scholar] [CrossRef]
- Dayan, F.E.; Duke, S.O. Porphyrin-generating herbicides. Pestic. Outlook 1996, 7, 22–27. [Google Scholar]
- Duke, S.O.; Lydon, J.; Becerril, J.M.; Sherman, T.D.; Lehnen Jr, L.P.; Matsumoto, H. Protoporphyrinogen oxidase-inhibiting herbicides. Weed Sci. 1991, 39, 465–473. [Google Scholar] [CrossRef]
- Lermontova, I.; Kruse, E.; Mock, H.P.; Grimm, B. Cloning and characterization of a plastidal and a mitochondrial isoform of tobacco protoporphyrinogen IX oxidase. Proc. Natl. Acad. Sci. USA 1997, 94, 8895–8900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matringe, M.; Camadro, J.M.; Labbe, P.; Scalla, R. Protoporphyrinogen oxidase as a molecular target for diphenyl ether herbicides. Biochem. J. 1989, 260, 231–235. [Google Scholar] [CrossRef]
- Orr, G.L.; Hess, F.D. Mechanism of Action of the Diphenyl Ether Herbicide Acifluorfen-Methyl in Excised Cucumber (Cucumis sativus L.) Cotyledons: Light activation and the subsequent formation of lipophilic free radicals. Plant Physiol. 1982, 69, 502–507. [Google Scholar] [CrossRef] [Green Version]
- Patzoldt, W.L.; Tranel, P.J.; Hager, A.G. A waterhemp (Amaranthus tuberculatus) biotype with multiple resistance across three herbicide sites of action. Weed Sci. 2005, 53, 30–36. [Google Scholar] [CrossRef]
- Watanabe, N.; Che, F.S.; Iwano, M.; Takayama, S.; Yoshida, S.; Isogai, A. Dual targeting of spinach protoporphyrinogen oxidase II to mitochondria and chloroplasts by alternative use of two in-frame initiation codons. J. Biol. Chem. 2001, 276, 20474–20481. [Google Scholar] [CrossRef] [Green Version]
- Dayan, F.E.; Barker, A.; Tranel, P.J. Origins and structure of chloroplastic and mitochondrial plant protoporphyrinogen oxidases: Implications for the evolution of herbicide resistance. Pest. Manag. Sci. 2018, 74, 2226–2234. [Google Scholar] [CrossRef] [PubMed]
- Mueller, T.C.; Boswell, B.W.; Mueller, S.S.; Steckel, L.E. Dissipation of fomesafen, saflufenacil, sulfentrazone, and flumioxazin from a Tennessee soil under field conditions. Weed Sci. 2014, 62, 664–671. [Google Scholar] [CrossRef]
- Shaner, D.L.E. Herbicide Handbook, 10th ed.; Weed Science Society of America: Westminster, CO, USA, 2014. [Google Scholar]
- Cobucci, T.; Prates, H.T.; Falcão, C.L.; Rezende, M.M. Effect of imazamox, fomesafen, and acifluorfen soil residue on rotational crops. Weed Sci. 1998, 46, 258–263. [Google Scholar] [CrossRef]
- Anonymous. Flexstar Herbicide Product Label; Syngenta Crop Protection, Inc.: Greensboro, NC, USA, 2022. [Google Scholar]
- Umphres, A.M.; Steckel, L.E.; Mueller, T.C. Control of protoporphyrinogen oxidase inhibiting herbicide resistant and susceptible Palmer amaranth (Amaranthus palmeri) with soil applied protoporphyrinogen oxi-dase-inhibiting herbicides. Weed Technol. 2018, 32, 95–100. [Google Scholar] [CrossRef]
- Patzoldt, W.L.; Hager, A.G.; McCormick, J.S.; Tranel, P.J. A codon deletion confers resistance to herbicides inhibiting protoporphyrinogen oxidase. Proc. Natl. Acad. Sci. USA 2006, 103, 12329–12334. [Google Scholar] [CrossRef] [Green Version]
- Salas-Perez, R.A.; Burgos, N.R.; Rangani, G.; Singh, S.; Refatti, J.P.; Piveta, L.; Tranel, P.J.; Mauromoustakos, A.; Scott, R.C. Frequency of Gly-210 deletion mutation among protoporphyrinogen oxidase inhibitor–resistant Palmer amaranth (Amaranthus palmeri) populations. Weed Sci. 2017, 65, 718–731. [Google Scholar] [CrossRef]
- Giacomini, D.A.; Umphres, A.M.; Nie, H.; Mueller, T.C.; Steckel, L.E.; Young, B.G.; Scott, R.C.; Tranel, P.J. Two new PPX2 mutations associated with resistance to PPO-inhibiting herbicides in Amaranthus palmeri. Pest. Manag. Sci. 2017, 73, 1559–1563. [Google Scholar] [CrossRef]
- Rangani, G.; Salas-Perez, R.A.; Aponte, R.A.; Knapp, M.; Craig, I.R.; Mietzner, T.; Langaro, A.C.; Noguera, M.M.; Porri, A.; Roma-Burgos, N. A Novel Single-Site Mutation in the Catalytic Domain of Protoporphyrinogen Oxidase IX (PPO) Confers Resistance to PPO-Inhibiting Herbicides. Front. Plant Sci. 2019, 10, 568. [Google Scholar] [CrossRef]
- Noguera, M.M.; Rangani, G.; Heiser, J.; Bararpour, T.; Steckel, L.E.; Betz, M.; Porri, A.; Lerchl, J.; Zimmermann, S.; Nichols, R.L. Functional PPO2 mutations: Co-occurrence in one plant or the same ppo2 allele of herbicide-resistant Amaranthus palmeri in the US mid-south. Pest. Manag. Sci. 2021, 77, 1001–1012. [Google Scholar] [CrossRef]
- Varanasi, V.K.; Brabham, C.; Norsworthy, J.K.; Nie, H.; Young, B.G.; Houston, M.; Barber, T.; Scott, R.C. A statewide survey of PPO-inhibitor resistance and the prevalent target-site mechanism in Palmer amaranth. Weed Sci. 2018, 66, 149–158. [Google Scholar] [CrossRef]
- Huang, Z.; Cui, H.; Wang, C.; Wu, T.; Zhang, C.; Huang, H.; Wei, S. Investigation of resistance mechanism to fomesafen in Amaranthus retroflexus L. Pestic. Biochem. Physiol. 2020, 165, 104560. [Google Scholar] [CrossRef] [PubMed]
- Rousonelos, S.L.; Lee, R.M.; Moreira, M.S.; VanGessel, M.J.; Tranel, P.J. Characterization of a common ragweed (Ambrosia artemisiifolia) population resistant to ALS- and PPO inhibiting herbicides. Weed Sci. 2012, 60, 335–344. [Google Scholar] [CrossRef]
- Nishimura, A.; Aichi, I.; Matsuoka, M. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 2006, 1, 2796–2802. [Google Scholar] [CrossRef] [PubMed]
- Counce, P.A.; Keisling, T.C.; Mitchell, A.J. A uniform, objective, and adaptative system for expressing rice development. Crop. Sci. 2000, 40, 436–443. [Google Scholar] [CrossRef] [Green Version]
- Burgos, N.R.; Tranel, P.J.; Streibig, J.C.; Davis, V.M.; Shaner, D.; Norsworthy, J.K.; Ritz, C. Confirmation of resistance to herbicides and evaluation of resistance levels. Weed Sci. 2013, 61, 4–20. [Google Scholar] [CrossRef]
- Frans, R.; Talbert, R.; Marx, D.; Crowley, H. Experimental design and techniques for measuring and analyzing plant responses to weed control practices. In Southern Weed Science Society, Research Methods in Weed Science, 1st ed.; Camper, N.D., Ed.; Southern Weed Science Society: Westminster, CO, USA, 1986; pp. 29–46. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Hongbao, M.; Young, J.; Shen, C. RNA, DNA and protein isolation using TRIzol reagent. Nat. Sci. 2008, 6, 66–75. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant. J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Gbur, E.E.; Stroup, W.W.; McCarter, K.S.; Durham, S.; Young, L.J.; Christman, M.; West, M.; Kramer, M. Analysis of Generalized Linear Mixed Models in the Agricultural and Natural Resources Sciences; American Society of Agronomy, Soil Science Society of America, Crop Science Society of America: Madison, WI, USA, 2012. [Google Scholar]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef] [Green Version]
- Ritz, C. Toward a unified approach to dose–response modeling in ecotoxicology. Environ. Toxicol. Chem. 2010, 29, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Carvalho de Lima, P. Resistance to Herbicides Conferred by Amaranthus palmeri Protoporphyrinogen IX Oxidase Mutations. Master Thesis, University of Arkansas, Fayetteville, AR, USA, 2020. [Google Scholar]
- Kim, E.; Park, H.; Yang, S.; Koo, Y. Current status of herbicide-tolerant protein research and strategy for development of herbicide-tolerant tomato using CRISPR-Cas9 system. Trends Agric. Life Sci. 2019, 57, 1–8. [Google Scholar] [CrossRef]
- Ishizuka, K.; Matsumoto, H.; Hyakutake, H. Selective inhibitory action of chlomethoxynil on rice and barnyardgrass [Echinochloa oryzicola] and its molecular fate in the light and dark. Weed Res. 1988, 33, 41–48. [Google Scholar]
- Niki, Y.; Kuwatsuka, S.; Yokomichi, I. Absorption, translocation and metabolism of chlomethoxynil (X-52) in plants. Agric. Biol. Chem. 1976, 40, 683–690. [Google Scholar]
- Capell, T.; Bassie, L.; Christou, P. Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc. Natl. Acad. Sci. USA 2004, 101, 9909–9914. [Google Scholar] [CrossRef] [Green Version]
- Toki, S.; Takamatsu, S.; Nojiri, C.; Ooba, S.; Anzai, H.; Iwata, M.; Christensen, A.H.; Quail, P.H.; Uchimiya, H. Expression of a maize ubiquitin gene promoter-bar chimeric gene in transgenic rice plants. Plant Physiol. 1992, 100, 1503–1507. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.I.; Kuk, Y.I.; Back, K.; Burgos, N.R. Resistance pattern and antioxidant enzyme profiles of protoporphyrinogen oxidase (PROTOX) inhibitor-resistant transgenic rice. Pesti. Biochem. Pysiol. 2008, 91, 53–65. [Google Scholar] [CrossRef]
- Jung, H.I.; Kuk, Y.I.; Kim, H.Y.; Back, K.; Lee, D.J.; Lee, S.; Burgos, N.R. Resistance levels and fitness of protoporphyrinogen oxidase (PROTOX) inhibitor-resistant transgenic rice in paddy fields. Field Crops Res. 2010, 115, 125–131. [Google Scholar] [CrossRef]
- McKenzie, K.S. Oxyfluorfen Resistant Rice Lines. U.S. Patent Application No. 2018/0070548, 15 March 2018. [Google Scholar]
- Bond, C.A.; Carter, C.A.; Farzin, Y.H. Medium grains, high stakes: Economics of genetically modified rice in California. AgBioForum 2003, 6, 146–154. [Google Scholar]
- Mulvaney, D.R.; Krupnik, T.J.; Koffler, K.B. Transgenic rice evaluated for risks to marketability. Calif. Agric. 2011, 65, 161–167. [Google Scholar] [CrossRef] [Green Version]
- Carvalho-Moore, P.; Rangani, G.; Heiser, J.; Findley, D.; Bowe, S.J.; Roma-Burgos, N. PPO2 Mutations in Amaranthus palmeri: Implications on cross-resistance. Agriculture 2021, 11, 760. [Google Scholar] [CrossRef]
- Vain, P.; James, V.; Worland, B.; Snape, J. Transgene behavior across two generations in a large random population of transgenic rice plants produced by particle bombardment. Theor. Appl. Genet. 2002, 105, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Day, C.D.; Lee, E.; Kobayashi, J.; Holappa, L.D.; Albert, H.; Ow, D.W. Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 2000, 14, 2869–2880. [Google Scholar] [CrossRef] [Green Version]
- Matzke, A.J.; Matzke, M.A. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1998, 1, 142–148. [Google Scholar] [CrossRef]
- McCabe, M.S.; Mohapatra, U.B.; Debnath, S.C.; Power, J.B.; Davey, M.R. Integration, expression, and inheritance of two linked T-DNA marker genes in transgenic lettuce. Mol. Breed 1999, 5, 329–344. [Google Scholar] [CrossRef]
- Lee, Y.; Jung, S.; Back, K. Expression of human protoporphyrinogen oxidase in transgenic rice induces both a photodynamic response and oxyfluorfen resistance. Pestic. Biochem. Physiol. 2004, 80, 65–74. [Google Scholar] [CrossRef]
- Jung, S.; Lee, Y.; Yang, K.; Lee, S.B.; Jang, S.M.; Ha, S.B.; Back, K. Dual targeting of Myxococcus xanthus protoporphyrinogen oxidase into chloroplasts and mitochondria and high level oxyfluorfen resistance. Plant Cell Environ. 2004, 27, 1436–1446. [Google Scholar] [CrossRef]
- Matsumoto, H.; Lee, J.; Ishizuka, K. Variation in crop response to protoporphyrinogen oxidase inhibitors. Porphyric Pesticides: Chemistry, Toxicology, and Pharmaceutical Applications. Am. Chem. Soc. Symp. Ser. 1995, 559, 120–132. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho-Moore, P.; Rangani, G.; Langaro, A.C.; Srivastava, V.; Porri, A.; Bowe, S.J.; Lerchl, J.; Roma-Burgos, N. Field-Evolved ΔG210-ppo2 from Palmer Amaranth Confers Pre-emergence Tolerance to PPO-Inhibitors in Rice and Arabidopsis. Genes 2022, 13, 1044. https://doi.org/10.3390/genes13061044
Carvalho-Moore P, Rangani G, Langaro AC, Srivastava V, Porri A, Bowe SJ, Lerchl J, Roma-Burgos N. Field-Evolved ΔG210-ppo2 from Palmer Amaranth Confers Pre-emergence Tolerance to PPO-Inhibitors in Rice and Arabidopsis. Genes. 2022; 13(6):1044. https://doi.org/10.3390/genes13061044
Chicago/Turabian StyleCarvalho-Moore, Pamela, Gulab Rangani, Ana Claudia Langaro, Vibha Srivastava, Aimone Porri, Steven J. Bowe, Jens Lerchl, and Nilda Roma-Burgos. 2022. "Field-Evolved ΔG210-ppo2 from Palmer Amaranth Confers Pre-emergence Tolerance to PPO-Inhibitors in Rice and Arabidopsis" Genes 13, no. 6: 1044. https://doi.org/10.3390/genes13061044
APA StyleCarvalho-Moore, P., Rangani, G., Langaro, A. C., Srivastava, V., Porri, A., Bowe, S. J., Lerchl, J., & Roma-Burgos, N. (2022). Field-Evolved ΔG210-ppo2 from Palmer Amaranth Confers Pre-emergence Tolerance to PPO-Inhibitors in Rice and Arabidopsis. Genes, 13(6), 1044. https://doi.org/10.3390/genes13061044






