Abstract
The early developmental phase is of critical importance for human health and disease later in life. To decipher the molecular mechanisms at play, current biomedical research is increasingly relying on large quantities of diverse omics data. The integration and interpretation of the different datasets pose a critical challenge towards the holistic understanding of the complex biological processes that are involved in early development. In this review, we outline the major transcriptomic and epigenetic processes and the respective datasets that are most relevant for studying the periconceptional period. We cover both basic data processing and analysis steps, as well as more advanced data integration methods. A particular focus is given to network-based methods. Finally, we review the medical applications of such integrative analyses.
1. Introduction
In recent decades, a digital revolution has taken place in many scientific fields. In biology, we are able to produce large amounts of omics data (e.g., genomics, transcriptomics, proteomics, epigenomics, and metabolomics), which allow us to describe biological events in all their complexity. This has also led to a shift in the way we study, diagnose, and treat diseases in medicine, moving away from focusing solely on symptoms and clinical signs towards more holistic and data-driven approaches [1].
Such approaches are also indispensable to reveal the developmental origins of health and disease (DOHaD) (Figure 1A). Indeed, the earliest period of life is especially complex: It involves several closely interacting individuals, first and foremost the baby and the mother, but the father and the environment also play important roles and may condition different growth trajectories [2] (Figure 1B). For example, the phenomenon of imprinting in embryo development, i.e., the silencing of specific genes, leads to different outcomes depending on whether it happens on the mother’s or the father’s allele [3]. Several events, such as hypomethylation and microRNA regulation, can cause the loss of imprinting in the insulin-like growth factor 2 (IGF2) [4]. Normally, this gene is expressed only from the paternal copy and silenced on the maternal one. The expression of IGF2 on both alleles leads to widespread genomic, proteomic, and metabolomic changes responsible for various pathological conditions, such as hyperplasia, cancer, and embryo developmental disorders [5] (Figure 1C). The complexity of this example illustrates the need for experts from different fields to join forces and for an integrative view and analysis of the numerous and complementary layers of information that are at play [6].
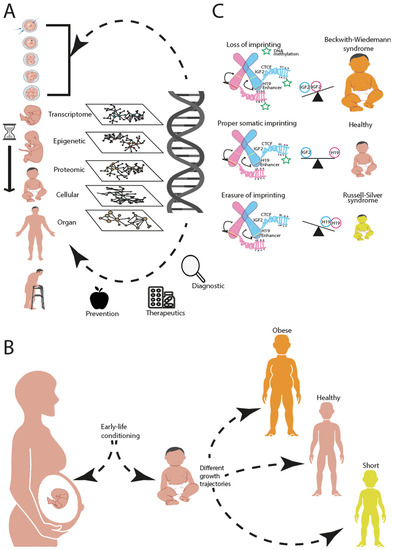
Figure 1.
Dissecting biological complexity in the different layers of biological organisation helps in prevention, diagnosis and treatment. (A) Genetic perturbation during the periconceptional period (first two weeks after conception) can propagate through the different layers of biological networks: transcriptome, epigenome, proteome, cellular level and organ level leading to predisposition for disease phenotypes later on in life. Dissecting and integrating these biological layers are crucial for prevention, early diagnosis and potential treatments. (B) Early life conditioning can influence growth trajectories in life, contributing in predisposition for different phenotypes (health, short, and obese). (C) Epigenetic modifications, such as DNA methylation in particular regions of the DNA containing imprinting genes, could alter the normal genetic balance of the maternal and paternal alleles. As an example, we show the consequences of alterations of the IGF2-H19 imprinting gene balance, which can lead to either gigantism (Beckwith–Wiedemann syndrome) or nanism (Russell–Silver syndrome).
The goal of this review is to facilitate the adoption of omics-based approaches in DOHaD research. We focus particularly on transcriptomics and epigenomics, given their widespread use and biological importance to DOHaD. We start with a brief introduction to transcriptomic data analysis. Next, we provide an overview of the plethora of epigenetic mechanisms at play in DOHaD. Finally, we introduce network-based methods as a tool for data integration from heterogeneous data sources and provide some concrete examples in the DOHaD context.
2. Transcriptomic View of Development and Analysis
Transcriptomics is the study of transcripts, or the expressed RNAs present in cells. From a transcriptomic perspective, human development can be seen as a continuous process in which fertilisation begins the transformation of the transcriptionally inactive oocyte into the active zygote, a process known as the maternal–zygotic transition [7,8]. This transition is but one of many windows of transcriptomic changes that the embryo undergoes as it develops and differentiates from a single cell to a complete organism [9,10]. These transition windows, often accompanied by epigenetic changes, have been suggested to be vulnerable periods in the embryo development, and disruptions during these periods can have lasting consequences in later life, as summarised in the DOHaD concept [11,12]. We describe how these transcripts (coding and non-coding RNA) regulate DNA expression and accompany epigenetic regulation mechanisms in the next section, while, in this section, we focus on the main principles of the bioinformatic pipeline used when facing a transcriptomic dataset and on the biological implications of these steps.
At its core, transcriptomic data analyses operate on the number of detected transcripts as a quantitative measure of gene expression. Historically, microarrays and RNA-seq have provided gene expression information for a group of cells. More recently, single-cell technologies allow us to measure transcript expression on a single-cell resolution [13,14]. The raw data of both bulk and single-cell technologies require several analytical steps, recapitulated in Figure 2 and described in the following sections.
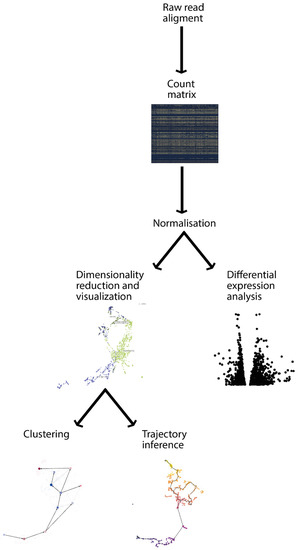
Figure 2.
Transcriptomic analysis pipeline. Starting from the raw read alignments, several steps are needed to obtain concrete biological results, such as the identification of differential expressed genes or cluster marker genes. The first step is to align gene sequences to a gene annotation reference to be able to count the number of reads for each gene. This will allow us to obtain a count matrix, which can be used for differential expression analysis, identifying genes that are significantly changed (up/down regulated) in certain conditions and visualising them for example in a volcano plot. In parallel, the dimensions of the count matrix can be reduced and visualised with several techniques (i.e., PCA, t-SNE, and UMAP). This allows to identify clusters and, in the context of single-cell experiments, also to infer developmental trajectories.
2.1. Data Preprocessing
Several pre-processing steps need to be performed before transcriptomic data can be analysed. The first step, common to all platforms, is the alignment of transcripts to their source in the DNA, so that we may identify the genes that are being expressed. Over the years, many tools have been developed to perform this task [15]; widely used examples include STAR [16], BWA [17], and HISAT2 [18]. Alignments have to be performed against a reference database, typically an assembled genome and its annotations, for which commonly used sources are UCSC, NCBI, and Ensembl [19,20,21]. When attempting to replicate an analysis, care must be taken to use the same set of annotations as there are significant differences between them [22].
Once the transcripts have been aligned, the number of transcripts present for each gene can be tallied up to measure their expression levels. However, this number needs to be normalised before further analysis can take place to account for noise and biases of sequencing technologies. Noise may vary between batches due to variations in sample and preparation quality, and in some technologies, such as microarrays, high expression levels may cause saturation, resulting in a non-linear response. Controls, such as spike-ins or housekeeping genes, may be employed to provide a stable baseline for normalisation, though purely mathematical approaches, such as quantile normalisation, are also used [23].
Background noise is especially pronounced in single cell RNA-seq (scRNA-seq) datasets, as these platforms operate on a small volume per sample. The nature of this technology requires a normalisation step for comparing the counts of gene values across samples, such as reads per million/counts per million (RPM/CPM), which simply normalises the detected features to the total number of counts within the sample. Further development resulted in reads/fragments per kilobase million (RPKM/FPKM) [24] and transcripts per million (TPM) [25], which also take the gene length into account to avoid the overrepresentation of longer genes with more numerous exons. The Trimmed Mean of M-values (TMM) and Relative Log Expression (RLE) take this a step further, by making the assumption that the majority of genes are similar across samples and using them as control to allow for a more accurate inter-sample comparison [26,27].
2.2. Dimensionality Reduction
Transcriptomic datasets contain expression levels of thousands of transcripts and are thus by nature very high-dimensional. To enable more efficient downstream analyses, such as the identification of clusters, dimensionality reduction is often applied. The most commonly used dimensionality reduction methods are principal component analysis (PCA) [28], t-stochastic neighbour embedding (t-SNE) [29], and uniform manifold approximation and projection (UMAP) [30]. PCA is by far the simplest and fastest method and works by calculating the vectors on which the dataset is most variable. Though it is quick and preserves global distances, PCA is offset by losing information from the culled components and the linear modelling inherent in the data processing, which often results in a lower ability to separate different classes that may be contained in the data. It is also not very useful for visualising datasets in which the variance is spread over a large number of components, as most visualisations can only display two-to-three axes.
In contrast to PCA, t-SNE is both slower and does not preserve the global structure of the data. When operating on large datasets, t-SNE is often run as a second stage after other dimensionality reduction techniques, such as PCA, though the more recent Fourier-interpolated (FIt-SNE) implementation provides significant improvements in run time [31]. t-SNE is sensitive to changes in its own hyperparameters, which in turn must be selected carefully for each analysis to make sure the visualisation is fit for purpose [29]. Despite its shortcomings, t-SNE has a greater ability to create separation between clusters. This is especially beneficial for exploratory visualisations, though the fact that it does not preserve global distances means that the interpretation of visually observed clusters needs to be conducted carefully as differences in separation between clusters may not contain any significant meaning. Finally, t-SNE works best when used for visualisation only, reducing the dimension of the data to two or three at most, which means it is not ideal for downstream unsupervised clustering as it will only produce clusters that agree with its visualisation, potentially losing the underlying structure and relation between them due to the loss of global structure information [32].
Comparatively recent to the other two approaches is UMAP. As in the case of t-SNE, UMAP is a non-linear technique. However, in addition to creating strong local clusters, UMAP also attempts to preserve the global structure in the data and has lower computational time requirements [30]. In terms of single-cell analysis in particular, it is capable of preserving the continuity of cell subsets, which provides a more meaningful visualisation compared to t-SNE [33]. UMAP is thus often regarded as an improvement for most use cases, though t-SNE still shows greater local structure separation and the FIt-SNE implementation, in particular, has comparable, if not superior, speed.
Dimensionality reduced data are prime for visualisation, as with reduced complexity they can be more easily projected to a two-dimensional plane. This is a very useful feature, as a quick visualisation process can be a powerful quality control tool [32]. Due to the nonlinearity of the most commonly used dimensionality reduction methods, some caution needs to be exercised for interpreting the graphs produced. For example, a t-SNE based scatterplot produces exaggerated distances between local groups. From a quality control perspective, however, it can be very useful to rapidly assess whether or not the data are behaving as expected, i.e., are the clusters sensible, or whether or not the distribution is as expected from what is known about the dataset. Should there be a mismatch, it may point to other issues that need to be corrected first, for example, in the normalisation procedure.
2.3. Clustering
One of the most basic techniques for interpreting transcriptomic data is clustering, which identifies groups of similar points within the dataset. With the availability of a variety of different approaches for visualising and analysing transcriptomic data, care must be taken to select the technique most suitable for enabling insights relevant to the goal of the study. In transcriptomic data, there are two basic ways to perform clustering, either on the level of samples and or on the level of genes. Clustering samples is useful for identifying samples with similar gene expression levels, while gene clustering identifies genes with similar expression profiles across samples. Gene clustering is especially helpful in bulk sequencing experiments using older methods, such as microarrays, where sample clustering is often limited by low resolution. Sample clustering is especially suitable for single-cell datasets, for example, for identifying different cell types within a tissue. Two common clustering methods applied to clustering transcriptomic data are hierarchical clustering and k-means [34].
Hierarchical clustering is one of the most basic clustering methods available, where the data are structured into a dendrogram based on a statistical distance metric, such as Euclidean distance, and the clusters are decided by splitting the dendrogram at a certain depth. The dendrogram can be constructed by linking each data point together from the bottom up (agglomerative) or by splitting the complete dataset into smaller subgroups based on dissimilarity (divisive). While simple to understand, naive hierarchical clustering algorithms are computationally intensive and are not suitable for larger datasets, for which heuristics are often employed to speed up the process [35,36]. This technique was largely used in embryo developmental studies to investigate relationships between different embryological phases based on their transcriptomic similarity [37,38,39].
K-means clustering provides a faster algorithm compared to hierarchical clustering, in which the dataset is split into k cluster centres and assigns each data point to a cluster based on which cluster centre they are the closest to. Classical k-means clustering is an efficient technique when the number of clusters to be expected in the dataset is already known [40]. For this reason, it was applied for studying embryo development on a morphological level by imaging analysis [41], but also on a molecular level [42]. Despite these successes, the number of clusters of a dataset is not always known a priori and, to overcome this limitation, a variety of other methods exist [32], including network-based strategies that operate on graphs of transcriptomic data points constructed using K-nearest neighbour methods [43]. This approach revealed its potential especially in single-cell RNA-seq experiments [44], as it was able to identify known and new cell groups in different biological contexts, including embryo development [45] and artificial reproductive treatments [46].
While clustering is a very valuable tool for exploration, it is also often used as a stepping stone for downstream analysis. For example, annotating the clusters with genes that are highly expressed can lead to a better identification of what cell types are represented by the clusters using reference databases of marker genes. The list of expressed genes can itself be used in gene ontology (GO) enrichment analysis, which provides a way for looking up gene functions and can then be followed up with network analysis on the pathways and relations of the expressed genes, as well as overrepresentation analysis to find prevalent GO terms, which can lead to the identification of overexpressed pathways.
Clustering can also be performed on a gene level instead of a cell level, in which case genes are clustered based on the similarity of their sequences. This is especially useful for identifying the functions of novel genes as they may share similarities to known genes [47], but also for phylogenetic tree reconstruction [48]. It is also possible to group genes based on known biological processes instead, and then testing for the overrepresentation of certain processes within the sample.
2.4. Differential Expression Analysis
Differential expression analysis is a basic method for comparing gene expression levels between samples, and is able to produce useful insight regardless of resolution as long as there is clear delineation between samples [14]. By finding which genes are expressed at different levels between samples of different phenotypes, correlations between phenotype and genotype can be drawn. This idea of comparing the difference in gene expression levels also forms the foundation of much of the techniques explored in the following sections. This basic concept of comparing two (or more) groups has guided the latest biological discoveries in embryology. For example, some studies have tried to establish which genes guide a competent embryo implantation through in vitro fertilization techniques [49], while others have tried to correlate embryo degeneration with specific protein families, such as the heat shock proteins (HSPs) [50]. Other authors have tried to use well-known animal models used in embryology, such as Xenopus, to create an atlas of differential expressed genes during embryogenesis [51] and, finally, other studies have tried to translate these findings from animal models to human [52].
2.5. Trajectory Analysis
Trajectory analysis methods can be used to characterise datasets containing samples taken from organism(s) at different points in development [53]. While this is simple for experimental techniques that allow for multiple measurements on the same sample over time, it is impossible in single-cell experiments as the samples are destroyed upon sequencing. For the latter case, the concept of pseudotime is often employed, where the trajectory is constructed over various samples, instead of reflecting a real time series. Trajectory analysis is closely linked to the concept of a “developmental landscape”, in which cell populations roll down a landscape representing the entropy of their current state, heading towards their terminally differentiated fate [54]. As cells form a continuous lineage as they traverse through this landscape, the trajectory of cell development can be inferred by comparing the data taken from two different points. This comparison is typically performed by representing a cell’s gene expression profile as a vector, which allows for distances between them to be calculated. A range of approaches and packages are available for this purpose [55,56,57].
A recent technique referred to as RNA velocity analysis provides a novel approach to trajectory inference from a single snapshot in time. The technique utilises the ratio of unspliced to spliced messenger RNA (mRNA) to quantify the rate of change in gene expression and fill in the gaps between pseudotime slices [58,59]. Bridging these gaps can be very useful as some trajectory inference methods only work under the assumption that there is only a small difference between the different states [56]. Similarly to RNA velocity analysis, partition-based graph abstraction (PAGA) aims to detect trajectories from a single snapshot; however, it does this by connecting similar cells to each other in a network [60]. This allows to combine trajectory inference with other methods, such as clustering for the same data. Inferring transcriptomic trajectories in embryological datasets corresponds to the identification of developmental trajectories, which map cell state evolution over time. This is very important to try to understand biological events that occur in a very small window of time, such as the periconceptional period. Studies in mice have revealed the key genes whose transcriptomic changes lead to differentiation and organogenesis [45,61]. Similar studies in zebrafish have looked at a larger time window and contextualised the impact of some gene knockouts on cell fates [62].
2.6. Expressed Variation Analysis
Transcriptomic data can also aid in the identification of relevant genetic variations, in particular expressed single nucleotide variations (eSNVs) [63,64,65]. Compared to genome-wide association studies (GWAS) [66], these methods do not rely on having a large sample population. By operating at a much more granular single-cell resolution, they allow for detecting variations that are not present in significant numbers within a sample population. These traits enable the application of variant analysis on a smaller scale, such as separating cells from different individuals [67] and cancer identification [68,69]. When applied at the population level, it is also possible to define expression quantitative trait loci (eQTL), genetic variations that cause changes in expression levels [70], for example, using data collected in the Genotype-Tissue Expression (GTEx) project [71].
In conjunction with trajectory analysis, variation data can reveal further information. It has been shown that SNVs, both expressed and not, affect not only a gene’s transcription level, but its changes at different points in development, suggesting complex interactions with underlying regulatory mechanisms within the genome [72,73]. eSNVs are particularly important in this context as these expressed variants implicate phenotypic effects [74]. Similarly, it is also possible to compare the variations present in different stages of development to analyse the rate at which mutations are accrued at different stages, such as in studying the mosaicism present in human prenatal development [75].
3. Epigenetic View of Development and Analysis
The term epigenome refers to all modifications to DNA and histone proteins that modulate chromatin structure and genome function [76]. The epigenome thus represents a crucial nexus between genetic variation, environment, health, and disease. Indeed, chemical compounds, such as environmental exposures, can cause (ir)reversible changes in DNA structure, such as chromatin unfolding, which allows transcription factors (TFs) to bind their target leading to an increased transcription of the genes localised in the respective DNA region. Another example showing the importance of epigenetics is the variability of cell states within the same individual. While all cells in an organism share the same genetic information, they differ largely in terms of their expression and phenotypic manifestations. Epigenomic and transcriptomic data thus convey two different types of information, but can also be seen as two sides of the same coin. Integrating these two sides into a single framework remains an important and challenging task in the biomedical field [77,78,79].
Epigenetic mechanisms have gained growing attention from the scientific community in recent decades. In contrast to genetic mutations, epigenetic changes are plastic events that may occur multiple times during the lifetime of a cell and that can be reversible. These mechanisms are involved in numerous pathological processes [80,81,82] and therapeutic outcomes [83]. In the following, we review major biological processes that collectively constitute the epigenome (Figure 3).
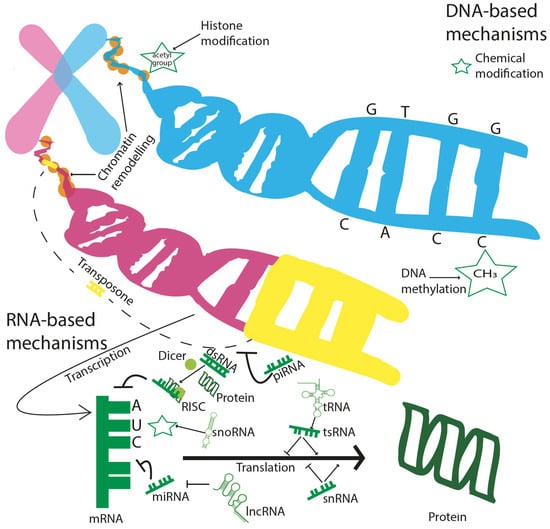
Figure 3.
Epigenetic modifications occur on different biological scales. DNA-based mechanisms are concerned with histone modifications, consisting of chemical modifications (i.e., acetylation), DNA methylation, chromatin remodelling, and transposons. RNA-based mechanisms are multiple, complex, and still only partially known: miRNA and the RISC complex can induce mRNA degradation; lncRNAs silence the activity of miRNA, while snRNA and tsRNA can both silence, but also induce, mRNA translation to protein. piRNAs can interact with DNA, interfering with the genetic movements of transposons; snoRNAs induce chemical modifications at the mRNA level.
3.1. DNA Methylation
DNA methylation is among the most studied and best understood epigenetic mechanisms. It contributes to gene expression regulation via an enzymatic reaction in which DNA methyltransferase (DNMT) catalyses the addition of a methyl group (-CH3) to a cytosine [84], causing DNA folding and the consequent inaccessibility by TFs, in turn silencing gene transcription in that particular region of DNA. In mammals, this process occurs only in dinucleotides CpG and is associated with gene inactivation [85,86]. Interestingly, each tissue seems to have a specific pattern of DNA methylation [87] that potentially changes over time, asserting the dynamic nature of this biological process [88,89] that can contribute to disease onset [90]. These dynamic methylation profile changes (epigenetic drift) contribute to cellular differentiation and tissue composition [91] and have been associated with development [92], ageing [93], and disease [94].
3.2. Non-Coding RNAs (ncRNAs)
The central dogma of molecular biology describes the flow of information from DNA to mRNA and finally to protein. Over the last decades, many additional mechanisms have been uncovered that can regulate and interfere with this linear process. Some of the main actors in this regulation are non-coding RNAs, which have been shown to regulate processes such as transcription, translation, and post-translational events [95,96]. There is a large variety of molecules that belong to this class and, surprisingly, some of them have been observed to be inheritable across generations [97,98]. They are often roughly classified based on their length into small (<200 nucleotides) and long (>200 nucleotides) non-coding RNAs. They can further be subdivided as follows:
siRNAs (small interfering RNAs) are very short sequences of RNA that are able to silence mRNA targets [99]. This phenomenon was first described in plants, fungi, and animals as RNA interference [100]. SiRNAs are generated from cutting long double-strand RNA (dsRNA), which can be generated from long hairpin RNA, genes, or pseudogenes. This process is executed by the biological machinery DICER.
snRNAs (small nuclear RNAs) are responsible for mRNA maturation [101] and participate in various fundamental biological mechanisms, such as splicing [102], TFs regulation [103], and the maintenance of telomeres [104].
SnoRNAs (small nucleolar RNA) are involved in chemical RNA modifications, such as methylation and pseudouridylation. The purpose and outcome of these modifications are still largely unknown, despite occurring in conserved regions of RNAs across species [105], as documented in recent databases that include information from seven different organisms [106], and integrate interaction information [107].
tsRNAs (transfer RNA derived small RNAs) are the most variable class of small non-coding RNA, having a repertoire of up to 150 modifications for each molecule [108]. Having been present since ancestral periods, they acquired different biological functions, ranging from bacterial development and viral infections to signalling molecules related to ageing, immunity, and disease [109]. Due to their cytoplasmic location and interaction with DICER, tsRNAs are regularly mis-annotated as miRNA [110], increasing the difficulties to fully understand their specific biological functions.
piRNAs (Piwi RNAs) are the largest class of small non-coding RNA molecules expressed in animal cells [111]. They are important to form protein complexes that silence transposons and other repetitive elements of the genome [112]. Estimates indicate that hundreds of thousands of different molecules belong to this class in mammals [113].
microRNAs (miRNAs) regulate gene expression by binding to mRNAs, thus suppressing their translation [114]. First described as early as 1993 [115], their basic mechanism of action has been known for some decades [116]. Still, many important questions remain unsolved [117], for example, how co-regulating miRNAs simultaneously regulate their target genes in different biological contexts.
Long non-coding RNAs (lncRNA) are RNAs with lengths exceeding 200 nucleotides and that are not translated into proteins. They have been implicated in many genomic processes, including parent-of-origin effects, alternative splicing, and tissue-specific gene expression [118,119]. In cancer, they have been observed to be particularly enriched for cis-expression quantitative trait loci (eQTL), which are often associated with genes regulating drug sensitivity [120]. LncRNAs are also associated with chromatin-modifying complexes [121] and histone methyltransferases [122].
3.3. Transposons
Transposons are regions of DNA that are repeated multiple times. They are also called “mobile elements” since they can change their position within the genome. They can be classified based on their mechanism of replication: Class I transposons, or retrotransposons, use a reverse transcriptase; and Class II transposons encode the protein transposase. Transposons play a major role in the diversification and evolution of the genome of different species, as well as individuals [123]. There are epigenetic mechanisms for avoiding unbalance in their transcription, for example, via methylation or zinc protein regulation [124]. However, despite their unpredictable jumps across the genome that may interrupt gene sequence and cause shifting mutations, certain diseases have shown specific associations with transposons, such as haemophilia [125] and Alzheimer’s disease [126].
3.4. Chromatin Modifications
With an estimated length of around 3 m [127], the DNA must be carefully folded to fit inside a cell. Together with histone proteins which aid in this process [128], it forms the chromatin complex. Chromatin is an extremely dynamic entity, whose changes lead to open or closed regions of the DNA, which in turn directly affects gene transcription. Chromatin modification is a set of epigenetic processes that govern many aspects of DNA replication, transcription, and repair. In eukaryotes, the basic unit of chromatin, the nucleosome, is comprised of 147 bp of DNA wrapped around a histone octamer made of two dimers of H2A and H2B and a tetramer of H3 and H4 proteins [129]. The interaction between DNA and histones occurs at the amino-terminal (N-terminal) tail of histone proteins and, for this reason, chemical modifications here, such as (de-) acetylation, phosphorylation and methylation, would change chromatin conformation, influencing various biological processes [130]. It is known that histone modifications are related to inheritance from mother to daughter cell and that this is influenced also by the environment, but the exact steps how this phenomenon occurs is still to be understood. These processes are crucial in development [131] and disease onset [132], but systematic large-scale studies remain scarce [133,134].
4. Network Models of the Epigenome
In light of the diversity of the biological mechanisms outlined above, it is clear that no isolated process or particular dataset alone will be able to provide a comprehensive picture of the developmental origins of health and disease. Indeed, after decades of biological research following a reductionist paradigm, a more holistic, systems-based framework is required [135]. Network theory can provide such a framework [136]. Networks are a general mathematical formalism for representing relationships (links) between objects (nodes). Important examples in biology and medicine range from protein–protein interaction (PPI) networks representing physical interactions between proteins [137] or gene regulatory networks representing transcription factors binding to DNA [138], to signalling networks of immune cells [139] or networks of organs linked by metabolism [140]. More generally speaking, we can distinguish between physical networks, where the links represent a direct physical relationship (e.g., protein interactions) and functional networks, where links represent indirect relationships (e.g., co-expression networks) [141] (Figure 4A).
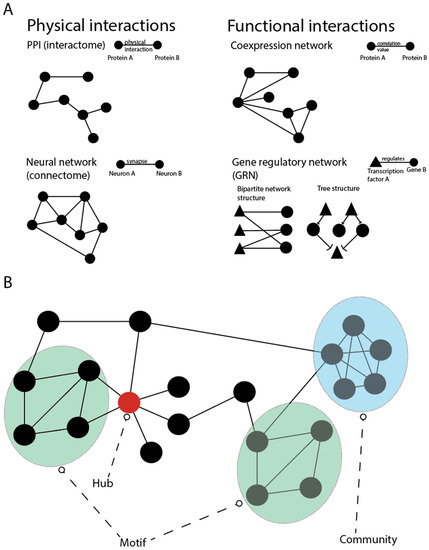
Figure 4.
Biological networks and their topological characteristics. (A) Classification of biological networks in two major categories: physical and functional interactions. The first category includes the protein–protein interaction network (interactome), which represents the map of the physical interactions of all proteins and the neural network (connectome), which shows synapses that connects neurons. Networks that are constituted by functional interactions have edges that represents functional relationships, such as the level of expression (co-expression network) or the gene regulation (gene regulatory network). (B) The most important features of a network are hub (a node connected to many others), motif (recurrent structures in different parts of the network), and community (group of densely interconnected nodes).
The abstraction of the complex biological machinery in terms of networks allows us to systematically investigate both global and local connection patterns and their respective biological implications [142]. For example, highly connected nodes (hubs) in PPI networks typically correspond to proteins with multiple biological functions. These proteins have been shown to be more likely essential [143,144], so that network analyses can help to identify the crucial genes involved in particular biological mechanisms [145]. Similarly, groups of densely interconnected nodes (network communities) and recurrent structures in different parts of the network (network motifs) have been shown to correspond to genes participating in the same biological process and allowed for the identification of fundamental building blocks of biological pathways, respectively [146,147,148] (Figure 4B). Network communities can also aid in the identification of genes that are involved in a particular disease and form a so-called disease module [149,150,151].
4.1. Gene Regulatory Networks (GRNs)
We can observe from the above that the relationship between genome and proteome is not a simple linear process, but that many factors and feedback mechanisms are involved. These can be external factors, internal molecules of the organism, and importantly also interactions among the genes themselves [152]. From a theoretical point of view, we can define the gene regulatory network as the wiring diagram that controls the collective gene expression [153]. In the early 1970s, Kauffmann and colleagues provided a first theory for GRNs [154]. Specifically, they considered Boolean networks and showed that a complex collective behaviour can emerge from simple logical operators among the individual components. The introduction of high-throughput technologies enabled the combination of theory and large-scale data [155,156,157]. In the last decade, various methods have been proposed to identify the (generally non-linear) functions that govern gene expression [158]. Today, we can incorporate a plethora of different types of omics data, such as RNA-seq, ChIP-seq (chromatin immunoprecipitation sequencing for identifying DNA binding sites), or ATAC-seq (assay for transposase accessible chromatin sequencing to identify open chromatin regions). This led to a somewhat broader definition of gene regulatory networks that includes various biological mechanisms that influence gene expression [159], such as transcriptional regulatory networks [160], protein interactions [161], microRNA networks [162], and metabolic networks [163].
GRNs can be used to better understand the molecular machinery governing cell states and to guide new screening experiments [164]. They may identify subnetworks and pathological pathways that can help to identify network-based biomarkers [165]. Gene regulatory networks also play an important role in development and ageing. In addition to dynamic changes over time, interindividual variation also needs to be considered. Methods that account for this include LIONESS (Linear Interpolation to Obtain Network Estimates for Single Samples), an approach able to distinguish the individual variability within a group [166]. This tool is part of a group of algorithms for GRN analysis called netZoo package [167] that also provides functionalities for investigating tissue specificity or multi-omic data integration.
4.2. Network Approaches for Interpreting DNA Methylation Profiles
Dynamic changes in methylation profiles (epigenetic drift) have been mapped on PPI networks, showing that mainly peripheral genes with low connectivity values are affected that fall into a number of connected network communities [168]. This enabled the identification of age-associated hot spots in stem-cell differentiation pathways [168]. Despite these successes, the mechanisms by which different methylated CpG regions influence remain poorly understood. To address this, the concept of Functional Epigenetic Modules (FEM) was proposed to identify gene modules of coordinated differential methylation and differential expression in the context of the human interactome [169].
To better understand how methylated genes are influencing each other, correlations between the demethylation status of a pair of genes can be considered. Computing all possible DNA methylation status comparisons leads to the so-called Co-occurrence and Mutual Exclusivity (COME), a table specifying for each gene pair whether their methylation status co-occurred or is mutually exclusive. This procedure was recently applied in cancer research using 14 main cancer types from The Cancer Genome Atlas (TCGA) [170], revealing a new way to stratify patients distinguishing several classes with distinct epigenetic trademarks that correspond to distinct clinical outcomes [171].
Similarly, DNA methylation correlation profiles were used to build a co-expression network (DNA methylation interaction network) in ovarian cancer, breast cancer, and glioblastoma multiforme, predicting new common prognostic genes [172]. Recently, the integration of differentially methylated genes and differentially expressed genes was used to identify new biomarkers in leukaemia [173]. Another approach for methylation and expression profile integration consisted of a multi-layer network approach called “Epigenetic Module based on Differential Networks (EMDN)”. The method first builds separate co-expression co-methylation networks. Then, the modules of densely connected node groups that are shared between the two are identified. The results indicate the potential of this procedure for finding new disease-associated genes in breast cancer [174].
Another challenge in the area of DNA methylation is to integrate large-scale population studies [175,176]. Similar to GWAS, which have been extensively used to find new genomic variants associated with specific phenotypes [177], epigenome-wide association studies (EWAS) have also been proposed to help to discover new aberrantly methylated genes [178]. In this context, a network-based algorithm (NEpiC) was proposed for combining methylation profiles from EWAS and PPI modularity. For each gene, the algorithm computes a score to identify differentially methylated genes that are then mapped on the PPI. Then, the modularity of these genes is evaluated and a prioritisation algorithm based on the connectivity is applied [179].
To encourage the usage of these and other tools and discoveries in a clinical setting and by people without highly specialised bioinformatics training, a number of user-friendly platforms have been recently developed. A prominent example is DNMIVD [180], an interactive DNA methylation visualisation resource providing information regarding DNA methylation-based diagnostic and prognostic models based on different cancer types from TCGA, expression-methylation quantitative trait loci (emQTL), pathway activity-methylation quantitative trait loci (pathway-meQTL), differentially variable and differentially methylated CpGs, survival analysis, and FEMs (from PPI and COMEs) [181].
4.3. Modelling Non-Coding RNA Interactions
As introduced above, many epigenetic molecules belong to the wide category of non-coding RNAs. For many non-coding RNAs, little is known about their biological activity or interactions with other biomolecules. The most information is available for miRNAs, with over 2000 miRNAs discovered in humans, many of which are associated with diseases [182,183]. Accordingly, miRNAs have also been the focus of network-based studies, although the general methodologies are likely to be applicable to all classes of non-coding RNAs.
An important challenge is to identify elements that jointly regulate their target genes in different biological contexts. For miRNAs, a solution has been proposed that starts from creating a network in which two miRNAs are connected when they share a significant number of gene targets, as determined by sequence complementarity and co-expression patterns [184]. The Molecular COmplex DEtection algorithm (MCODE) was then used to identify 12 different miRNA modules in this network. The cooperativity of miRNAs within a module was evaluated by their shared TFs and the functional similarity of their target genes. Similarly, synergistic miRNA-miRNA networks have been proposed, in which connections are based on common targets with similar biological functions and close proximity in the PPI network [185]. It was shown that disease associated miRNAs are located at central positions in the resulting network and that miRNAs associated with the same disease tend to form connected clusters [186].
Network science also provides an arsenal of tools for finding new miRNA–diseases associations [187]. For example, one can construct a bipartite network in which miRNAs are connected to diseases based on their reported associations. New miRNA–disease associations can then be predicted using a combined score that takes functional similarity among miRNAs into account, as well as similarity among diseases [188]. These predictions can be further improved by including network topology features, such as neighbour similarities and network distance [189]. In this fashion, new functional annotations and similarities between miRNA pairs were discovered in the context of prostate cancer [190].
Another important class of non-coding RNAs are lncRNAs. One of the first attempts to study the biological functions of lncRNA in a large-scale fashion was presented in [191]. In that study, the authors built a coding–non-coding gene (CNC) co-expression network for predicting the biological role of 340 lncRNAs. The function of a particular lncRNA was inferred based on patterns of co-expression and genomic co-location, using the gene ontology annotations of the coding genes in its immediate neighbourhood in the CNC network. The concept of predicting biological functions based on the interactions between lncRNAs and proteins has further been applied using a random walk with a restart algorithm on a lncRNA–protein interaction network [192].
Despite many promising initial results, the predictive power of these methods remains limited. One reason for this is likely that they rely on a direct link between non-coding elements and coding genes, for which data are scarce. This can be mitigated by considering additional datasets, for example, phenotypic similarities [193]. Similarly, the focus on a single class of biomolecules is often a limitation. Indeed, many lncRNAs have been shown to interact with other non-coding elements, such as miRNA [194]. An approach using a combined lncRNA–miRNA–mRNA interaction network found that predictions based on all three data performed better than using only lncRNA–protein interactions and can be used to identify clinical biomarkers in the context of breast cancer [195]. Along the same lines, the integration of pre- and post-transcriptional information into a lncRNA functional similarity score was used to predict disease associations [196]. These approaches were later expanded to include both human and mouse data [197,198].
4.4. Network Approaches for Chromatin Modifications and Transposons
To date, more than a hundred enzyme complexes, grouped in at least eight different classes, are known to catalyse enzymatic reactions in histones and cause changes in the DNA structure [199]. A collection of manually curated genes–diseases associations related to chromatin modifications is available from [200,201]. These resources can also be used to construct networks for elucidating the relationship between chromatin modifications and disease [202]. For example, the conformational state of chromatin was shown to be responsible for the switching to an inflammatory phenotype in macrophages and that the underlying mechanism that regulates this process is governed by a transcriptional regulator network [203]. While only a few network studies have been performed in this area of epigenetic regulation, the growing amount of related genes–disease annotation opens up new doors for systematic investigations [204]. The potential of investigating genetic mobile elements was touched only on the surface and it could unravel many open biological questions. For example, Levy et al. have developed a computational framework that is able to identify retrotransposons playing a key role in species evolution [205].
5. Towards an Integrative Analysis across Biological Hierarchies
Several of the examples above showed the potential of combining different data sources and corresponding biological mechanisms. The ultimate goal, of course, is to integrate all relevant layers of biological information into a single comprehensive picture [206]. Indeed, no single dataset can capture the complex and high-dimensional nature of the biological processes involved in health and disease [207]. The integration of data from various sources poses both technical and conceptual challenges [208]. In the following, we highlight recent developments in this area, with a particular focus on examples that involve the various mechanisms introduced above.
From a data science perspective, data integration can often be boiled down to finding correlations and corresponding trends between different datasets. A basic way to achieve this is through a particular type of matrix operations, so called matrix factorisation, which provides a dimensionality-reduced model of the relation between features in different data. A widely used method for this is joint non-negative matrix factorisation. While simple to implement, being based only on matrix multiplication, it requires large amounts of computing resources and proper care needs to be taken during normalisation. More advanced variations of this technique that pose less restrictions on the data that can be used include the iCluster package [209].
There are also network-based methods for data integration. These methods build on the fact that many biological processes have a direct network representation, but also on the interpretation of networks as a visual and abstract representation of data matrices, allowing an intuitive, but also formal, approach to biological data analysis. For example, MpDisNet is a network-based methodology for identifying disease–disease relationships by integrating four different biological networks: disease–miRNA network, miRNA–gene network, disease–gene network, and the human interactome [210]. The methodology revealed that comorbidities for pulmonary diseases were driven by miRNA-mediated pathobiological pathways. It thus identified a new type of disease–disease relationship that filled a gap between genome and phenotype using miRNA data as a bridge.
Another approach for integrating different types of biomolecules uses cooperative networks, in which the different components contribute to a common goal. This concept was applied in cancer research to elucidate the mechanisms responsible for the upregulation of oncogenes [211]. Here, different components contribute to the cancerogenic phenotype: chromatin opening, the recognition of the gene-specific DNA motif, the creation of a scaffold between histones, and the constitution of the transcriptional complex. The method involves several types of enrichment analysis to obtain a prioritisation of the key elements that regulate each epigenetic step of transcription regulation. For example, motif enrichment was used to determine which transcription factor motifs are significant for certain promoters, while the transcription factor target analysis defined the transcription factors that govern a set of target genes.
Network-based data integration has been successfully applied not only in oncology, but also in other medical fields. In virology, for example, genetic–epigenetic interactions were used in a cross-species integration between Epstein–Barr virus and human cells [212]. The resulting network was obtained by merging PPIs, gene–miRNA interactions, gene–lncRNA interactions, and host–virus cross talk networks. This allows for the identification of dynamic epigenetic patterns, suggesting an initial epigenetic inhibition by viral proteins that results in an immune response dysregulation of the host. The analysis further uncovered the most active viral proteins and miRNAs responsible for resistance mechanisms against the host’s defence. In cardiology, analogous methods were used to distinguish different heart failure phenotypes, proposing the “EPi-transgeneratIonal network mOdeling for STratificatiOn of heaRt Morbidity” (EPIKO-STORM) [80].
While this review mainly focuses on network approaches to integrative analysis, other methods for data integration are also of interest. An approach that is currently undergoing rapid development is the use of machine learning. This computational approach is based on the capability of the algorithm to identify patterns from a part of the dataset (training set) that is used to learn fundamental characteristics of the shape of the data that can be used for classification and integration tasks [213]. Such an approach is able to integrate data beyond transcriptomic and epigenomic data, having been applied to other omic data types, such as genomic, proteomic, and metabolomic data [214,215]. As such, machine learning presents itself as a promising approach to integration, though having the limitation that the training dataset should be large enough to contain all the representative features of the real dataset, which is not always the case [216].
In conclusion, using these integrative approaches in a plethora of transcriptomic (Table 1) and epigenetic data (Table 2) can help to understand the pathophysiological mechanisms behind common conditions of the periconceptional period and pregnancy, such as placenta percreta [217], pre-eclampsia [218], and the pregnancy-induced hypertension syndrome [219].

Table 1.
Published transcriptomic datasets within the context of early human development.

Table 2.
Resources containing epigenic data.
6. Conclusions
Multi-omics data are becoming more and more prevalent. Even nowadays, there is a massive amount of transcriptomic data as well as a growing amount of diverse epigenetics data publicly available to the community. These data represent a treasure trove for the community, aiming to unravel the developmental origins of health and disease. Mining these data requires approaches for their systematic integration and interpretation. Network-based methods are particularly promising for these purposes. We hope that the examples highlighted in this review may serve as an inspiration and motivation for this exciting area of research, showing that integrative analysis enables insights that are not accessible from any one dataset or biological process alone. In the future, the scale and diversity of relevant dataset will only grow, enabling, for example, to connect molecular data with epidemiological findings. The insights that could be gleaned from such datasets have potential to impact large scale populations, and to help in the prevention, prediction of disease onset and intervention.
Author Contributions
Conceptualisation and writing—original draft preparation, S.D.L. and I.F.W.; writing—review and editing, S.D.L., I.F.W., A.S. and J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 812660 (DohART-NET) to A.S. and J.M., and by the Vienna Science and Technology Fund (WWTF) through project VRG15-005 granted to J.M.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
In this section, you can acknowledge any support given which is not covered by the author contribution or funding sections. This may include administrative and technical support, or donations in kind (e.g., materials used for experiments).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, Transcriptome and Proteome: The Rise of Omics Data and Their Integration in Biomedical Sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Eidem, H.R.; McGary, K.L.; Capra, J.A.; Abbot, P.; Rokas, A. The Transformative Potential of an Integrative Approach to Pregnancy. Placenta 2017, 57, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kumar, K.; Jain, S.; Hassan, T.; Dada, R. Novel Insights into the Genetic and Epigenetic Paternal Contribution to the Human Embryo. Clinics 2013, 68 (Suppl. S1), 5–14. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Shin, D.-M.; Schneider, G.; Ratajczak, J.; Kucia, M. Parental Imprinting Regulates Insulin-like Growth Factor Signaling: A Rosetta Stone for Understanding the Biology of Pluripotent Stem Cells, Aging and Cancerogenesis. Leukemia 2013, 27, 773–779. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joyce, J.A.; Lam, W.K.; Catchpoole, D.J.; Jenks, P.; Reik, W.; Maher, E.R.; Schofield, P.N. Imprinting of IGF2 and H19: Lack of Reciprocity in Sporadic Beckwith-Wiedemann Syndrome. Hum. Mol. Genet. 1997, 6, 1543–1548. [Google Scholar] [CrossRef]
- Zapalska-Sozoniuk, M.; Chrobak, L.; Kowalczyk, K.; Kankofer, M. Is It Useful to Use Several “Omics” for Obtaining Valuable Results? Mol. Biol. Rep. 2019, 46, 3597–3606. [Google Scholar] [CrossRef] [PubMed]
- Niakan, K.K.; Han, J.; Pedersen, R.A.; Simon, C.; Pera, R.A.R. Human Pre-Implantation Embryo Development. Development 2012, 139, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Niakan, K.K.; Eggan, K. Analysis of Human Embryos from Zygote to Blastocyst Reveals Distinct Gene Expression Patterns Relative to the Mouse. Dev. Biol. 2013, 375, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, P.L. Building Principles for Constructing a Mammalian Blastocyst Embryo. Biology 2018, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Turco, M.Y.; Moffett, A. Development of the Human Placenta. Development 2019, 146, dev163428. [Google Scholar] [CrossRef]
- Barker, D.J.; Osmond, C. Infant Mortality, Childhood Nutrition, and Ischaemic Heart Disease in England and Wales. Lancet 1986, 1, 1077–1081. [Google Scholar] [CrossRef]
- Hochberg, Z.; Feil, R.; Constancia, M.; Fraga, M.; Junien, C.; Carel, J.-C.; Boileau, P.; Le Bouc, Y.; Deal, C.L.; Lillycrop, K.; et al. Child Health, Developmental Plasticity, and Epigenetic Programming. Endocr. Rev. 2011, 32, 159–224. [Google Scholar] [CrossRef] [PubMed]
- Mantione, K.J.; Kream, R.M.; Kuzelova, H.; Ptacek, R.; Raboch, J.; Samuel, J.M.; Stefano, G.B. Comparing Bioinformatic Gene Expression Profiling Methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014, 20, 138–142. [Google Scholar]
- McGettigan, P.A. Transcriptomics in the RNA-Seq Era. Curr. Opin. Chem. Biol. 2013, 17, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Baruzzo, G.; Hayer, K.E.; Kim, E.J.; Di Camillo, B.; FitzGerald, G.A.; Grant, G.R. Simulation-Based Comprehensive Benchmarking of RNA-Seq Aligners. Nat. Methods 2017, 14, 135–139. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Navarro Gonzalez, J.; Zweig, A.S.; Speir, M.L.; Schmelter, D.; Rosenbloom, K.R.; Raney, B.J.; Powell, C.C.; Nassar, L.R.; Maulding, N.D.; Lee, C.M.; et al. The UCSC Genome Browser Database: 2021 Update. Nucleic Acids Res. 2021, 49, D1046–D1057. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, B. A Comprehensive Evaluation of Ensembl, RefSeq, and UCSC Annotations in the Context of RNA-Seq Read Mapping and Gene Quantification. BMC Genom. 2015, 16, 97. [Google Scholar] [CrossRef]
- Kepler, T.B.; Crosby, L.; Morgan, K.T. Normalization and Analysis of DNA Microarray Data by Self-Consistency and Local Regression. Genome Biol. 2002, 3, RESEARCH0037. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Harold, H. Relations between Two Sets of Variates. Biometrika 1936, 28, 321–377. [Google Scholar]
- Van der Maaten, L.; Hinton, G. Visualizing Data Using T-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Linderman, G.C.; Rachh, M.; Hoskins, J.G.; Steinerberger, S.; Kluger, Y. Fast Interpolation-Based t-SNE for Improved Visualization of Single-Cell RNA-Seq Data. Nat. Methods 2019, 16, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Luecken, M.D.; Theis, F.J. Current Best Practices in Single-Cell RNA-Seq Analysis: A Tutorial. Mol. Syst. Biol. 2019, 15, e8746. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality Reduction for Visualizing Single-Cell Data Using UMAP. Nat. Biotechnol. 2019, 37, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Si, Y. Cluster Analysis of RNA-Sequencing Data. In Statistical Analysis of Next Generation Sequencing Data; Datta, S., Nettleton, D., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 191–217. ISBN 9783319072128. [Google Scholar]
- Murtagh, F. A Survey of Recent Advances in Hierarchical Clustering Algorithms. Comput. J. 1983, 26, 354–359. [Google Scholar] [CrossRef]
- Murtagh, F.; Contreras, P. Algorithms for Hierarchical Clustering: An Overview. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2012, 2, 86–97. [Google Scholar] [CrossRef]
- Xiang, D.; Venglat, P.; Tibiche, C.; Yang, H.; Risseeuw, E.; Cao, Y.; Babic, V.; Cloutier, M.; Keller, W.; Wang, E.; et al. Genome-Wide Analysis Reveals Gene Expression and Metabolic Network Dynamics during Embryo Development in Arabidopsis. Plant Physiol. 2011, 156, 346–356. [Google Scholar] [CrossRef]
- Vallée, M.; Dufort, I.; Desrosiers, S.; Labbe, A.; Gravel, C.; Gilbert, I.; Robert, C.; Sirard, M.-A. Revealing the Bovine Embryo Transcript Profiles during Early in Vivo Embryonic Development. Reproduction 2009, 138, 95–105. [Google Scholar] [CrossRef]
- Yan, J.; Buer, H.; Wang, Y.P.; Zhula, G.; Bai, Y.E. Transcriptomic Time-Series Analyses of Gene Expression Profile during Zygotic Embryo Development in Picea Mongolica. Front. Genet. 2021, 12, 738649. [Google Scholar] [CrossRef]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Oakland, CA, USA, 1 January 1967; Volume 1, pp. 281–297. [Google Scholar]
- Lumchanow, W.; Udomsiri, S. Chicken Embryo Development Detection Using Self-Organizing Maps and K-Mean Clustering. In Proceedings of the 2017 International Electrical Engineering Congress (iEECON), Pattaya, Thailand, 8–10 March 2017; pp. 1–4. [Google Scholar]
- Balsor, J.L.; Arbabi, K.; Singh, D.; Kwan, R.; Zaslavsky, J.; Jeyanesan, E.; Murphy, K.M. A Practical Guide to Sparse K-Means Clustering for Studying Molecular Development of the Human Brain. Front. Neurosci. 2021, 15, 668293. [Google Scholar] [CrossRef]
- Liu, Z. Clustering Single-Cell RNA-Seq Data with Regularized Gaussian Graphical Model. Genes 2021, 12, 311. [Google Scholar] [CrossRef]
- Xu, C.; Su, Z. Identification of Cell Types from Single-Cell Transcriptomes Using a Novel Clustering Method. Bioinformatics 2015, 31, 1974–1980. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Cao, J.; Martin, B.K.; Li, T.; Welsh, I.C.; Srivatsan, S.; Huang, X.; Calderon, D.; Noble, W.S.; Disteche, C.M.; et al. Systematic Reconstruction of Cellular Trajectories across Mouse Embryogenesis. Nat. Genet. 2022, 54, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Milewski, R.; Malinowski, P.; Milewska, A.; Czerniecki, J.; Ziniewicz, P.; Wołczyński, S. Nearest Neighbor Concept in the Study of IVF ICSI/ET Treatment Effectiveness. Stud. Log. Gramm. Rhetor. Log. Stat. Comput. Methods Med. 2011, 25, 49–57. [Google Scholar]
- Wei, D.; Jiang, Q.; Wei, Y.; Wang, S. A Novel Hierarchical Clustering Algorithm for Gene Sequences. BMC Bioinform. 2012, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; He, L.; He, R.L.; Yau, S.S.-T. A Novel Approach to Clustering Genome Sequences Using Inter-Nucleotide Covariance. Front. Genet. 2019, 10, 234. [Google Scholar] [CrossRef] [PubMed]
- Groff, A.F.; Resetkova, N.; DiDomenico, F.; Sakkas, D.; Penzias, A.; Rinn, J.L.; Eggan, K. RNA-Seq as a Tool for Evaluating Human Embryo Competence. Genome Res. 2019, 29, 1705–1718. [Google Scholar] [CrossRef]
- Zhang, B.; Peñagaricano, F.; Driver, A.; Chen, H.; Khatib, H. Differential Expression of Heat Shock Protein Genes and Their Splice Variants in Bovine Preimplantation Embryos. J. Dairy Sci. 2011, 94, 4174–4182. [Google Scholar] [CrossRef]
- Pollet, N.; Muncke, N.; Verbeek, B.; Li, Y.; Fenger, U.; Delius, H.; Niehrs, C. An Atlas of Differential Gene Expression during Early Xenopus Embryogenesis. Mech. Dev. 2005, 122, 365–439. [Google Scholar] [CrossRef]
- Hu, B.; Zheng, L.; Long, C.; Song, M.; Li, T.; Yang, L.; Zuo, Y. EmExplorer: A Database for Exploring Time Activation of Gene Expression in Mammalian Embryos. Open Biol. 2019, 9, 190054. [Google Scholar] [CrossRef]
- Reid, J.E.; Wernisch, L. Pseudotime Estimation: Deconfounding Single Cell Time Series. Bioinformatics 2016, 32, 2973–2980. [Google Scholar] [CrossRef]
- Waddington, C.H. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology; George Allen & Unwin: Crows Nest, Australia, 1957. [Google Scholar]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Schiebinger, G.; Shu, J.; Tabaka, M.; Cleary, B.; Subramanian, V.; Solomon, A.; Gould, J.; Liu, S.; Lin, S.; Berube, P.; et al. Optimal-Transport Analysis of Single-Cell Gene Expression Identifies Developmental Trajectories in Reprogramming. Cell 2019, 176, 1517. [Google Scholar] [CrossRef] [PubMed]
- Saelens, W.; Cannoodt, R.; Saeys, Y. A Comprehensive Evaluation of Module Detection Methods for Gene Expression Data. Nat. Commun. 2018, 9, 1090. [Google Scholar] [CrossRef] [PubMed]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA Velocity of Single Cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef]
- Bergen, V.; Soldatov, R.A.; Kharchenko, P.V.; Theis, F.J. RNA Velocity—Current Challenges and Future Perspectives. Mol. Syst. Biol. 2021, 17, e10282. [Google Scholar] [CrossRef]
- Wolf, F.A.; Hamey, F.K.; Plass, M.; Solana, J.; Dahlin, J.S.; Göttgens, B.; Rajewsky, N.; Simon, L.; Theis, F.J. PAGA: Graph Abstraction Reconciles Clustering with Trajectory Inference through a Topology Preserving Map of Single Cells. Genome Biol. 2019, 20, 59. [Google Scholar] [CrossRef]
- Mittnenzweig, M.; Mayshar, Y.; Cheng, S.; Ben-Yair, R.; Hadas, R.; Rais, Y.; Chomsky, E.; Reines, N.; Uzonyi, A.; Lumerman, L.; et al. A Single-Embryo, Single-Cell Time-Resolved Model for Mouse Gastrulation. Cell 2021, 184, 2825–2842. [Google Scholar] [CrossRef]
- Farrell, J.A.; Wang, Y.; Riesenfeld, S.J.; Shekhar, K.; Regev, A.; Schier, A.F. Single-Cell Reconstruction of Developmental Trajectories during Zebrafish Embryogenesis. Science 2018, 360, eaar3131. [Google Scholar] [CrossRef]
- Xu, C. A Review of Somatic Single Nucleotide Variant Calling Algorithms for Next-Generation Sequencing Data. Comput. Struct. Biotechnol. J. 2018, 16, 15–24. [Google Scholar] [CrossRef]
- Pirooznia, M.; Kramer, M.; Parla, J.; Goes, F.S.; Potash, J.B.; McCombie, W.R.; Zandi, P.P. Validation and Assessment of Variant Calling Pipelines for Next-Generation Sequencing. Hum. Genom. 2014, 8, 14. [Google Scholar] [CrossRef]
- NM, P.; Liu, H.; Bousounis, P.; Spurr, L.; Alomran, N.; Ibeawuchi, H.; Sein, J.; Reece-Stremtan, D.; Horvath, A. Estimating the Allele-Specific Expression of SNVs From 10× Genomics Single-Cell RNA-Sequencing Data. Genes 2020, 11, 240. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular Profiling for Precision Cancer Therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271–281.e7. [Google Scholar] [CrossRef]
- Fasterius, E.; Uhlén, M.; Al-Khalili Szigyarto, C. Single-Cell RNA-Seq Variant Analysis for Exploration of Genetic Heterogeneity in Cancer. Sci. Rep. 2019, 9, 9524. [Google Scholar] [CrossRef]
- Nica, A.C.; Dermitzakis, E.T. Expression Quantitative Trait Loci: Present and Future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120362. [Google Scholar] [CrossRef]
- Aguet, F.; Anand, S.; Ardlie, K.G.; Gabriel, S.; Getz, G.A.; Graubert, A.; Hadley, K.; Handsaker, R.E.; Huang, K.H.; Kashin, S.; et al. The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330. [Google Scholar]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic Analyses Identify Molecular Subtypes of Pancreatic Cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Haraksingh, R.R.; Snyder, M.P. Impacts of Variation in the Human Genome on Gene Regulation. J. Mol. Biol. 2013, 425, 3970–3977. [Google Scholar] [CrossRef]
- Spurr, L.F.; Alomran, N.; Bousounis, P.; Reece-Stremtan, D.; Prashant, N.M.; Liu, H.; Słowiński, P.; Li, M.; Zhang, Q.; Sein, J.; et al. ReQTL: Identifying Correlations between Expressed SNVs and Gene Expression Using RNA-Sequencing Data. Bioinformatics 2020, 36, 1351–1359. [Google Scholar] [CrossRef]
- Bae, T.; Tomasini, L.; Mariani, J.; Zhou, B.; Roychowdhury, T.; Franjic, D.; Pletikos, M.; Pattni, R.; Chen, B.-J.; Venturini, E.; et al. Different Mutational Rates and Mechanisms in Human Cells at Pregastrulation and Neurogenesis. Science 2018, 359, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Meissner, A.; Lander, E.S. The Mammalian Epigenome. Cell 2007, 128, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Perrino, C.; Barabási, A.-L.; Condorelli, G.; Davidson, S.M.; De Windt, L.; Dimmeler, S.; Engel, F.B.; Hausenloy, D.J.; Hill, J.A.; Van Laake, L.W.; et al. Epigenomic and Transcriptomic Approaches in the Post-Genomic Era: Path to Novel Targets for Diagnosis and Therapy of the Ischaemic Heart? Position Paper of the European Society of Cardiology Working Group on Cellular Biology of the Heart. Cardiovasc. Res. 2017, 113, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-G.; Tan, L.-J.; Xu, C.; He, H.; Tian, Q.; Zhou, Y.; Qiu, C.; Chen, X.-D.; Deng, H.-W. Integrative Analysis of Transcriptomic and Epigenomic Data to Reveal Regulation Patterns for BMD Variation. PLoS ONE 2015, 10, e0138524. [Google Scholar] [CrossRef]
- Sun, X.; Yi, J.; Yang, J.; Han, Y.; Qian, X.; Liu, Y.; Li, J.; Lu, B.; Zhang, J.; Pan, X.; et al. An Integrated Epigenomic-Transcriptomic Landscape of Lung Cancer Reveals Novel Methylation Driver Genes of Diagnostic and Therapeutic Relevance. Theranostics 2021, 11, 5346–5364. [Google Scholar] [CrossRef]
- Napoli, C.; Benincasa, G.; Donatelli, F.; Ambrosio, G. Precision Medicine in Distinct Heart Failure Phenotypes: Focus on Clinical Epigenetics. Am. Heart J. 2020, 224, 113–128. [Google Scholar] [CrossRef]
- Gluckman, P.D.; Hanson, M.A.; Buklijas, T.; Low, F.M.; Beedle, A.S. Epigenetic Mechanisms That Underpin Metabolic and Cardiovascular Diseases. Nat. Rev. Endocrinol. 2009, 5, 401–408. [Google Scholar] [CrossRef]
- Urdinguio, R.G.; Sanchez-Mut, J.V.; Esteller, M. Epigenetic Mechanisms in Neurological Diseases: Genes, Syndromes, and Therapies. Lancet Neurol. 2009, 8, 1056–1072. [Google Scholar] [CrossRef]
- Kelly, T.K.; De Carvalho, D.D.; Jones, P.A. Epigenetic Modifications as Therapeutic Targets. Nat. Biotechnol. 2010, 28, 1069–1078. [Google Scholar] [CrossRef]
- Lyko, F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Li, E.; Zhang, Y. DNA Methylation in Mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef] [PubMed]
- Mohandas, T.; Sparkes, R.S.; Shapiro, L.J. Reactivation of an Inactive Human X Chromosome: Evidence for X Inactivation by DNA Methylation. Science 1981, 211, 393–396. [Google Scholar] [CrossRef]
- Razin, A. Tissue Specific DNA Methylation Patterns: Biochemistry of Formation and Possible Role. In Biological Methylation and Drug Design; Humana Press: Totowa, NJ, USA, 1986; pp. 127–137. [Google Scholar]
- Kass, S.U.; Landsberger, N.; Wolffe, A.P. DNA Methylation Directs a Time-Dependent Repression of Transcription Initiation. Curr. Biol. 1997, 7, 157–165. [Google Scholar] [CrossRef]
- Luo, C.; Hajkova, P.; Ecker, J.R. Dynamic DNA Methylation: In the Right Place at the Right Time. Science 2018, 361, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D.; Wolffe, A.P. DNA Methylation in Health and Disease. Nat. Rev. Genet. 2000, 1, 11–19. [Google Scholar] [CrossRef]
- Yuan, T.; Jiao, Y.; de Jong, S.; Ophoff, R.A.; Beck, S.; Teschendorff, A.E. An Integrative Multi-Scale Analysis of the Dynamic DNA Methylation Landscape in Aging. PLoS Genet. 2015, 11, e1004996. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, A.R.; Larrick, J.W. Epigenetic Drift Is a Determinant of Mammalian Lifespan. Rejuvenation Res. 2017, 20, 430–436. [Google Scholar] [CrossRef]
- Issa, J.-P. Aging and Epigenetic Drift: A Vicious Cycle. J. Clin. Investig. 2014, 124, 24–29. [Google Scholar] [CrossRef]
- Lara, E.; Calvanese, V.; Fraga, M.F. Epigenetic Drift and Aging. In Epigenetics of Aging; Springer: New York, NY, USA, 2010; pp. 257–273. [Google Scholar]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small Non-Coding RNA and Cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef]
- Kato, M.; Slack, F.J. Ageing and the Small, Non-Coding RNA World. Ageing Res. Rev. 2013, 12, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Schuster, A.; Skinner, M.K.; Yan, W. Ancestral Vinclozolin Exposure Alters the Epigenetic Transgenerational Inheritance of Sperm Small Noncoding RNAs. In Environmental Epigenetics; Springer: Berlin/Heidelberg, Germany, 2016; Volume 2. [Google Scholar] [CrossRef]
- Houri-Zeevi, L.; Korem Kohanim, Y.; Antonova, O.; Rechavi, O. Three Rules Explain Transgenerational Small RNA Inheritance in C. Elegans. Cell 2020, 182, 1186–1197.e12. [Google Scholar] [CrossRef]
- Ye, K.; Malinina, L.; Patel, D.J. Recognition of Small Interfering RNA by a Viral Suppressor of RNA Silencing. Nature 2003, 426, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Non–Coding RNA Genes and the Modern RNA World. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef]
- Sperling, R. Small Non-Coding RNA within the Endogenous Spliceosome and Alternative Splicing Regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194406. [Google Scholar] [CrossRef]
- Goodrich, J.A.; Kugel, J.F. Non-Coding-RNA Regulators of RNA Polymerase II Transcription. Nat. Rev. Mol. Cell Biol. 2006, 7, 612–616. [Google Scholar] [CrossRef]
- Cusanelli, E.; Romero, C.A.P.; Chartrand, P. Telomeric Noncoding RNA TERRA Is Induced by Telomere Shortening to Nucleate Telomerase Molecules at Short Telomeres. Mol. Cell 2013, 51, 780–791. [Google Scholar] [CrossRef]
- Mitchell, J.R.; Cheng, J.; Collins, K. A Box H/ACA Small Nucleolar RNA-like Domain at the Human Telomerase RNA 3’ End. Mol. Cell. Biol. 1999, 19, 567–576. [Google Scholar] [CrossRef]
- Yoshihama, M.; Nakao, A.; Kenmochi, N. SnOPY: A Small Nucleolar RNA Orthological Gene Database. BMC Res. Notes 2013, 6, 426. [Google Scholar] [CrossRef]
- Bouchard-Bourelle, P.; Desjardins-Henri, C.; Mathurin-St-Pierre, D.; Deschamps-Francoeur, G.; Fafard-Couture, É.; Garant, J.-M.; Elela, S.A.; Scott, M.S. SnoDB: An Interactive Database of Human SnoRNA Sequences, Abundance and Interactions. Nucleic Acids Res. 2020, 48, D220–D225. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Oberbauer, V.; Schaefer, M.R. TRNA-Derived Small RNAs: Biogenesis, Modification, Function and Potential Impact on Human Disease Development. Genes 2018, 9, 607. [Google Scholar] [CrossRef] [PubMed]
- Schopman, N.C.T.; Heynen, S.; Haasnoot, J.; Berkhout, B. A MiRNA-TRNA Mix-up: TRNA Origin of Proposed MiRNA. RNA Biol. 2010, 7, 573–576. [Google Scholar] [CrossRef]
- Seto, A.G.; Kingston, R.E.; Lau, N.C. The Coming of Age for Piwi Proteins. Mol. Cell 2007, 26, 603–609. [Google Scholar] [CrossRef]
- Aravin, A.A.; Sachidanandam, R.; Girard, A.; Fejes-Toth, K.; Hannon, G.J. Developmentally Regulated PiRNA Clusters Implicate MILI in Transposon Control. Science 2007, 316, 744–747. [Google Scholar] [CrossRef]
- Das, P.P.; Bagijn, M.P.; Goldstein, L.D.; Woolford, J.R.; Lehrbach, N.J.; Sapetschnig, A.; Buhecha, H.R.; Gilchrist, M.J.; Howe, K.L.; Stark, R.; et al. Piwi and PiRNAs Act Upstream of an Endogenous SiRNA Pathway to Suppress Tc3 Transposon Mobility in the Caenorhabditis Elegans Germline. Mol. Cell 2008, 31, 79–90. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Plasterk, R.H.A. The Diverse Functions of MicroRNAs in Animal Development and Disease. Dev. Cell 2006, 11, 441–450. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Vanderburg, C.; Beheshti, A. MicroRNAs (MiRNAs), the Final Frontier: The Hidden Master Regulators Impacting Biological Response in All Organisms Due to Spaceflight. 2020. Available online: https://three.jsc.nasa.gov/articles/miRNA_Beheshti.pdf (accessed on 16 September 2020).
- Mattick, J.S. The State of Long Non-Coding RNA Biology. Non-Coding RNA 2018, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M. Genome-Wide Analysis of Long Noncoding RNA Stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, C.; Guo, J.; Liu, K.; Hu, Y.; Wu, D.; Fang, H.; Zou, Y.; Wei, Z.; Wang, Z.; et al. Cis- and Trans-Acting Expression Quantitative Trait Loci of Long Non-Coding RNA in 2,549 Cancers With Potential Clinical and Therapeutic Implications. Front. Oncol. 2020, 10, 602104. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Schauer, T.; Krause, S.; Villa, R.; Thomae, A.W.; Becker, P.B. Two-Step Mechanism for Selective Incorporation of LncRNA into a Chromatin Modifier. Nucleic Acids Res. 2020, 48, 7483–7501. [Google Scholar] [CrossRef]
- Li, Y.-Q.; Sun, N.; Zhang, C.-S.; Li, N.; Wu, B.; Zhang, J.-L. Inactivation of LncRNA HOTAIRM1 Caused by Histone Methyltransferase RIZ1 Accelerated the Proliferation and Invasion of Liver Cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8767–8777. [Google Scholar]
- Munoz-Lopez, M.; Garcia-Perez, J.L. DNA Transposons: Nature and Applications in Genomics. Curr. Genom. 2010, 11, 115–128. [Google Scholar] [CrossRef]
- Yang, P.; Wang, Y.; Macfarlan, T.S. The Role of KRAB-ZFPs in Transposable Element Repression and Mammalian Evolution. Trends Genet. 2017, 33, 871–881. [Google Scholar] [CrossRef]
- Kazazian, H.H.; Wong, C.; Youssoufian, H.; Scott, A.F.; Phillips, D.G.; Antonarakis, S.E. Haemophilia A Resulting from de Novo Insertion of L1 Sequences Represents a Novel Mechanism for Mutation in Man. Nature 1988, 332, 164–166. [Google Scholar] [CrossRef]
- Sun, W.; Samimi, H.; Gamez, M.; Zare, H.; Frost, B. Pathogenic Tau-Induced PiRNA Depletion Promotes Neuronal Death through Transposable Element Dysregulation in Neurodegenerative Tauopathies. Nat. Neurosci. 2018, 21, 1038–1048. [Google Scholar] [CrossRef]
- Campbell, N.A. Biology: Concepts & Connections; Pearson/Benjamin Cummings: San Francisco, CA, USA, 2009; ISBN 9780321489845. [Google Scholar]
- Doenecke, D.; Gallwitz, D. Acetylation of Histones in Nucleosomes. Mol. Cell. Biochem. 1982, 44, 113–128. [Google Scholar] [CrossRef]
- Zhang, T.; Cooper, S.; Brockdorff, N. The Interplay of Histone Modifications—Writers That Read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Li, E. Chromatin Modification and Epigenetic Reprogramming in Mammalian Development. Nat. Rev. Genet. 2002, 3, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.A. Chromatin Modification and Disease. J. Med. Genet. 2000, 37, 905–915. [Google Scholar] [CrossRef][Green Version]
- Schones, D.E.; Zhao, K. Genome-Wide Approaches to Studying Chromatin Modifications. Nat. Rev. Genet. 2008, 9, 179–191. [Google Scholar] [CrossRef]
- Villaseñor, R.; Pfaendler, R.; Ambrosi, C.; Butz, S.; Giuliani, S.; Bryan, E.; Sheahan, T.W.; Gable, A.L.; Schmolka, N.; Manzo, M.; et al. ChromID Identifies the Protein Interactome at Chromatin Marks. Nat. Biotechnol. 2020, 38, 728–736. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Mapstone, M.; Connors, E.; Jacobson, M.; Monuki, E.S.; Malik, S.; Macciardi, F.; Federoff, H.J. Systems Healthcare: A Holistic Paradigm for Tomorrow. BMC Syst. Biol. 2017, 11, 142. [Google Scholar] [CrossRef]
- Silverman, E.K.; Schmidt, H.H.H.; Anastasiadou, E.; Altucci, L.; Angelini, M.; Badimon, L.; Balligand, J.-L.; Benincasa, G.; Capasso, G.; Conte, F.; et al. Molecular Networks in Network Medicine: Development and Applications. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1489. [Google Scholar] [CrossRef]
- Caldera, M.; Buphamalai, P.; Müller, F.; Menche, J. Interactome-Based Approaches to Human Disease. Curr. Opin. Syst. Biol. 2017, 3, 88–94. [Google Scholar] [CrossRef]
- Davidson, E.H. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution; Elsevier: Amsterdam, The Netherlands, 2010; ISBN 9780080455570. [Google Scholar]
- Bergthaler, A.; Menche, J. The Immune System as a Social Network. Nat. Immunol. 2017, 18, 481–482. [Google Scholar] [CrossRef]
- Schmidt, H.H.H.W.; Menche, J. The Regulatory Network Architecture of Cardiometabolic Diseases. Nat. Genet. 2022, 54, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Pržulj, N. Analyzing Network Data in Biology and Medicine: An Interdisciplinary Textbook for Biological, Medical and Computational Scientists; Cambridge University Press: Cambridge, UK, 2019; ISBN 9781108432238. [Google Scholar]
- Loan Vulliard, J.M. Complex Networks in Health and Disease. Syst. Med. 2021, 26–33. [Google Scholar] [CrossRef]
- Goymer, P. Why Do We Need Hubs? Nat. Rev. Genet. 2008, 9, 651. [Google Scholar] [CrossRef]
- Zotenko, E.; Mestre, J.; O’Leary, D.P.; Przytycka, T.M. Why Do Hubs in the Yeast Protein Interaction Network Tend to Be Essential: Reexamining the Connection between the Network Topology and Essentiality. PLoS Comput. Biol. 2008, 4, e1000140. [Google Scholar] [CrossRef]
- Muetze, T.; Goenawan, I.H.; Wiencko, H.L.; Bernal-Llinares, M.; Bryan, K.; Lynn, D.J. Contextual Hub Analysis Tool (CHAT): A Cytoscape App for Identifying Contextually Relevant Hubs in Biological Networks. F1000Research 2016, 5, 1745. [Google Scholar] [CrossRef]
- Sah, P.; Singh, L.O.; Clauset, A.; Bansal, S. Exploring Community Structure in Biological Networks with Random Graphs. BMC Bioinform. 2014, 15, 220. [Google Scholar] [CrossRef]
- Wilson, S.J.; Wilkins, A.D.; Lin, C.-H.; Lua, R.C.; Lichtarge, O. Discovery of functional and disease pathways by community detection in protein-protein interaction networks. Pac. Symp. Biocomput. 2017, 22, 336–347. [Google Scholar]
- Kim, W.; Li, M.; Wang, J.; Pan, Y. Biological Network Motif Detection and Evaluation. BMC Syst. Biol. 2011, 5, S5. [Google Scholar] [CrossRef]
- Tripathi, B.; Parthasarathy, S.; Sinha, H.; Raman, K.; Ravindran, B. Adapting Community Detection Algorithms for Disease Module Identification in Heterogeneous Biological Networks. Front. Genet. 2019, 10, 164. [Google Scholar] [CrossRef]
- Ghiassian, S.D.; Menche, J.; Barabási, A.-L. A DIseAse MOdule Detection (DIAMOnD) Algorithm Derived from a Systematic Analysis of Connectivity Patterns of Disease Proteins in the Human Interactome. PLoS Comput. Biol. 2015, 11, e1004120. [Google Scholar] [CrossRef]
- Choobdar, S.; Ahsen, M.E.; Crawford, J.; Tomasoni, M.; Fang, T.; Lamparter, D.; Lin, J.; Hescott, B.; Hu, X.; Mercer, J.; et al. Assessment of Network Module Identification across Complex Diseases. Nat. Methods 2019, 16, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. The Molecular and Mathematical Basis of Waddington’s Epigenetic Landscape: A Framework for Post-Darwinian Biology? Bioessays 2012, 34, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Davidson, E.H.; Erwin, D.H. Gene Regulatory Networks and the Evolution of Animal Body Plans. Science 2006, 311, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, S.A. Metabolic Stability and Epigenesis in Randomly Constructed Genetic Nets. J. Theor. Biol. 1969, 22, 437–467. [Google Scholar] [CrossRef]
- Arkin, A.; Ross, J.; McAdams, H.H. Stochastic Kinetic Analysis of Developmental Pathway Bifurcation in Phage Lambda-Infected Escherichia Coli Cells. Genetics 1998, 149, 1633–1648. [Google Scholar] [CrossRef]
- Hasty, J.; McMillen, D.; Isaacs, F.; Collins, J.J. Computational Studies of Gene Regulatory Networks: In Numero Molecular Biology. Nat. Rev. Genet. 2001, 2, 268–279. [Google Scholar] [CrossRef]
- de Jong, H. Modeling and Simulation of Genetic Regulatory Systems: A Literature Review. J. Comput. Biol. 2002, 9, 67–103. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Dehmer, M.; Haibe-Kains, B. Gene Regulatory Networks and Their Applications: Understanding Biological and Medical Problems in Terms of Networks. Front. Cell Dev. Biol. 2014, 2, 38. [Google Scholar] [CrossRef]
- Blais, A.; Dynlacht, B.D. Constructing Transcriptional Regulatory Networks. Genes Dev. 2005, 19, 1499–1511. [Google Scholar] [CrossRef]
- Vlaic, S.; Conrad, T.; Tokarski-Schnelle, C.; Gustafsson, M.; Dahmen, U.; Guthke, R.; Schuster, S. ModuleDiscoverer: Identification of Regulatory Modules in Protein-Protein Interaction Networks. Sci. Rep. 2018, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory Network of MiRNA on Its Target: Coordination between Transcriptional and Post-Transcriptional Regulation of Gene Expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Watson, E.; Yilmaz, L.S.; Walhout, A.J.M. Understanding Metabolic Regulation at a Systems Level: Metabolite Sensing, Mathematical Predictions, and Model Organisms. Annu. Rev. Genet. 2015, 49, 553–575. [Google Scholar] [CrossRef] [PubMed]
- Benes, B.; Guan, K.; Lang, M.; Long, S.P.; Lynch, J.P.; Marshall-Colón, A.; Peng, B.; Schnable, J.; Sweetlove, L.J.; Turk, M.J. Multiscale Computational Models Can Guide Experimentation and Targeted Measurements for Crop Improvement. Plant J. 2020, 103, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Dehmer, M.; Mueller, L.A.J.; Emmert-Streib, F. Quantitative Network Measures as Biomarkers for Classifying Prostate Cancer Disease States: A Systems Approach to Diagnostic Biomarkers. PLoS ONE 2013, 8, e77602. [Google Scholar] [CrossRef]
- Kuijjer, M.L.; Tung, M.G.; Yuan, G.; Quackenbush, J.; Glass, K. Estimating Sample-Specific Regulatory Networks. IScience 2019, 14, 226–240. [Google Scholar] [CrossRef]
- Available online: https://netzoo.github.io (accessed on 3 March 2022).
- West, J.; Widschwendter, M.; Teschendorff, A.E. Distinctive Topology of Age-Associated Epigenetic Drift in the Human Interactome. Proc. Natl. Acad. Sci. USA 2013, 110, 14138–14143. [Google Scholar] [CrossRef]
- Jiao, Y.; Widschwendter, M.; Teschendorff, A.E. A Systems-Level Integrative Framework for Genome-Wide DNA Methylation and Gene Expression Data Identifies Differential Gene Expression Modules under Epigenetic Control. Bioinformatics 2014, 30, 2360–2366. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An Immeasurable Source of Knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Ding, W.; Feng, G.; Hu, Y.; Chen, G.; Shi, T. Co-Occurrence and Mutual Exclusivity Analysis of DNA Methylation Reveals Distinct Subtypes in Multiple Cancers. Front. Cell Dev. Biol. 2020, 8, 20. [Google Scholar] [CrossRef]
- Hu, W.-L.; Zhou, X.-H. Identification of Prognostic Signature in Cancer Based on DNA Methylation Interaction Network. BMC Med. Genom. 2017, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, R.; Mackenzie, S.A. Integrative Network Analysis of Differentially Methylated and Expressed Genes for Biomarker Identification in Leukemia. Sci. Rep. 2020, 10, 2123. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Z.; Zhang, Z.; Huang, X.; Tang, W. Multiple Network Algorithm for Epigenetic Modules via the Integration of Genome-Wide DNA Methylation and Gene Expression Data. BMC Bioinform. 2017, 18, 72. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B. Overview of Statistical Methods for Genome-Wide Association Studies (GWAS). Methods Mol. Biol. 2013, 1019, 149–169. [Google Scholar] [PubMed]
- Michels, K.B.; Binder, A.M.; Dedeurwaerder, S.; Epstein, C.B.; Greally, J.M.; Gut, I.; Houseman, E.A.; Izzi, B.; Kelsey, K.T.; Meissner, A.; et al. Recommendations for the Design and Analysis of Epigenome-Wide Association Studies. Nat. Methods 2013, 10, 949–955. [Google Scholar] [CrossRef]
- Cantor, R.M.; Lange, K.; Sinsheimer, J.S. Prioritizing GWAS Results: A Review of Statistical Methods and Recommendations for Their Application. Am. J. Hum. Genet. 2010, 86, 6–22. [Google Scholar] [CrossRef]
- Birney, E.; Smith, G.D.; Greally, J.M. Epigenome-Wide Association Studies and the Interpretation of Disease-Omics. PLoS Genet. 2016, 12, e1006105. [Google Scholar] [CrossRef]
- Ruan, P.; Shen, J.; Santella, R.M.; Zhou, S.; Wang, S. NEpiC: A Network-Assisted Algorithm for Epigenetic Studies Using Mean and Variance Combined Signals. Nucleic Acids Res. 2016, 44, e134. [Google Scholar] [CrossRef][Green Version]
- Available online: http://www.unimd.org/dnmivd/ (accessed on 3 March 2022).
- Ding, W.; Chen, J.; Feng, G.; Chen, G.; Wu, J.; Guo, Y.; Ni, X.; Shi, T. DNMIVD: DNA Methylation Interactive Visualization Database. Nucleic Acids Res. 2020, 48, D856–D862. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between MiRNAs and Human Diseases. J. Cell. Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef]
- Li, Y.; Xu, J.; Chen, H.; Bai, J.; Li, S.; Zhao, Z.; Shao, T.; Jiang, T.; Ren, H.; Kang, C.; et al. Comprehensive Analysis of the Functional MicroRNA–MRNA Regulatory Network Identifies MiRNA Signatures Associated with Glioma Malignant Progression. Nucleic Acids Res. 2013, 41, e203. [Google Scholar] [CrossRef] [PubMed]
- Na, Y.-J.; Kim, J.H. Understanding Cooperativity of MicroRNAs via MicroRNA Association Networks. BMC Genom. 2013, 14 (Suppl. S5), S17. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, C.-X.; Li, Y.-S.; Lv, J.-Y.; Ma, Y.; Shao, T.-T.; Xu, L.-D.; Wang, Y.-Y.; Du, L.; Zhang, Y.-P.; et al. MiRNA–MiRNA Synergistic Network: Construction via Co-Regulating Functional Modules and Disease MiRNA Topological Features. Nucleic Acids Res. 2010, 39, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Cui, Q. An Analysis of Human MicroRNA and Disease Associations. PLoS ONE 2008, 3, e3420. [Google Scholar] [CrossRef]
- Parikh, V.N.; Jin, R.C.; Rabello, S.; Gulbahce, N.; White, K.; Hale, A.; Cottrill, K.A.; Shaik, R.S.; Waxman, A.B.; Zhang, Y.-Y.; et al. MicroRNA-21 Integrates Pathogenic Signaling to Control Pulmonary Hypertension: Results of a Network Bioinformatics Approach. Circulation 2012, 125, 1520–1532. [Google Scholar] [CrossRef]
- Chen, X.; Xie, D.; Wang, L.; Zhao, Q.; You, Z.-H.; Liu, H. BNPMDA: Bipartite Network Projection for MiRNA–Disease Association Prediction. Bioinformatics 2018, 34, 3178–3186. [Google Scholar] [CrossRef]
- Zhao, H.; Kuang, L.; Feng, X.; Zou, Q.; Wang, L. A Novel Approach Based on a Weighted Interactive Network to Predict Associations of MiRNAs and Diseases. Int. J. Mol. Sci. 2018, 20, 110. [Google Scholar] [CrossRef]
- Wei, J.; Yin, Y.; Deng, Q.; Zhou, J.; Wang, Y.; Yin, G.; Yang, J.; Tang, Y. Integrative Analysis of MicroRNA and Gene Interactions for Revealing Candidate Signatures in Prostate Cancer. Front. Genet. 2020, 11, 176. [Google Scholar] [CrossRef]
- Liao, Q.; Liu, C.; Yuan, X.; Kang, S.; Miao, R.; Xiao, H.; Zhao, G.; Luo, H.; Bu, D.; Zhao, H.; et al. Large-Scale Prediction of Long Non-Coding RNA Functions in a Coding–Non-Coding Gene Co-Expression Network. Nucleic Acids Res. 2011, 39, 3864–3878. [Google Scholar] [CrossRef]
- Li, A.; Ge, M.; Zhang, Y.; Peng, C.; Wang, M. Predicting Long Noncoding RNA and Protein Interactions Using Heterogeneous Network Model. Biomed Res. Int. 2015, 2015, 671950. [Google Scholar] [CrossRef]
- Chen, X. Predicting LncRNA-Disease Associations and Constructing LncRNA Functional Similarity Network Based on the Information of MiRNA. Sci. Rep. 2015, 5, 13186. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Nangia-Makker, P.; Farhana, L.; Majumdar, A.P.N. A Novel Mechanism of LncRNA and MiRNA Interaction: CCAT2 Regulates MiR-145 Expression by Suppressing Its Maturation Process in Colon Cancer Cells. Mol. Cancer 2017, 16, 155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Wang, Q.; Zhang, X.; Wang, D.; Tang, H.C.; Meng, X.; Ding, X. Identification of an LncRNA-miRNA-mRNA Interaction Mechanism in Breast Cancer Based on Bioinformatic Analysis. Mol. Med. Rep. 2017, 16, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Shi, H.; Wang, Z.; Hu, Y.; Yang, H.; Zhou, C.; Sun, J.; Zhou, M. IntNetLncSim: An Integrative Network Analysis Method to Infer Human LncRNA Functional Similarity. Oncotarget 2016, 7, 47864–47874. [Google Scholar] [CrossRef]
- LncRNA2Target. Available online: http://123.59.132.21/lncrna2target/ (accessed on 3 March 2022).
- Cheng, L.; Wang, P.; Tian, R.; Wang, S.; Guo, Q.; Luo, M.; Zhou, W.; Liu, G.; Jiang, H.; Jiang, Q. LncRNA2Target v2.0: A Comprehensive Database for Target Genes of LncRNAs in Human and Mouse. Nucleic Acids Res. 2019, 47, D140–D144. [Google Scholar] [CrossRef]
- DesJarlais, R.; Tummino, P.J. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry 2016, 55, 1584–1599. [Google Scholar] [CrossRef]
- Turinsky, A.L.; Turner, B.; Borja, R.C.; Gleeson, J.A.; Heath, M.; Pu, S.; Switzer, T.; Dong, D.; Gong, Y.; On, T.; et al. DAnCER: Disease-Annotated Chromatin Epigenetics Resource. Nucleic Acids Res. 2011, 39, D889–D894. [Google Scholar] [CrossRef][Green Version]
- DAnCER. Available online: http://wodaklab.org/dancer/ (accessed on 3 March 2022).
- Lundberg, S.M.; Tu, W.B.; Raught, B.; Penn, L.Z.; Hoffman, M.M.; Lee, S.-I. ChromNet: Learning the Human Chromatin Network from All ENCODE ChIP-Seq Data. Genome Biol. 2016, 17, 82. [Google Scholar] [CrossRef]
- Schmidt, S.V.; Krebs, W.; Ulas, T.; Xue, J.; Baßler, K.; Günther, P.; Hardt, A.-L.; Schultze, H.; Sander, J.; Klee, K.; et al. The Transcriptional Regulator Network of Human Inflammatory Macrophages Is Defined by Open Chromatin. Cell Res. 2016, 26, 151–170. [Google Scholar] [CrossRef]
- Helin, K.; Dhanak, D. Chromatin Proteins and Modifications as Drug Targets. Nature 2013, 502, 480–488. [Google Scholar] [CrossRef]
- Levy, O.; Knisbacher, B.A.; Levanon, E.Y.; Havlin, S. Integrating Networks and Comparative Genomics Reveals Retroelement Proliferation Dynamics in Hominid Genomes. Sci. Adv. 2017, 3, e1701256. [Google Scholar] [CrossRef] [PubMed]
- Buphamalai, P.; Kokotovic, T.; Nagy, V.; Menche, J. Network Analysis Reveals Rare Disease Signatures across Multiple Levels of Biological Organization. Nat. Commun. 2021, 12, 6306. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Snyder, M.P. Integrative Omics for Health and Disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Stuart, T.; Satija, R. Integrative Single-Cell Analysis. Nat. Rev. Genet. 2019, 20, 257–272. [Google Scholar] [CrossRef]
- Shen, R.; Mo, Q.; Schultz, N.; Seshan, V.E.; Olshen, A.B.; Huse, J.; Ladanyi, M.; Sander, C. Integrative Subtype Discovery in Glioblastoma Using ICluster. PLoS ONE 2012, 7, e35236. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zeng, X.; Fang, J.; Lin, J.; Chan, S.Y.; Erzurum, S.C.; Cheng, F. A Network-Based Approach to Uncover MicroRNA-Mediated Disease Comorbidities and Potential Pathobiological Implications. NPJ Syst. Biol. Appl. 2019, 5, 41. [Google Scholar] [CrossRef]
- Wilson, S.; Filipp, F.V. A Network of Epigenomic and Transcriptional Cooperation Encompassing an Epigenomic Master Regulator in Cancer. NPJ Syst. Biol. Appl. 2018, 4, 24. [Google Scholar] [CrossRef]
- Li, C.-W.; Jheng, B.-R.; Chen, B.-S. Investigating Genetic-and-Epigenetic Networks, and the Cellular Mechanisms Occurring in Epstein-Barr Virus-Infected Human B Lymphocytes via Big Data Mining and Genome-Wide Two-Sided NGS Data Identification. PLoS ONE 2018, 13, e0202537. [Google Scholar] [CrossRef]
- Picard, M.; Scott-Boyer, M.-P.; Bodein, A.; Périn, O.; Droit, A. Integration Strategies of Multi-Omics Data for Machine Learning Analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef]
- Grapov, D.; Fahrmann, J.; Wanichthanarak, K.; Khoomrung, S. Rise of Deep Learning for Genomic, Proteomic, and Metabolomic Data Integration in Precision Medicine. OMICS 2018, 22, 630–636. [Google Scholar] [CrossRef]
- Lewis, J.E.; Kemp, M.L. Integration of Machine Learning and Genome-Scale Metabolic Modeling Identifies Multi-Omics Biomarkers for Radiation Resistance. Nat. Commun. 2021, 12, 2700. [Google Scholar] [CrossRef] [PubMed]
- Okoro, P.C.; Schubert, R.; Guo, X.; Johnson, W.C.; Rotter, J.I.; Hoeschele, I.; Liu, Y.; Im, H.K.; Luke, A.; Dugas, L.R.; et al. Transcriptome Prediction Performance across Machine Learning Models and Diverse Ancestries. HGG Adv. 2021, 2, 100019. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Dai, L.; Chen, N.; Li, J.; Gao, Y.; Zhao, J.; Ding, L.; Xie, C.; Yi, X.; Deng, H.; et al. Integrative Analysis Provides Multi-Omics Evidence for the Pathogenesis of Placenta Percreta. J. Cell. Mol. Med. 2020, 24, 13837–13852. [Google Scholar] [CrossRef] [PubMed]
- Benny, P.A.; Alakwaa, F.M.; Schlueter, R.J.; Lassiter, C.B.; Garmire, L.X. A Review of Omics Approaches to Study Preeclampsia. Placenta 2020, 92, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Yang, K.; Ge, J. Uncovering the Pharmacological Mechanism of Astragalus Salvia Compound on Pregnancy-Induced Hypertension Syndrome by a Network Pharmacology Approach. Sci. Rep. 2017, 7, 16849. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez, M.G.; Wells, D.; Malter, H.; Munné, S.; Cohen, J.; Steuerwald, N.M. Expression Profiles of Individual Human Oocytes Using Microarray Technology. Reprod. Biomed. Online 2004, 8, 325–337. [Google Scholar] [CrossRef]
- Assou, S.; Anahory, T.; Pantesco, V.; Le Carrour, T.; Pellestor, F.; Klein, B.; Reyftmann, L.; Dechaud, H.; De Vos, J.; Hamamah, S. The Human Cumulus--Oocyte Complex Gene-Expression Profile. Hum. Reprod. 2006, 21, 1705–1719. [Google Scholar] [CrossRef]
- Kocabas, A.M.; Crosby, J.; Ross, P.J.; Otu, H.H.; Beyhan, Z.; Can, H.; Tam, W.-L.; Rosa, G.J.M.; Halgren, R.G.; Lim, B.; et al. The Transcriptome of Human Oocytes. Proc. Natl. Acad. Sci. USA 2006, 103, 14027–14032. [Google Scholar] [CrossRef]
- Zhang, P.; Kerkelä, E.; Skottman, H.; Levkov, L.; Kivinen, K.; Lahesmaa, R.; Hovatta, O.; Kere, J. Distinct Sets of Developmentally Regulated Genes That Are Expressed by Human Oocytes and Human Embryonic Stem Cells. Fertil. Steril. 2007, 87, 677–690. [Google Scholar] [CrossRef]
- Wood, J.R.; Dumesic, D.A.; Abbott, D.H.; Strauss, J.F. 3rd Molecular Abnormalities in Oocytes from Women with Polycystic Ovary Syndrome Revealed by Microarray Analysis. J. Clin. Endocrinol. Metab. 2007, 92, 705–713. [Google Scholar] [CrossRef]
- Gasca, S.; Pellestor, F.; Assou, S.; Loup, V.; Anahory, T.; Dechaud, H.; De Vos, J.; Hamamah, S. Identifying New Human Oocyte Marker Genes: A Microarray Approach. Reprod. Biomed. Online 2007, 14, 175–183. [Google Scholar] [CrossRef]
- Gasca, S.; Reyftmann, L.; Pellestor, F.; Rème, T.; Assou, S.; Anahory, T.; Dechaud, H.; Klein, B.; De Vos, J.; Hamamah, S. Total Fertilization Failure and Molecular Abnormalities in Metaphase II Oocytes. Reprod. Biomed. Online 2008, 17, 772–781. [Google Scholar] [CrossRef]
- Jones, G.M.; Cram, D.S.; Song, B.; Magli, M.C.; Gianaroli, L.; Lacham-Kaplan, O.; Findlay, J.K.; Jenkin, G.; Trounson, A.O. Gene Expression Profiling of Human Oocytes Following in Vivo or in Vitro Maturation. Hum. Reprod. 2008, 23, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.; Patrizio, P. Gene Expression Profiling of Human Oocytes at Different Maturational Stages and after in Vitro Maturation. Am. J. Obstet. Gynecol. 2008, 198, e1–e9. [Google Scholar] [CrossRef] [PubMed]
- Grøndahl, M.L.; Yding Andersen, C.; Bogstad, J.; Nielsen, F.C.; Meinertz, H.; Borup, R. Gene Expression Profiles of Single Human Mature Oocytes in Relation to Age. Hum. Reprod. 2010, 25, 957–968. [Google Scholar] [CrossRef]
- Dobson, A.T.; Raja, R.; Abeyta, M.J.; Taylor, T.; Shen, S.; Haqq, C.; Pera, R.A.R. The Unique Transcriptome through Day 3 of Human Preimplantation Development. Hum. Mol. Genet. 2004, 13, 1461–1470. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.-L.; Liu, Y.-H.; Tseng, C.-N.; Singh, S. Analysis of Gene Expression in Single Human Oocytes and Preimplantation Embryos. Biochem. Biophys. Res. Commun. 2006, 340, 48–53. [Google Scholar] [CrossRef]
- Jaroudi, S.; Kakourou, G.; Cawood, S.; Doshi, A.; Ranieri, D.M.; Serhal, P.; Harper, J.C.; SenGupta, S.B. Expression Profiling of DNA Repair Genes in Human Oocytes and Blastocysts Using Microarrays. Hum. Reprod. 2009, 24, 2649–2655. [Google Scholar] [CrossRef]
- Zhang, P.; Zucchelli, M.; Bruce, S.; Hambiliki, F.; Stavreus-Evers, A.; Levkov, L.; Skottman, H.; Kerkelä, E.; Kere, J.; Hovatta, O. Transcriptome Profiling of Human Pre-Implantation Development. PLoS ONE 2009, 4, e7844. [Google Scholar] [CrossRef]
- Smith, H.L.; Stevens, A.; Minogue, B.; Sneddon, S.; Shaw, L.; Wood, L.; Adeniyi, T.; Xiao, H.; Lio, P.; Kimber, S.J.; et al. Systems Based Analysis of Human Embryos and Gene Networks Involved in Cell Lineage Allocation. BMC Genom. 2019, 20, 171. [Google Scholar] [CrossRef]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-Cell RNA-Seq Profiling of Human Preimplantation Embryos and Embryonic Stem Cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Blakeley, P.; Fogarty, N.; Del Valle, I.; Wamaitha, S.; Hu, T.X.; Elder, K.; Snell, P.; Christie, L.; Robson, P.; Niakan, K. Defining the Three Cell Lineages of the Human Blastocyst by Single-Cell RNA-Seq. Mech. Dev. 2017, 145, S26. [Google Scholar] [CrossRef]
- Petropoulos, S.; Edsgärd, D.; Reinius, B.; Deng, Q.; Panula, S.P.; Codeluppi, S.; Plaza Reyes, A.; Linnarsson, S.; Sandberg, R.; Lanner, F. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016, 165, 1012–1026. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Wang, R.; Yuan, P.; Ren, Y.; Mao, Y.; Li, R.; Lian, Y.; Li, J.; Wen, L.; Yan, L.; et al. Reconstituting the Transcriptome and DNA Methylome Landscapes of Human Implantation. Nature 2019, 572, 660–664. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).