Cytosolic and Nucleosolic Calcium-Regulated Molecular Networks in Response to Long-Term Treatment with Abscisic Acid and Methyl Jasmonate in Arabidopsis thaliana
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Imaging of [Ca2+]cyt and [Ca2+]nuc in A. thaliana Roots
2.3. Stomatal Aperture Bioassay
2.4. Total RNA Isolation and Analysis of Microarray Data
2.5. Construction of the A. thaliana Gene Co-Expression Network
2.6. Firefly Luciferase Complementation Imaging (LCI) Assay
2.7. qRT-PCR
2.8. Statistical Analysis
3. Results
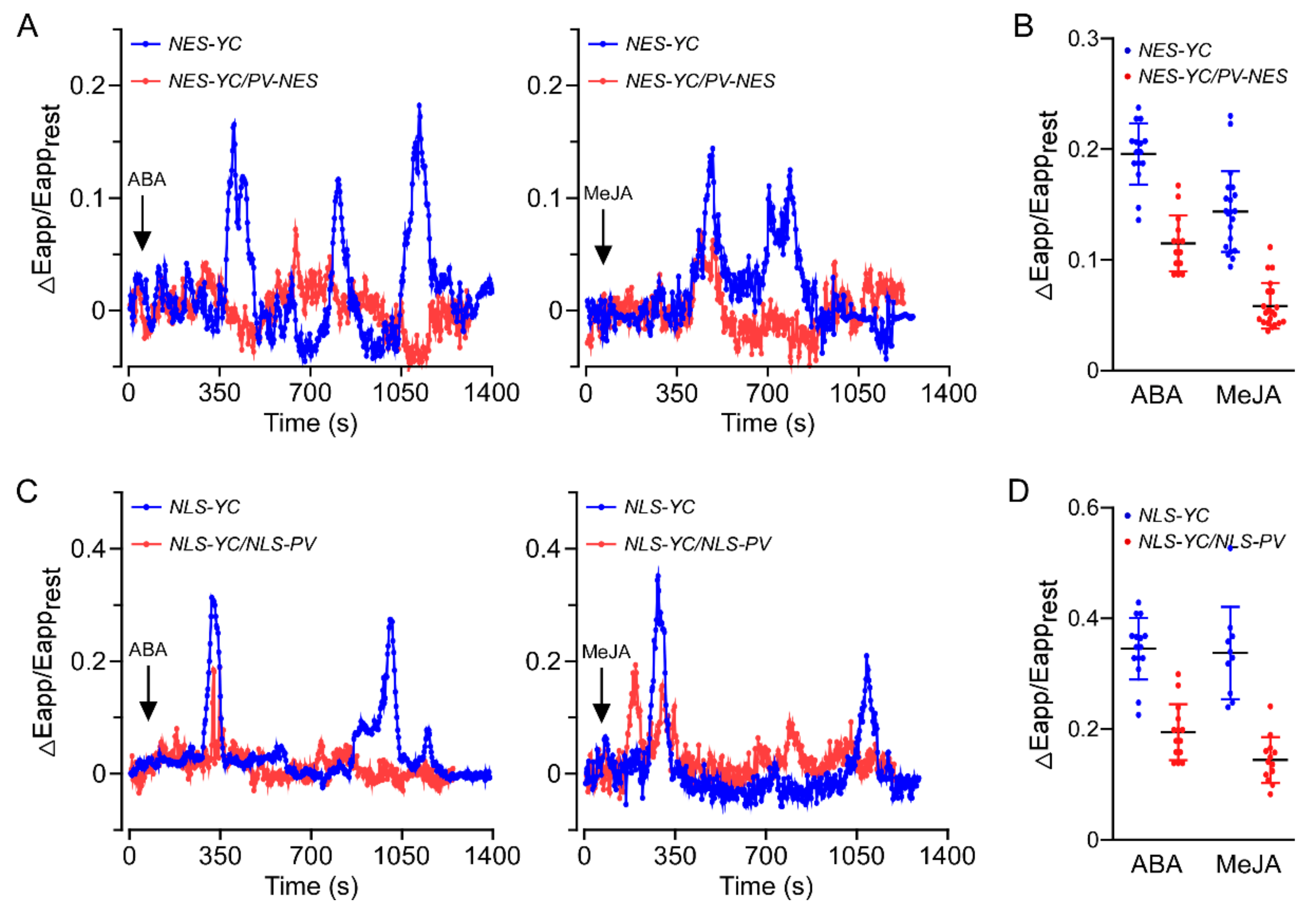
3.1. Both ABA and MeJA Triggered [Ca2+]cyt and [Ca2+]nuc Increases in A. thaliana Roots
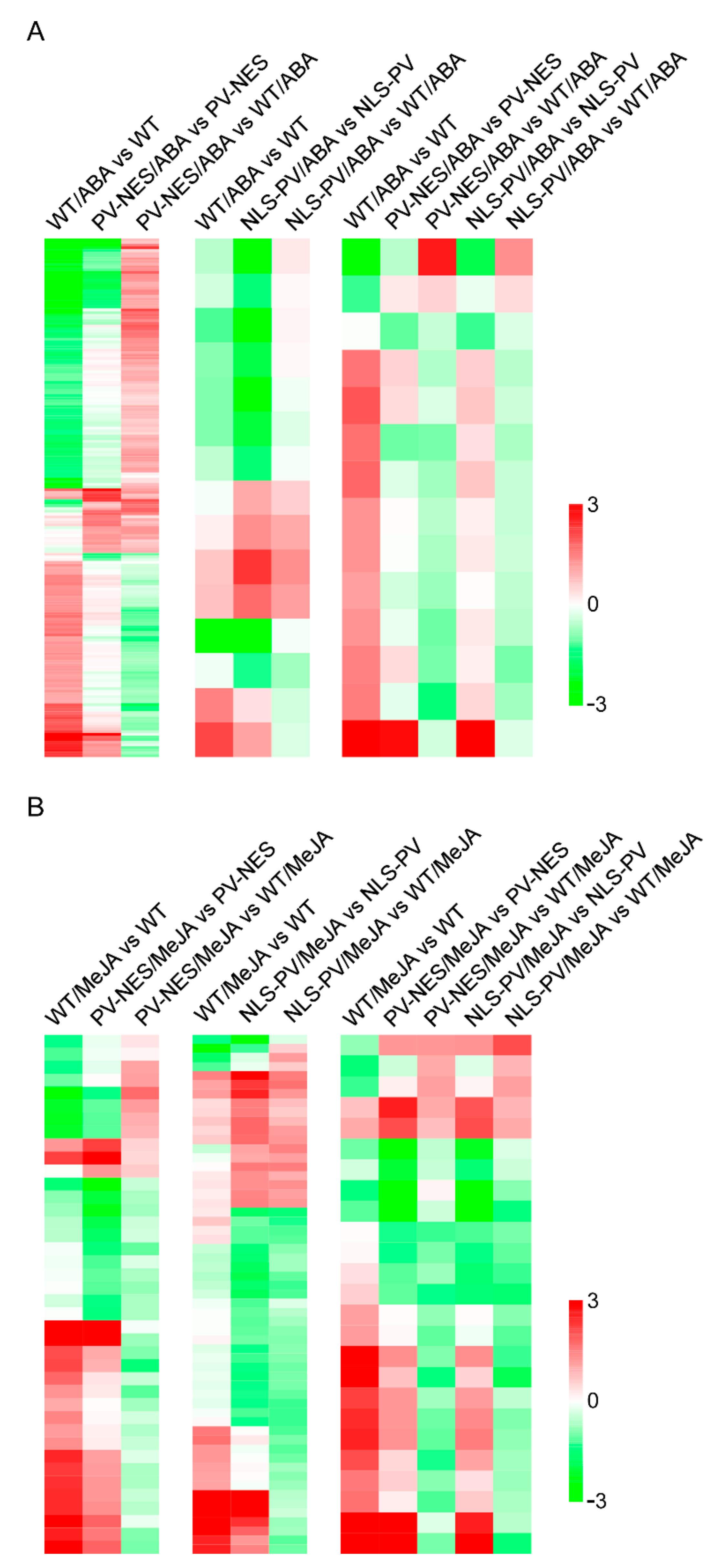
3.2. Elevated [Ca2+]cyt and [Ca2+]nuc Contributed to Significant Changes in ABA- or MeJA-Responsive Gene Expression Profiles in A. thaliana
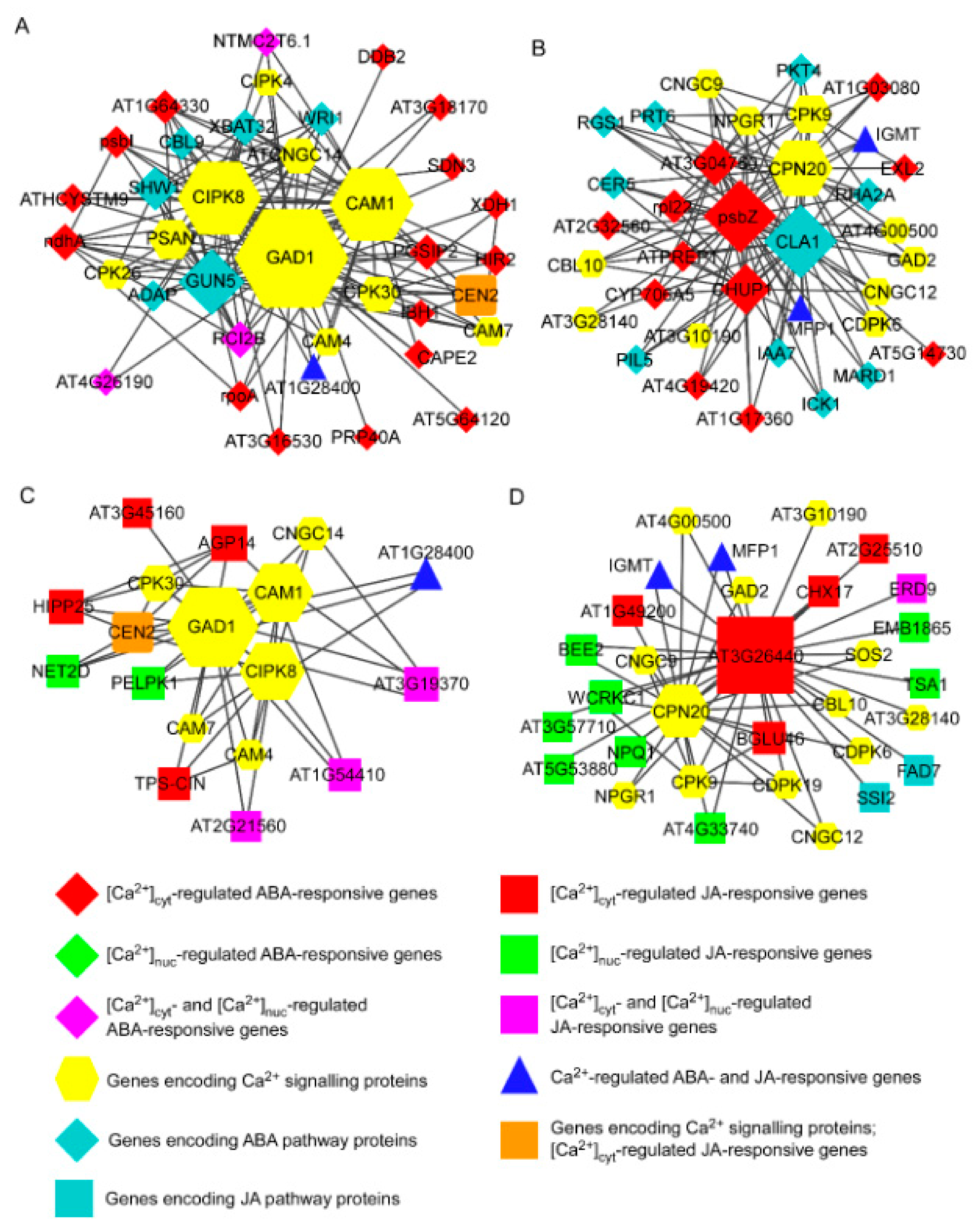
3.3. Integrative Co-Expression Analysis to Identify Hub Genes within Calcium-Regulated Transcriptional Modules
3.4. Interactions among the Hub Proteins CAM1, CIPK8, and GAD1 and with Proteins Encoded by Ca2+-Regulated Hormone-Responsive Genes
3.5. CAM1 and CIPK8 Are Required for MeJA-Induced Stomatal Closure and Are Associated with ABA-Inhibited Seed Germination
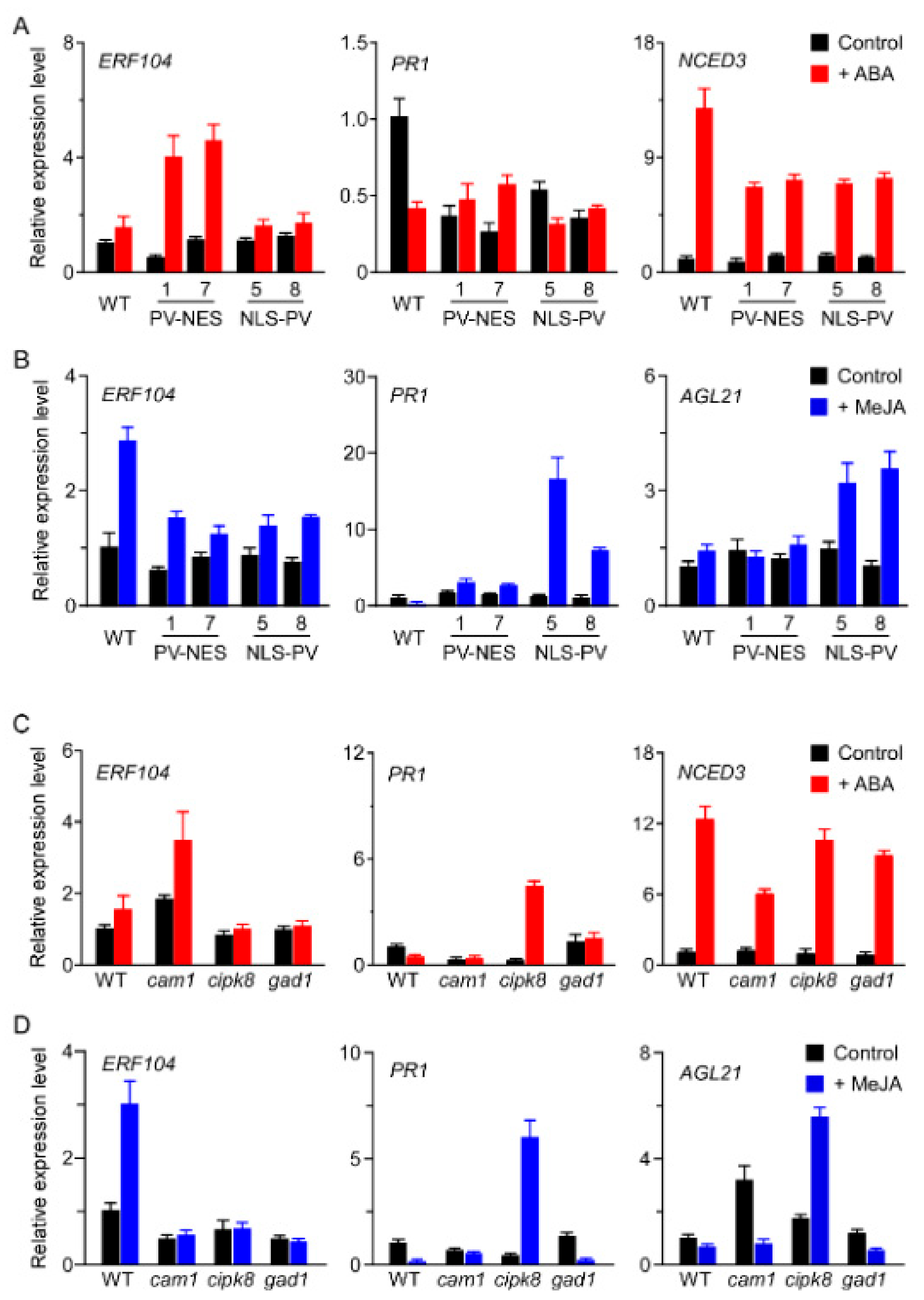
3.6. CAM1, CIPK8, and GAD1 Regulate the Transcription of [Ca2+]-Regulated Genes in Response to ABA or MeJA Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, s401–s417. [Google Scholar] [CrossRef] [PubMed]
- McAinsh, M.R.; Pittman, J.K. Shaping the calcium signature. New Phytol. 2009, 181, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.M.; Gucel, S. Jasmonates: Multifunctional roles in stress tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Allen, G.J.; Kwak, J.M.; Chu, S.P.; Llopis, J.; Tsien, R.Y.; Harper, J.F.; Schroeder, J.I. Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J. 1999, 19, 735–747. [Google Scholar] [CrossRef]
- Bai, L.; Zhang, G.; Zhou, Y.; Zhang, Z.; Wang, W.; Du, Y.; Wu, Z.; Song, C.P. Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 2009, 60, 314–327. [Google Scholar] [CrossRef]
- Islam, M.M.; Hossain, M.A.; Jannat, R.; Munemasa, S.; Nakamura, Y.; Mori, I.C.; Murata, Y. Cytosolic alkalization and cytosolic calcium oscillation in Arabidopsis guard cells response to ABA and MeJA. Plant Cell Physiol. 2010, 51, 1721–1730. [Google Scholar] [CrossRef]
- Walter, A.; Mazars, C.; Maitrejean, M.; Hopke, J.; Ranjeva, R.; Boland, W.; Mithöfer, A. Structural requirements of jasmonates and synthetic analogues as inducers of Ca2+ signals in the nucleus and the cytosol of plant cells. Angew. Chem. Int. Ed. 2007, 46, 4783–4785. [Google Scholar] [CrossRef]
- Krebs, M.; Held, K.; Binder, A.; Hashimoto, K.; Herder, G.D.; Parniske, M.; Kudla, J.; Schumacher, K. FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J. 2012, 69, 181–192. [Google Scholar] [CrossRef]
- Huang, F.; Luo, J.; Ning, T.; Cao, W.; Jin, X.; Zhao, H.; Wang, Y.; Han, S. Cytosolic and nucleosolic calcium signaling in response to osmotic and salt stresses are independent of each other in roots of Arabidopsis seedlings. Front. Plant Sci. 2017, 8, 1648. [Google Scholar] [CrossRef]
- Ranty, B.; Aldon, D.; Galaud, J.-P. Plant calmodulins and calmodulin-related proteins. Plant Signal. Behav. 2006, 1, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Gleason, C.; Chaudhuri, S.; Yang, T.; Muñoz, A.; Poovaiah, B.W.; Oldroyd, G.E.D. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 2006, 441, 1149–1152. [Google Scholar] [CrossRef]
- Tirichine, L.; Imaizumi-Anraku, H.; Yoshida, S.; Murakami, Y.; Madsen, L.H.; Miwa, H.; Nakagawa, T.; Sandal, N.; Albrektsen, A.S.; Kawaguchi, M.; et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 2006, 441, 1153–1156. [Google Scholar] [CrossRef] [PubMed]
- Batistic, O.; Kudla, J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 2009, 1793, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Day, I.S.; Reddy, V.S.; Shad, A.G.; Reddy, A.S. Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 2002, 3, 51–56. [Google Scholar] [CrossRef]
- Harper, J.F.; Harmon, A. Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell Biol. 2005, 6, 555–566. [Google Scholar] [CrossRef]
- McCormack, E.; Braam, J. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 2003, 159, 585–598. [Google Scholar] [CrossRef]
- Kolukisaoglu, Ü.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice cbl-cipk signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef]
- Batistic, O.; Kudla, J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 2012, 1820, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, R.; Singh, A.; Chattopadhyay, S. Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 2008, 20, 1747–1759. [Google Scholar] [CrossRef] [PubMed]
- Abbas, N.; Maurya, J.P.; Senapati, D.; Gangappa, S.N.; Chattopadhyay, S. Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 2014, 26, 1036–1052. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, H.-Y. Functional analysis of a calcium-binding transcription factor involved in plant salt stress signaling. FEBS Lett. 2006, 580, 5251–5256. [Google Scholar] [CrossRef]
- Gilroy, S.; Hughes, W.A.; Trewavas, A.J. Calmodulin antagonists increase free cytosolic calcium levels in plant protoplasts in vivo. FEBS Lett. 1987, 212, 133–137. [Google Scholar] [CrossRef]
- Kaplan, B.; Davydov, O.; Knight, H.; Galon, Y.; Knight, M.R.; Fluhr, R.; Fromm, H. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 2006, 18, 2733–2748. [Google Scholar] [CrossRef]
- Whalley, H.J.; Sargeant, A.W.; Steele, J.F.; Lacoere, T.; Lamb, R.; Saunders, N.J.; Knight, H.; Knight, M.R. Transcriptomic analysis reveals calcium regulation of specific promoter motifs in Arabidopsis. Plant Cell 2011, 23, 4079–4095. [Google Scholar] [CrossRef]
- Han, S.C.; Tang, R.H.; Anderson, L.K.; Woerner, T.E.; Pei, Z.M. A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature 2003, 425, 196–200. [Google Scholar] [CrossRef]
- Hu, P.; Zhou, W.; Cheng, Z.; Fan, M.; Wang, L.; Xie, D. JAV1 controls jasmonate-regulated plant defense. Molecular Cell 2013, 50, 504–515. [Google Scholar] [CrossRef]
- Goda, H.; Sasaki, E.; Akiyama, K.; Maruyama-Nakashita, A.; Nakabayashi, K.; Li, W.; Ogawa, M.; Yamauchi, Y.; Preston, J.; Aoki, K.; et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J 2008, 55, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Yun, J.; Likhacheva, A.V.; Alonso, J.M. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant cell 2007, 19, 2169–2185. [Google Scholar] [CrossRef] [PubMed]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 2007, 50, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A.; Horvath, S.; Geschwind, D.H. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 12698–12703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005, 4, 1–45. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef]
- Munemasa, S.; Hossain, M.A.; Nakamura, Y.; Mori, I.C.; Murata, Y. The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol. 2011, 155, 553–561. [Google Scholar] [CrossRef]
- Oliveira, A.G.; Guimarães, E.S.; Andrade, L.M.; Menezes, G.B.; Leite, F.M. Decoding calcium signaling across the nucleus. Physiology 2014, 29, 361–368. [Google Scholar] [CrossRef][Green Version]
- Hardingham, G.E.; Chawla, S.; Johnson, C.M.; Bading, H. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature 1997, 385, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; Andrade, V.A.; Andrade, S.J.; Pusl, T.; Ortega, J.M.; Goes, A.M.; Leite, M.F. Inhibition of the TEF/TEAD transcription factor activity by nuclear calcium and distinct kinase pathways. Biochem. Biophy. Res. Comm. 2003, 301, 267–274. [Google Scholar] [CrossRef]
- Charpentier, M.; Sun, J.; Martins, T.V.; Radhakrishnan, G.V.; Findlay, K.; Soumpourou, E.; Thouin, J.; Véry, A.-A.; Sanders, D.; Morris, R.J.; et al. Nuclear-localized cyclic nucleotide–gated channels mediate symbiotic calcium oscillations. Science 2016, 352, 1102–1105. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef]
- Salvi, P.; Manna, M.; Kaur, H.; Thakur, T.; Gandass, N.; Bhatt, D.; Muthamilarasan, M. Phytohormone signaling and crosstalk in regulating drought stress response in plants. Plant Cell Rep. 2021, 40, 1305–1329. [Google Scholar] [CrossRef]
- Liang, X.; Ma, M.; Zhou, Z.; Wang, J.; Yang, X.; Rao, S.; Bi, G.; Li, L.; Zhang, X.; Chai, J.; et al. Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res. 2018, 28, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Chi, W.; Sun, X.; Zhang, L. Intracellular signaling from plastid to nucleus. Ann. Rev. Plant Biol. 2013, 64, 559–582. [Google Scholar] [CrossRef]
- Bräutigam, K.; Dietzel, L.; Pfannschmidt, T. Plastid-nucleus communication: Anterograde and retrograde signalling in the development and function of plastids. In Cell and Molecular Biology of Plastids; Bock, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 409–455. [Google Scholar]
- Wang, M.; Lee, J.; Choi, B.; Park, Y.; Sim, H.J.; Kim, H.; Hwang, I. Physiological and molecular processes associated with long duration of ABA treatment. Front. Plant Sci. 2018, 9, 176. [Google Scholar] [CrossRef]
- Kmiecik, P.; Leonardelli, M.; Teige, M. Novel connections in plant organellar signalling link different stress responses and signalling pathways. J. Exp. Bot. 2016, 67, 3793–3807. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef]
- Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of endogenous abscisic acid in methyl jasmonate-induced stomatal closure in Arabidopsis. Plant Physiol. 2011, 156, 430–438. [Google Scholar] [CrossRef] [PubMed]


| nLUC-GAD1 | nLUC-CAM1 | nLUC-CIPK8 | ||
|---|---|---|---|---|
| [Ca2+]cyt-regulated ABA-responsive genes | cLUC-NDHA | − | − | − |
| cLUC-PSBI | − | + | + | |
| cLUC-PGSIP2 | − | + | − | |
| cLUC-CAPE2 | − | − | − | |
| cLUC-AT1G64330 | + | + | − | |
| cLUC-HIR2 | − | − | − | |
| [Ca2+]cyt-regulated JA-responsive genes | cLUC-AT3G45160 | + | + | − |
| [Ca2+]cyt- and [Ca2+]nuc-regulated ABA-responsive genes | cLUC-RCI2B | + | + | − |
| cLUC-NTMC2T6.1 | − | + | − | |
| [Ca2+]cyt- and [Ca2+]nuc-regulated JA-responsive genes | cLUC-AT1G54410 | − | − | − |
| cLUC-AT3G19370 | + | + | − | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Huang, F.; Yan, P.; Nie, Y.; Chen, L.; Luo, J.; Zhao, H.; Wang, Y.; Han, S. Cytosolic and Nucleosolic Calcium-Regulated Molecular Networks in Response to Long-Term Treatment with Abscisic Acid and Methyl Jasmonate in Arabidopsis thaliana. Genes 2022, 13, 524. https://doi.org/10.3390/genes13030524
Wang D, Huang F, Yan P, Nie Y, Chen L, Luo J, Zhao H, Wang Y, Han S. Cytosolic and Nucleosolic Calcium-Regulated Molecular Networks in Response to Long-Term Treatment with Abscisic Acid and Methyl Jasmonate in Arabidopsis thaliana. Genes. 2022; 13(3):524. https://doi.org/10.3390/genes13030524
Chicago/Turabian StyleWang, Doudou, Feifei Huang, Pengcheng Yan, Yanli Nie, Lvli Chen, Jin Luo, Heping Zhao, Yingdian Wang, and Shengcheng Han. 2022. "Cytosolic and Nucleosolic Calcium-Regulated Molecular Networks in Response to Long-Term Treatment with Abscisic Acid and Methyl Jasmonate in Arabidopsis thaliana" Genes 13, no. 3: 524. https://doi.org/10.3390/genes13030524
APA StyleWang, D., Huang, F., Yan, P., Nie, Y., Chen, L., Luo, J., Zhao, H., Wang, Y., & Han, S. (2022). Cytosolic and Nucleosolic Calcium-Regulated Molecular Networks in Response to Long-Term Treatment with Abscisic Acid and Methyl Jasmonate in Arabidopsis thaliana. Genes, 13(3), 524. https://doi.org/10.3390/genes13030524






