miR-185-5p Regulates Inflammation and Phagocytosis through CDC42/JNK Pathway in Macrophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. miRNA Microarrays and Screening of DEMs
2.2. Cell Culture and Treatment
2.3. RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.4. Cell Transfection
2.5. Prediction for Target Genes of mmu-miR-185-5p
2.6. Dual Luciferase Reporter Assays
2.7. ELISA Assays
2.8. Phagocytosis Assays
2.9. Western Blot
2.10. Statistical Analyses
3. Results
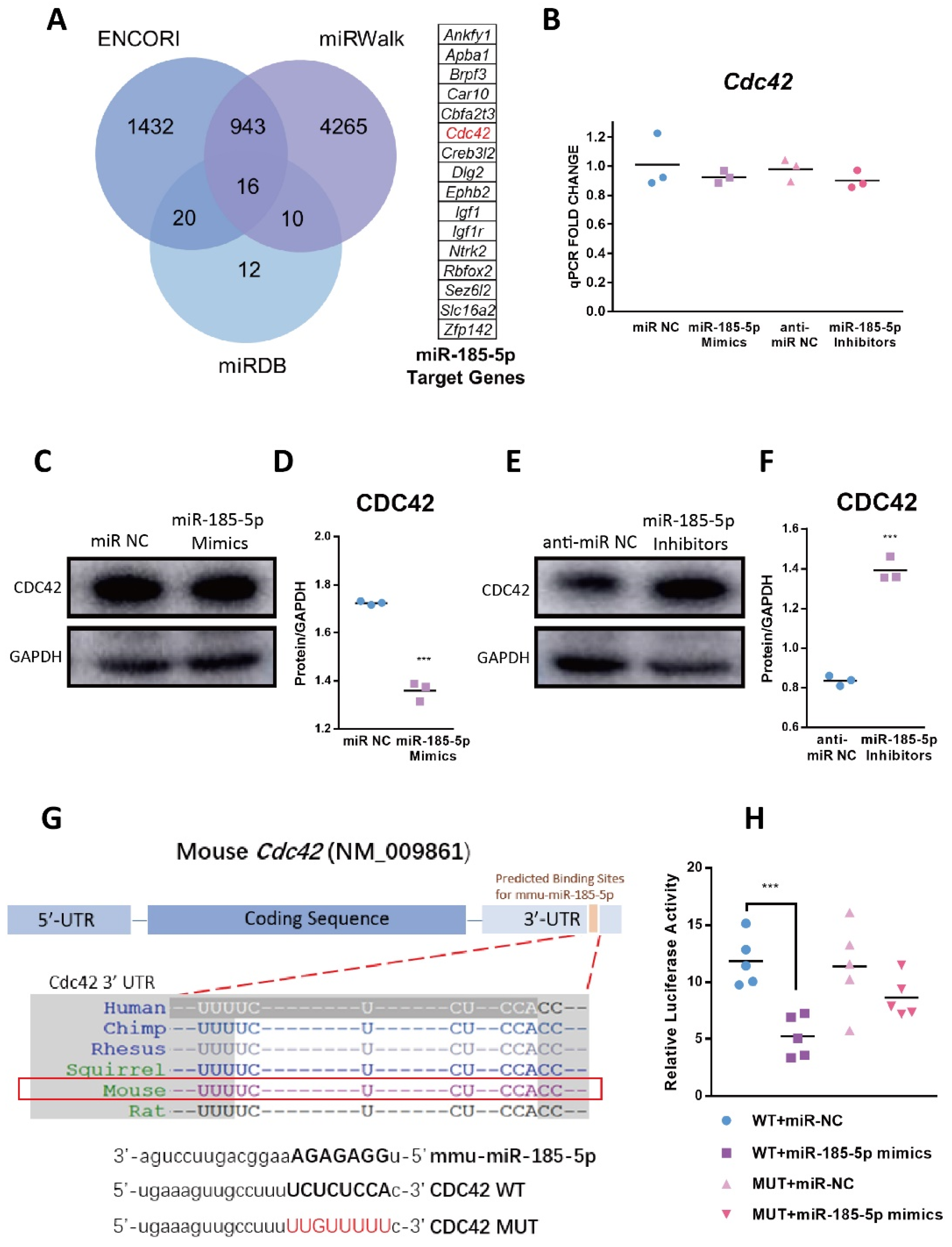
3.1. Identification of the Candidate DEM
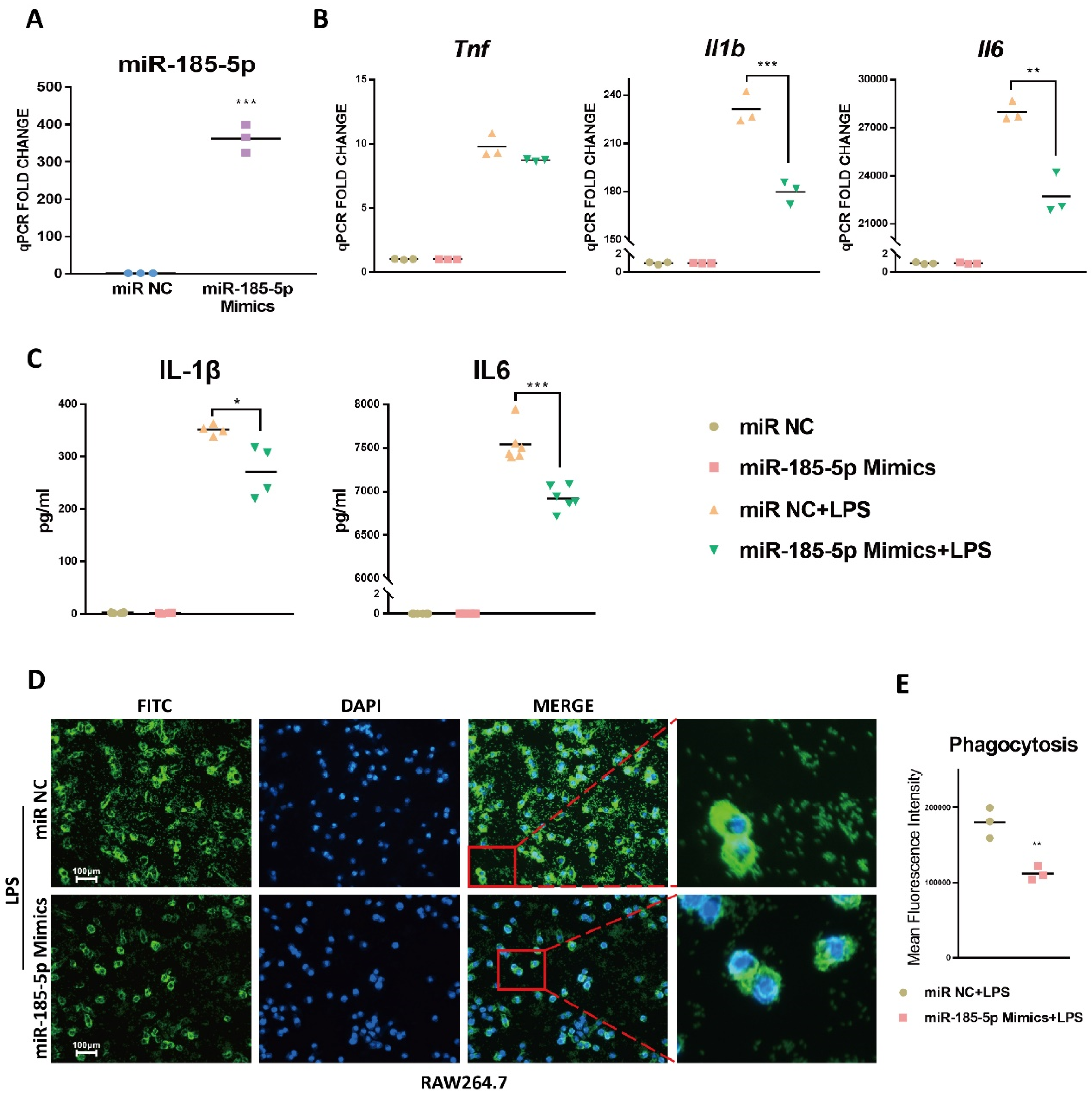
3.2. miR-185-5p Overexpression Inhibits LPS-Induced Inflammation and Phagocytosis
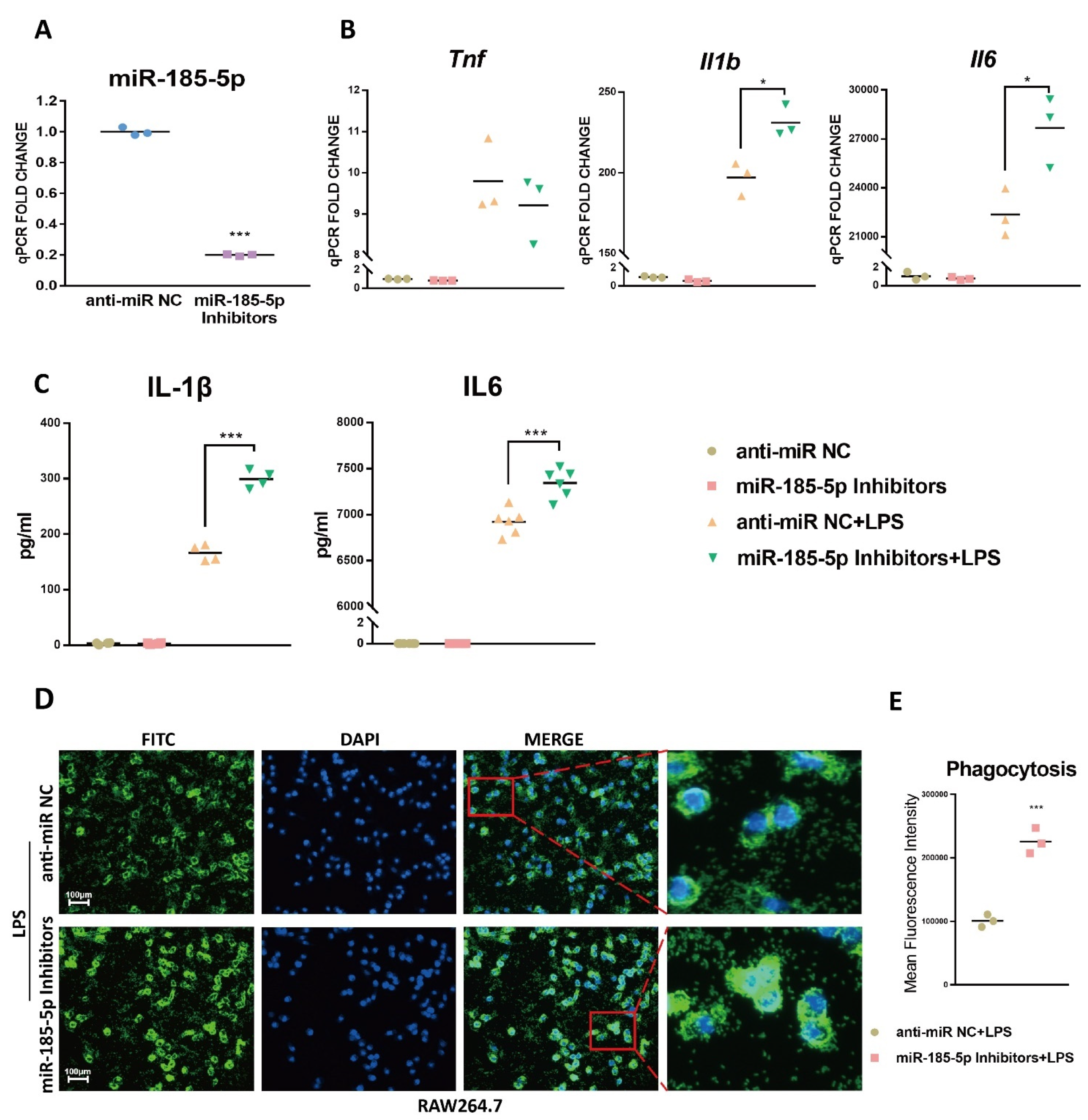
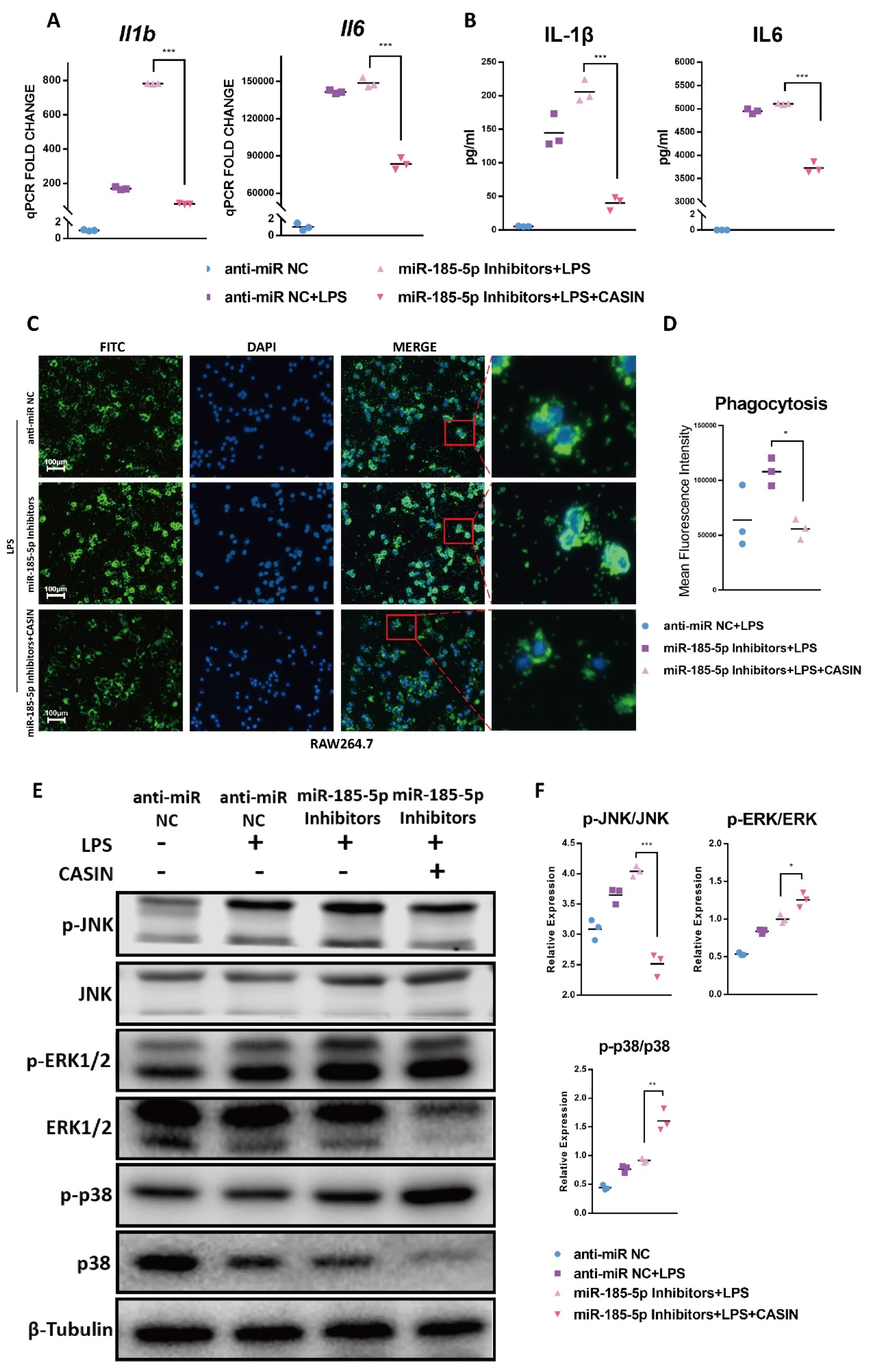
3.3. miR-185-5p Suppression Promotes LPS-Induced Inflammation and Phagocytosis
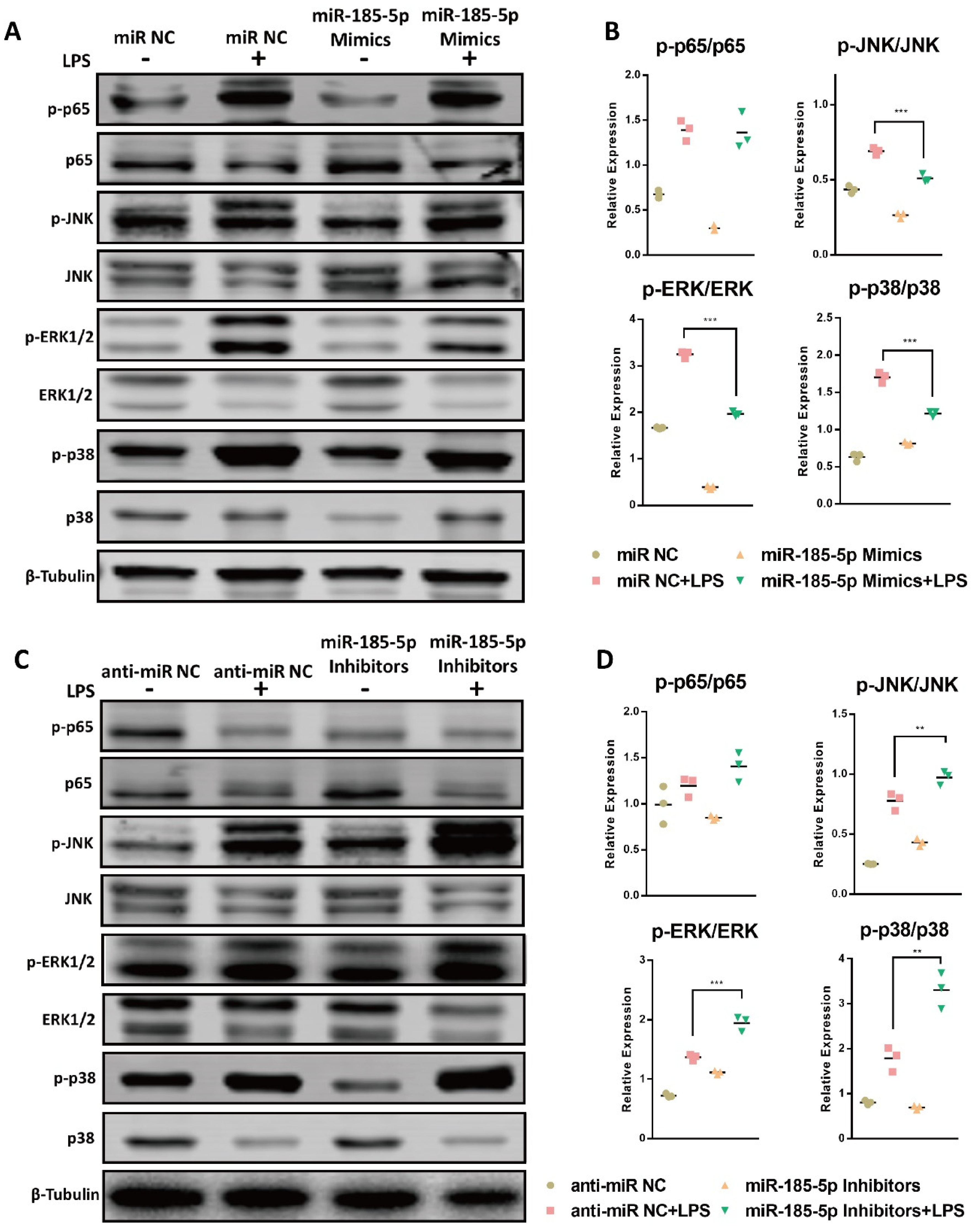
3.4. miR-185-5p Regulates the Activation of MAPK Pathways
3.5. CDC42 Is a Direct Target of miR-185-5p
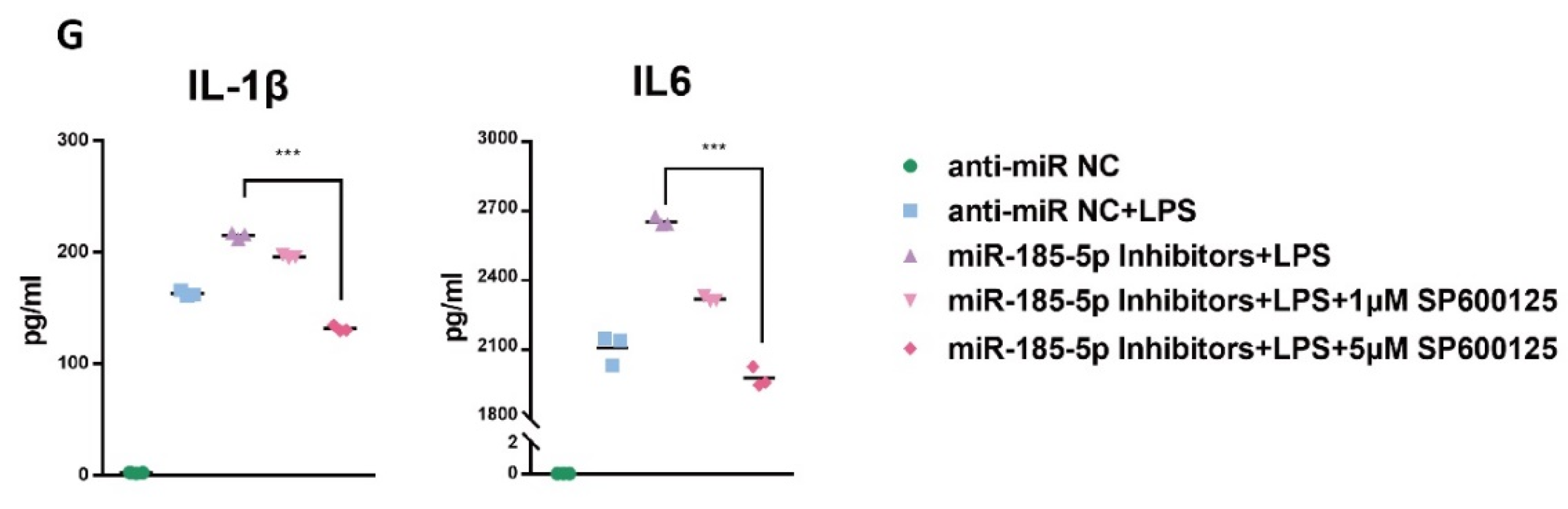
3.6. CASIN Modulates miR-185-5p-Mediated Pro-Inflammatory Phenotype Partially through Targeting MAPK JNK Pathways
4. Discussion
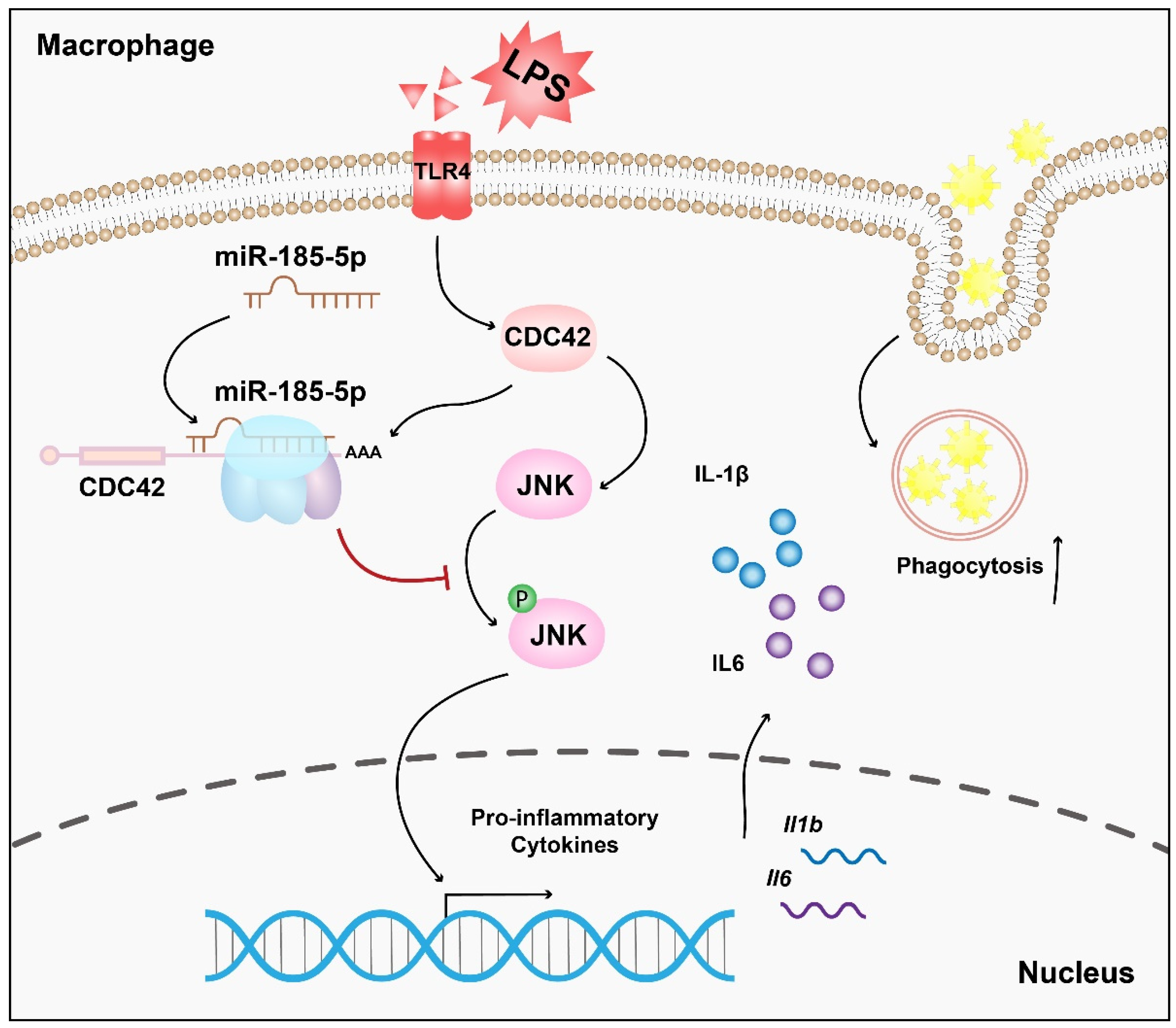
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BMDM | Bone Marrow–derived Macrophages |
| CASIN | CDC42 Activity-Specific Inhibitor |
| CCL2 | C-C Motif Chemokine Ligand 2 |
| CDC42 | Cell Division Cycle 42 |
| CRP | C-reactive Protein |
| DEM | Differentially Expressed miRNA |
| DMSO | Dimethyl Sulfoxide |
| ERK | Extracellular-signal-regulated Kinase |
| FC | Fold Change |
| HUVEC | Human Umbilical Vein Endothelial Cell |
| IL-1β | Interleukin-1β |
| IL-6 | Interleukin-6 |
| INF-γ | Interferon-γ |
| JNK | Jun amino-terminal Kinase |
| KO | Knockout |
| LPS | Lipopolysaccharide |
| MAPK | Mitogen-activated Protein Kinase |
| miRNAs | MicroRNAs |
| MUT | Mutated |
| NAFLD | Non-alcoholic Fatty Liver Disease |
| NC | Normal Control |
| p38/SAPK | Stress-activated Protein Kinase |
| PI3K | Phosphatidylin-ositol-3-kinase |
| SD | Standard Deviation |
| TLR4 | Toll-like Receptor 4 |
| TNF | Tumor Necrosis Factor |
| WT | Wild Type |
References
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Hansson, G.K.; Hermansson, A. The immune system in atherosclerosis. Nat. Immunol. 2011, 12, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, L.; de Winther, M.P. Macrophage subsets in atherosclerosis as defined by single-cell technologies. J. Pathol. 2020, 250, 705–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, K.J.; Tabas, I. Macrophages in the pathogenesis of atherosclerosis. Cell 2011, 145, 341–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabas, I.; Bornfeldt, K.E. Macrophage phenotype and function in different stages of atherosclerosis. Circ. Res. 2016, 118, 653–667. [Google Scholar] [CrossRef] [Green Version]
- Shirai, T.; Nazarewicz, R.R.; Wallis, B.B.; Yanes, R.E.; Watanabe, R.; Hilhorst, M.; Tian, L.; Harrison, D.G.; Giacomini, J.C.; Assimes, T.L.; et al. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J. Exp. Med. 2016, 213, 337–354. [Google Scholar] [CrossRef] [Green Version]
- Koelwyn, G.J.; Corr, E.M.; Erbay, E.; Moore, K.J. Regulation of macrophage immunometabolism in atherosclerosis. Nat. Immunol. 2018, 19, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Bode, J.G.; Ehlting, C.; Häussinger, D. The macrophage response towards LPS and its control through the p38 (MAPK)-STAT3 axis. Cell. Signal. 2012, 24, 1185–1194. [Google Scholar] [CrossRef]
- Morrison, D.K. MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.M.; Wang, J.; Zhang, B.; Fang, L.; Xu, K.; Liu, R.Y. CpG-ODN promotes phagocytosis and autophagy through JNK/P38 signal pathway in Staphylococcus aureus-stimulated macrophage. Life Sci. 2016, 161, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wen, C.; Chen, S.; Li, W.; Qin, Q.; He, L.; Wang, F.; Chen, J.; Ye, W.; Li, W.; et al. ROS/JNK/C-jun pathway is involved in chaetocin induced colorectal cancer cells apoptosis and macrophage phagocytosis enhancement. Front. Pharmacol. 2021, 12, 729367. [Google Scholar] [CrossRef] [PubMed]
- Rincón, M.; Davis, R.J. Regulation of the immune response by stress-activated protein kinases. Immunol. Rev. 2009, 228, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Nofer, J.R.; Feuerborn, R.; Levkau, B.; Sokoll, A.; Seedorf, U.; Assmann, G. Involvement of Cdc42 signaling in apoA-I-induced cholesterol efflux. J. Biol. Chem. 2003, 278, 53055–53062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heasman, S.J.; Ridley, A.J. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008, 9, 690–701. [Google Scholar] [CrossRef]
- Zhang, X.; Bandyopadhyay, S.; Araujo, L.P.; Tong, K.; Flores, J.; Laubitz, D.; Zhao, Y.; Yap, G.; Wang, J.; Zou, Q.; et al. Elevating EGFR-MAPK program by a nonconventional Cdc42 enhances intestinal epithelial survival and regeneration. JCI Insight 2020, 5, e135923. [Google Scholar] [CrossRef]
- Johnson, G.L.; Nakamura, K. The c-jun kinase/stress-activated pathway: Regulation, function and role in human disease. Biochim. Biophys. Acta 2007, 1773, 1341–1348. [Google Scholar] [CrossRef] [Green Version]
- Komaravolu, R.K.; Adam, C.; Moonen, J.R.; Harmsen, M.C.; Goebeler, M.; Schmidt, M. Erk5 inhibits endothelial migration via KLF2-dependent down-regulation of PAK1. Cardiovasc. Res. 2015, 105, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.S.; Wang, H.Y.; Liao, Y.C.; Tsai, P.C.; Chen, K.C.; Cheng, H.Y.; Lin, R.T.; Juo, S.H. MicroRNA-195 regulates vascular smooth muscle cell phenotype and prevents neointimal formation. Cardiovasc. Res. 2012, 95, 517–526. [Google Scholar] [CrossRef] [Green Version]
- Florian, M.C.; Leins, H.; Gobs, M.; Han, Y.; Marka, G.; Soller, K.; Vollmer, A.; Sakk, V.; Nattamai, K.J.; Rayes, A.; et al. Inhibition of Cdc42 activity extends lifespan and decreases circulating inflammatory cytokines in aged female C57BL/6 mice. Aging Cell 2020, 19, e13208. [Google Scholar] [CrossRef]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 2019, 20, 1836–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Jia, X.J.; Jiang, H.J.; Du, Y.; Yang, F.; Si, S.Y.; Hong, B. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol. Cell. Biol. 2013, 33, 1956–1964. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.C.; Zhan, X.R.; Li, X.Y.; Yu, J.J.; Liu, X.M. MicroRNA-185 regulates expression of lipid metabolism genes and improves insulin sensitivity in mice with non-alcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 17914–17923. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Fu, X.; Si, M.; Wang, Y.; Ma, R.; Ren, X.; Lv, H. MicroRNA-185 targets SOCS3 to inhibit β-cell dysfunction in diabetes. PLoS ONE 2015, 10, e0116067. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, J.; Du, Y.; Jia, X.; Yang, F.; Si, S.; Wang, L.; Hong, B. MicroRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis 2015, 243, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Corbalán-Campos, J.; Gurung, R.; Natarelli, L.; Zhu, M.; Exner, N.; Erhard, F.; Greulich, F.; Geißler, C.; Uhlenhaut, N.H.; et al. Dicer in macrophages prevents atherosclerosis by promoting mitochondrial oxidative metabolism. Circulation 2018, 138, 2007–2020. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Guo, Q.; Zou, J.; Wang, B.; Zhang, Z.; Wei, R.; Zhao, L.; Zhang, Y.; Chu, C.; Fu, X.; et al. MiR-19a-3p suppresses M1 macrophage polarization by inhibiting STAT1/IRF1 pathway. Front. Pharmacol. 2021, 12, 614044. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sahlin, S.; Hed, J.; Rundquist, I. Differentiation between attached and ingested immune complexes by a fluorescence quenching cytofluorometric assay. J. Immunol. Methods 1983, 60, 115–124. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef]
- Das, S.; Reddy, M.A.; Senapati, P.; Stapleton, K.; Lanting, L.; Wang, M.; Amaram, V.; Ganguly, R.; Zhang, L.; Devaraj, S.; et al. Diabetes mellitus-induced long noncoding RNA Dnm3os regulates macrophage functions and inflammation via nuclear mechanisms. Arter. Thromb. Vasc. Biol. 2018, 38, 1806–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, P.; Chakrabarti, J.; Li, Y.; Kalim, K.W.; Zhang, M.; Zhang, L.; Zheng, Y.; Guo, F. Rational targeting of Cdc42 overcomes drug resistance of multiple myeloma. Front. Oncol. 2019, 9, 958. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Schober, A. MicroRNA regulation of macrophages in human pathologies. Cell. Mol. Life Sci. 2016, 73, 3473–3495. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Rothe, K.; Chen, M.; Wu, A.; Babaian, A.; Yen, R.; Zeng, J.; Ruschmann, J.; Petriv, O.I.; O’Neill, K.; et al. The miR-185/PAK6 axis predicts therapy response and regulates survival of drug-resistant leukemic stem cells in CML. Blood 2020, 136, 596–609. [Google Scholar] [CrossRef]
- Libby, P. Interleukin-1 β as a target for atherosclerosis therapy: Biological basis of CANTOS and beyond. J. Am. Coll. Cardiol. 2017, 70, 2278–2289. [Google Scholar] [CrossRef]
- Lutgens, E.; Atzler, D.; Döring, Y.; Duchene, J.; Steffens, S.; Weber, C. Immunotherapy for cardiovascular disease. Eur. Heart J. 2019, 40, 3937–3946. [Google Scholar] [CrossRef]
- Quinn, S.R.; Mangan, N.E.; Caffrey, B.E.; Gantier, M.P.; Williams, B.R.; Hertzog, P.J.; McCoy, C.E.; O’Neill, L.A. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J. Biol. Chem. 2014, 289, 4316–4325. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Ma, L.; Parrini, M.C.; Mao, X.; Lopez, M.; Wu, C.; Marks, P.W.; Davidson, L.; Kwiatkowski, D.J.; Kirchhausen, T.; et al. Cdc42 is required for PIP (2)-induced actin polymerization and early development but not for cell viability. Curr. Biol. 2000, 10, 758–765. [Google Scholar] [CrossRef] [Green Version]
- Deroanne, C.F.; Hamelryckx, D.; Ho, T.T.; Lambert, C.A.; Catroux, P.; Lapière, C.M.; Nusgens, B.V. Cdc42 downregulates MMP-1 expression by inhibiting the ERK1/2 pathway. J. Cell Sci. 2005, 118, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, S.; Isberg, R.R. Cdc42 interacts with the exocyst complex to promote phagocytosis. J. Cell Biol. 2013, 200, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Cox, D.; Li, J.; Greenberg, S. Rac1 and Cdc42 are required for phagocytosis, but not NF-kappaB-dependent gene expression, in macrophages challenged with Pseudomonas aeruginosa. J. Biol. Chem. 2000, 275, 141–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlam, D.; Bagshaw, R.D.; Freeman, S.A.; Collins, R.F.; Pawson, T.; Fairn, G.D.; Grinstein, S. Phosphoinositide 3-kinase enables phagocytosis of large particles by terminating actin assembly through Rac/Cdc42 GTPase-activating proteins. Nat. Commun. 2015, 6, 8623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Liu, H.; Chen, F. Functioning of long noncoding RNAs expressed in macrophage in the development of atherosclerosis. Front. Pharmacol. 2020, 11, 567582. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, X.; Wei, N.; Liu, S.; Ling, Y.; Wang, H. P21-activated kinase 4 as a switch between caspase-8 apoptosis and NF-κB survival signals in response to TNF-α in hepatocarcinoma cells. Biochem. Biophys. Res. Commun. 2018, 503, 3003–3010. [Google Scholar] [CrossRef]
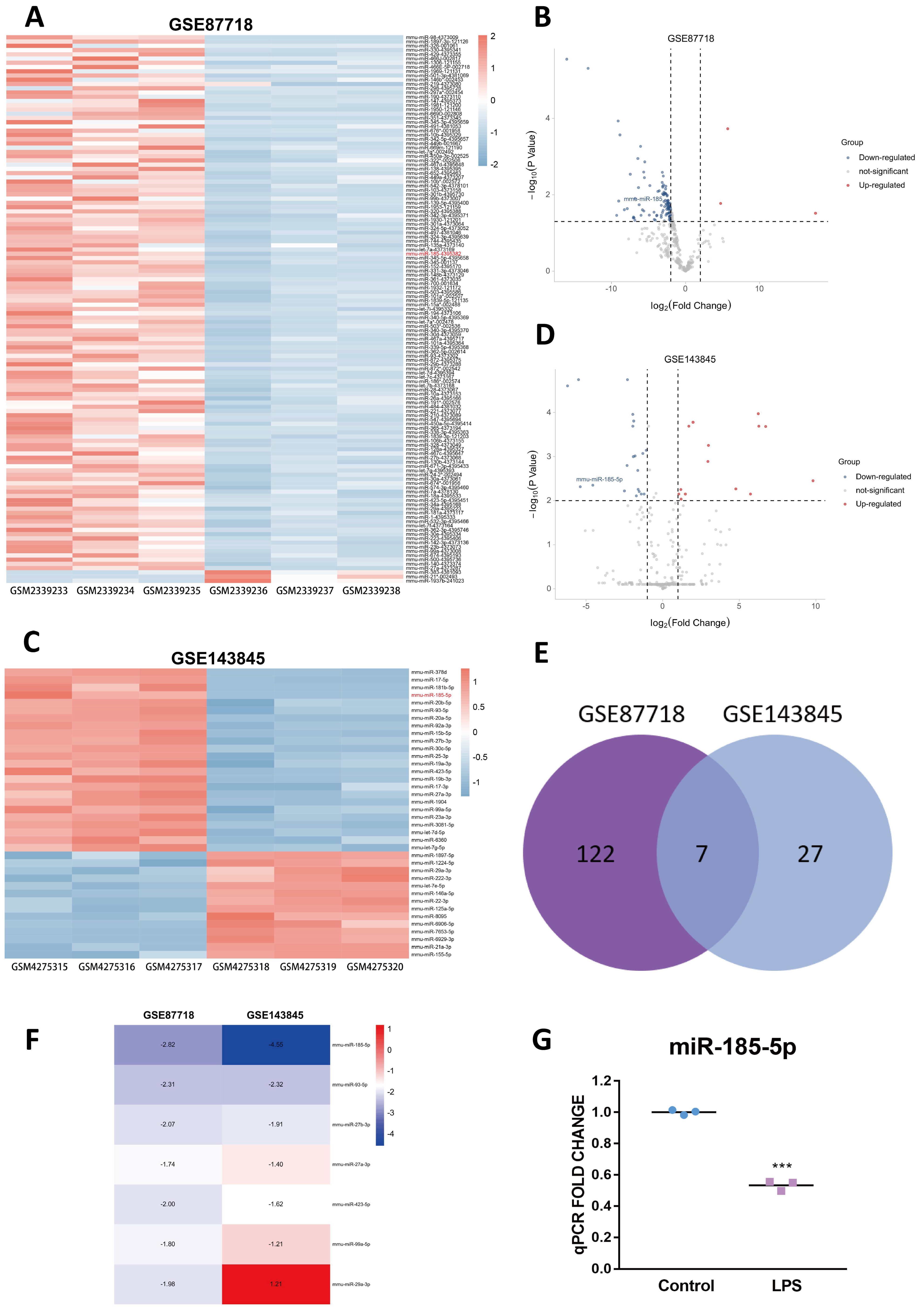
| Accession | Platform | Sample | Control Group | Treated Group |
|---|---|---|---|---|
| GSE87718 | GPL18082 | 6 | Dicer Wild Type BMDMs (n = 3) | Dicer Knock Out BMDMs (n = 3) |
| GSE143845 | GPL21265 | 6 | RAW264.7 untreated (n = 3) | RAW264.7 treated with LPS (100ng/mL) and INF-γ (20ng/mL) for 24 h (n = 3) |
| RT-qPCR | ||
|---|---|---|
| Il6 | Forward Primer | acaaagccagagtccttcagag |
| Il6 | Reverse Primer | accacagtgaggaatgtccac |
| Il1b | Forward Primer | tgccaccttttgacagtgatg |
| Il1b | Reverse Primer | tgatactgcctgcctgaagc |
| Tnf | Forward Primer | tgttgcctcctcttttgctt |
| Tnf | Reverse Primer | tggtcaccaaatcagcgtta |
| Cdc42 | Forward Primer | cggagaagctgaggacaagatctaa |
| Cdc42 | Reverse Primer | aggagacatgttttaccaacagc |
| Actb | Forward Primer | accttctacaatgagctgcg |
| Actb | Reverse Primer | ctggatggctacgtacatgg |
| U6 | Forward Primer | ctcgcttcggcagcaca |
| U6 | Reverse Primer | aacgcttcacgaatttgcgt |
| mmu-miR-185-5p | gtcgtatccagtgcagggtccgaggtattcgcactggatacgactcagga | |
| miRNA transfection | ||
| miR NC | Sense (5′-3′) | UUUGUACUACACAAAAGUACUG |
| miR NC | Anti-sense (5′-3′) | CAGUACUUUUGUGUAGUACAAA |
| miR-185-5p mimics | Sense (5′-3′) | UGGAGAGAAAGGCAGUUCCUGA |
| miR-185-5p mimics | Anti-sense (5′-3′) | UCAGGAACUGCCUUUCUCUCCA |
| Anti-miR NC | CAGUACUUUUGUGUAGUACAAA | |
| miR-185-5p inhibitors | UCAGGAACUGCCUUUCUCUCCA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Liu, H.; Zhu, J.; Zhang, C.; Peng, Y.; Mao, Z.; Jing, Y.; Chen, F. miR-185-5p Regulates Inflammation and Phagocytosis through CDC42/JNK Pathway in Macrophages. Genes 2022, 13, 468. https://doi.org/10.3390/genes13030468
Ma X, Liu H, Zhu J, Zhang C, Peng Y, Mao Z, Jing Y, Chen F. miR-185-5p Regulates Inflammation and Phagocytosis through CDC42/JNK Pathway in Macrophages. Genes. 2022; 13(3):468. https://doi.org/10.3390/genes13030468
Chicago/Turabian StyleMa, Xirui, Huifang Liu, Jing Zhu, Caoxu Zhang, Yajie Peng, Ziming Mao, Yu Jing, and Fengling Chen. 2022. "miR-185-5p Regulates Inflammation and Phagocytosis through CDC42/JNK Pathway in Macrophages" Genes 13, no. 3: 468. https://doi.org/10.3390/genes13030468
APA StyleMa, X., Liu, H., Zhu, J., Zhang, C., Peng, Y., Mao, Z., Jing, Y., & Chen, F. (2022). miR-185-5p Regulates Inflammation and Phagocytosis through CDC42/JNK Pathway in Macrophages. Genes, 13(3), 468. https://doi.org/10.3390/genes13030468






