Overexpression of PvFAD3 Gene from Plukenetia volubilis Promotes the Biosynthesis of α-Linolenic Acid in Transgenic Tobacco Seeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cloning of the DNA Sequence of the PvFAD3 Gene
2.3. Bioinformatics Analysis
2.4. Expression Pattern of PvFAD3 Gene
2.5. Construction of Fusion Vector and Overexpression Vector
2.6. Subcellular Localization
2.7. Generation of Transgenic Tobacco and the Determination of Fatty Acid Composition
3. Results
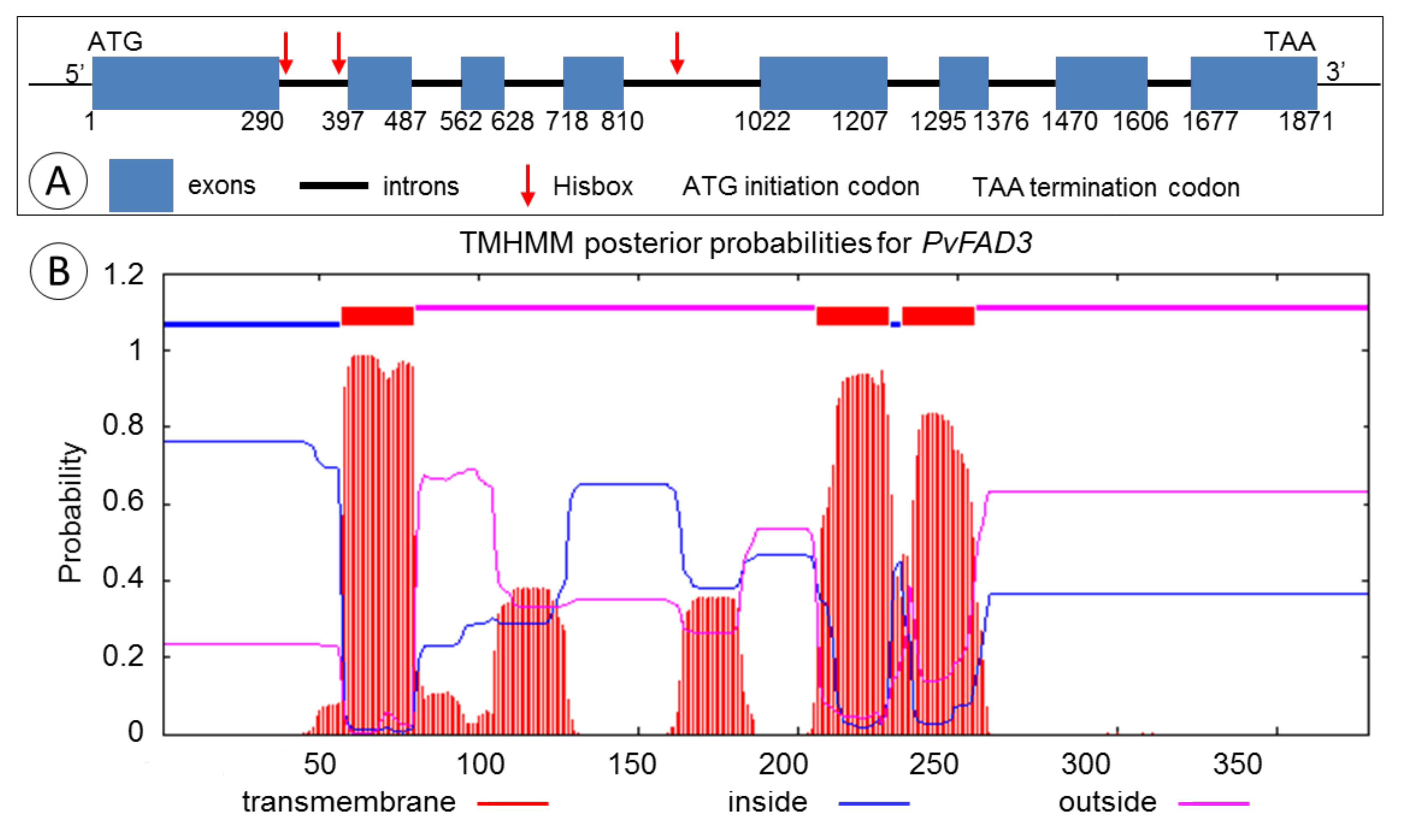
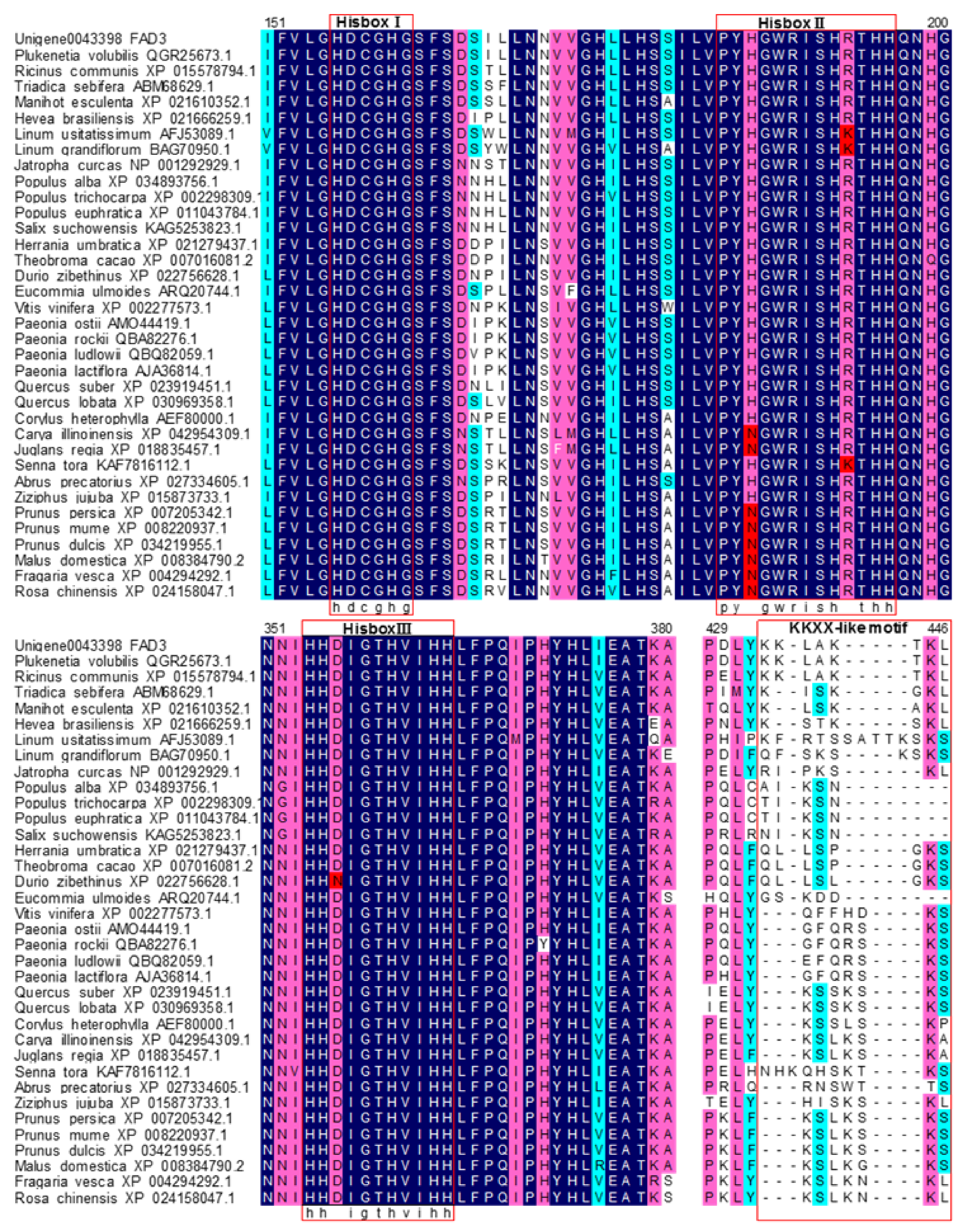
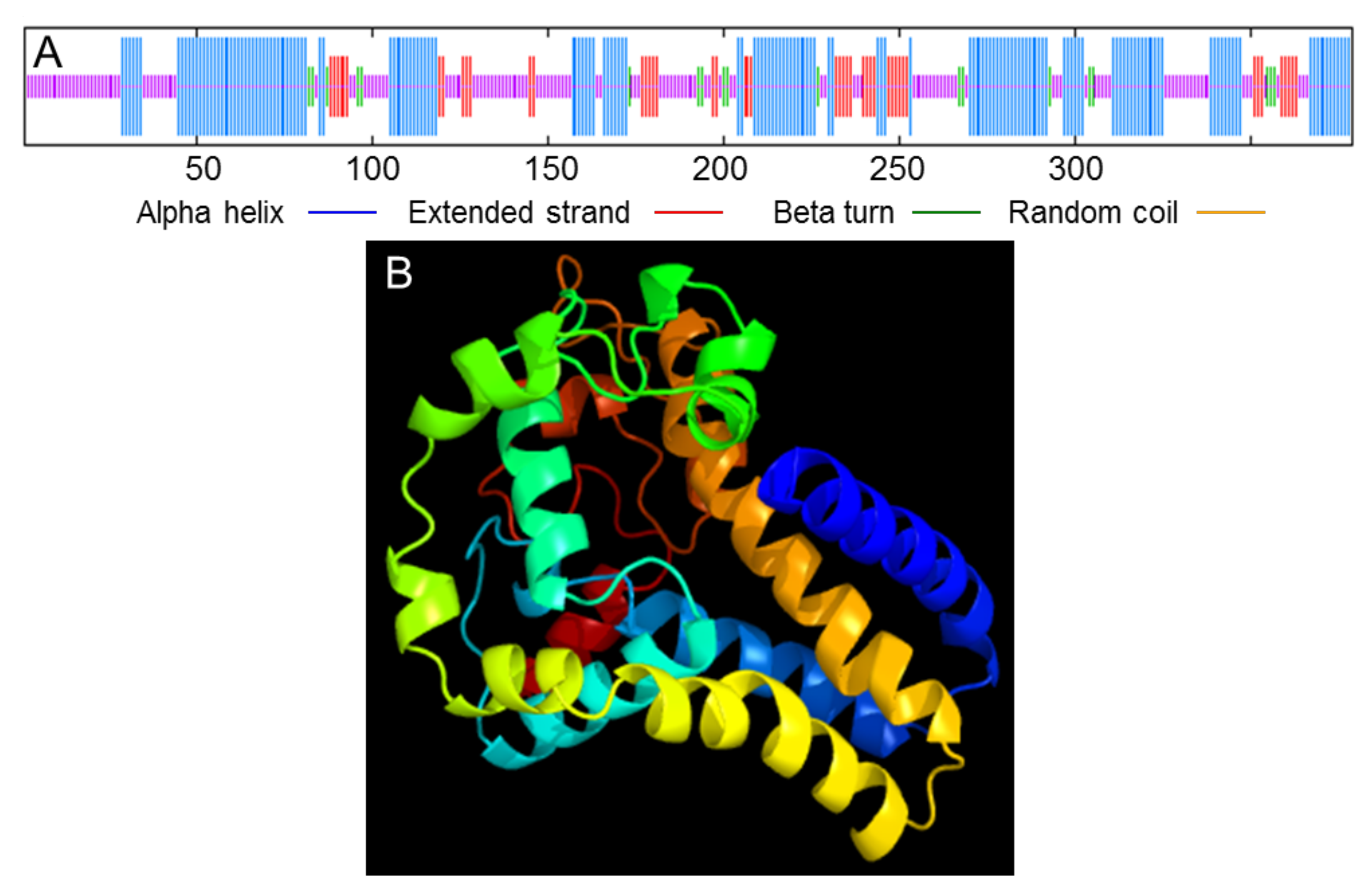
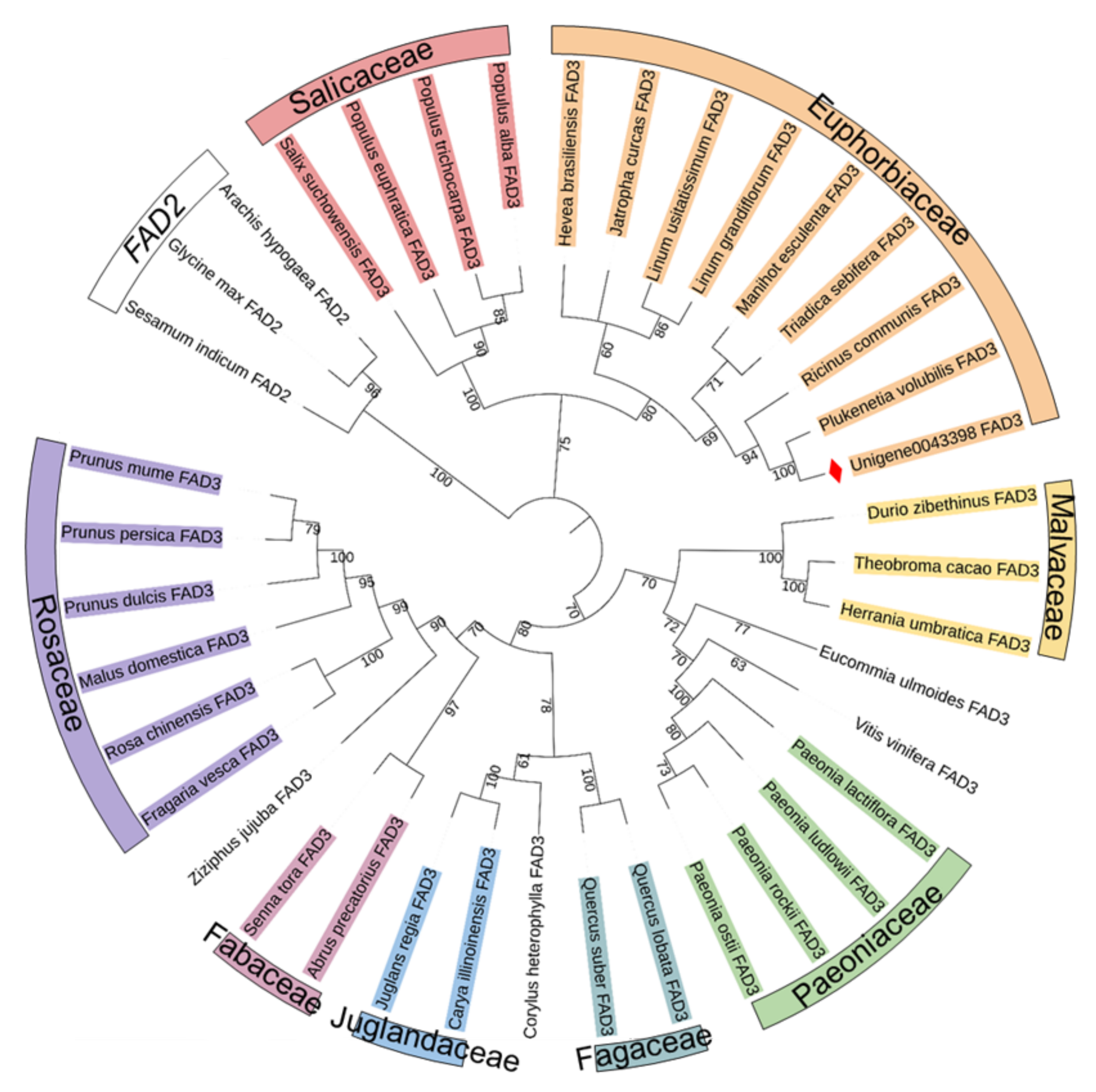
3.1. Cloning and Sequence Analysis of the PvFAD3
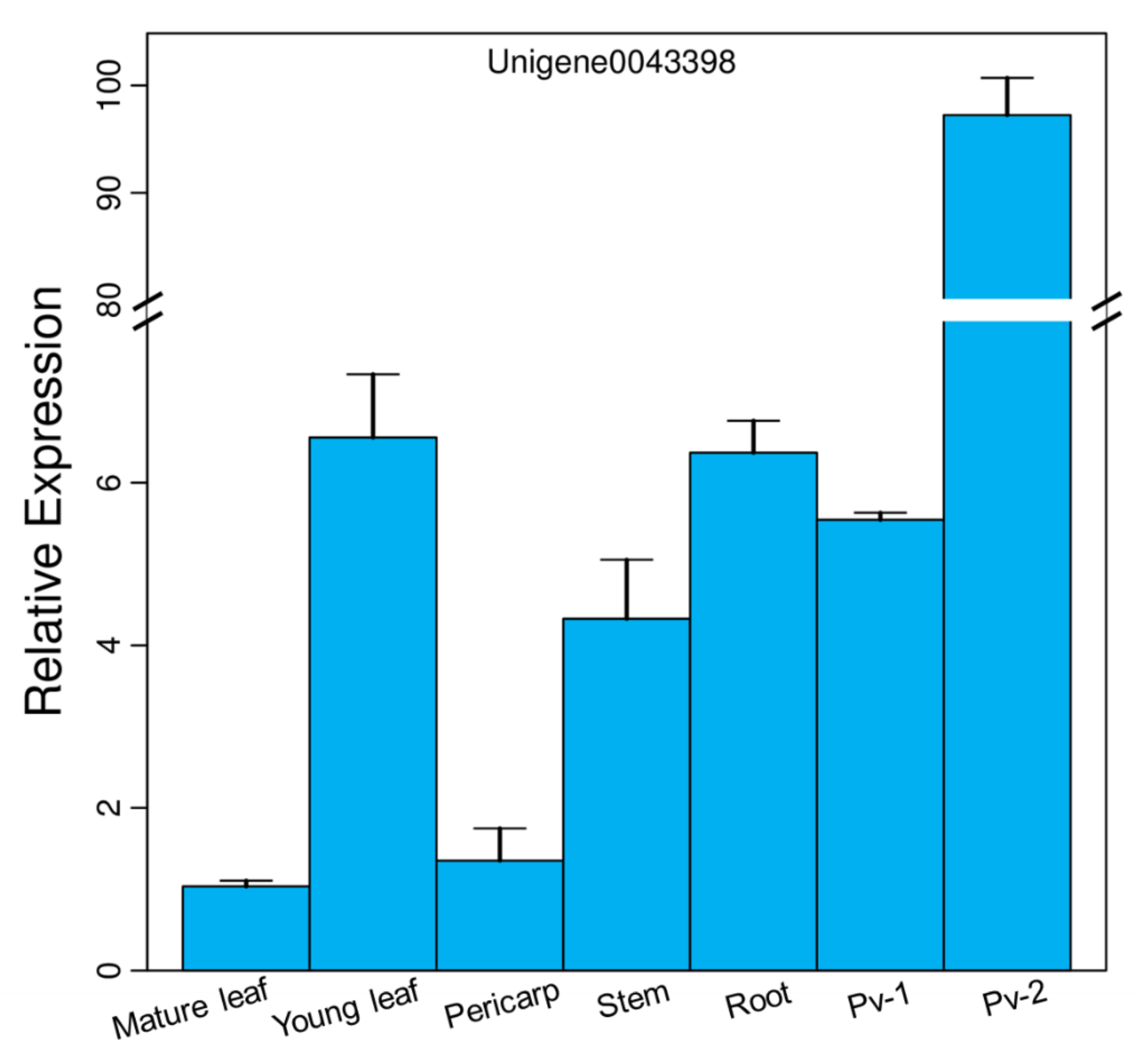
3.2. Expression Pattern of PvFAD3 in Different Tissues
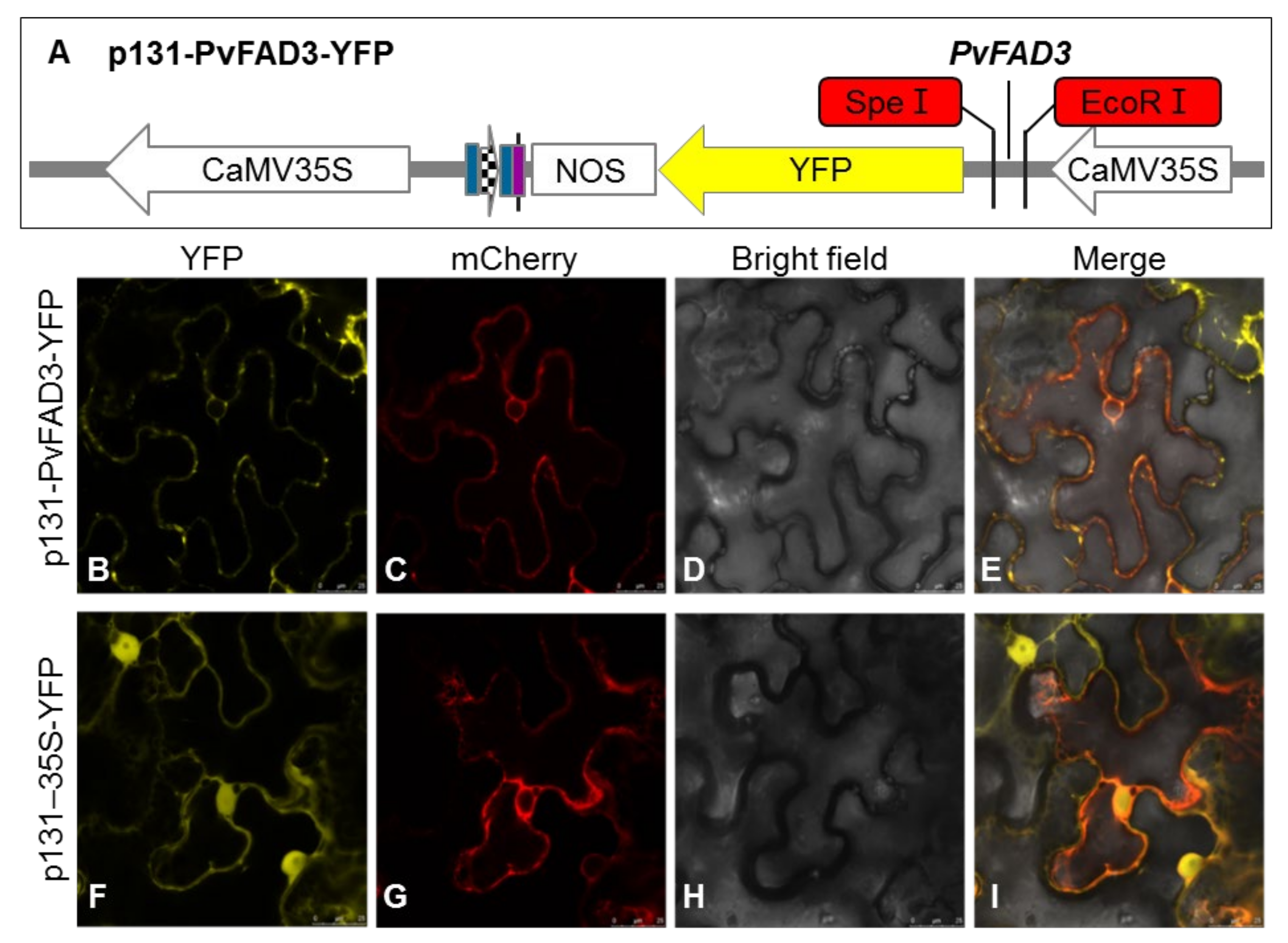
3.3. Subcellular Localization of PvFAD3
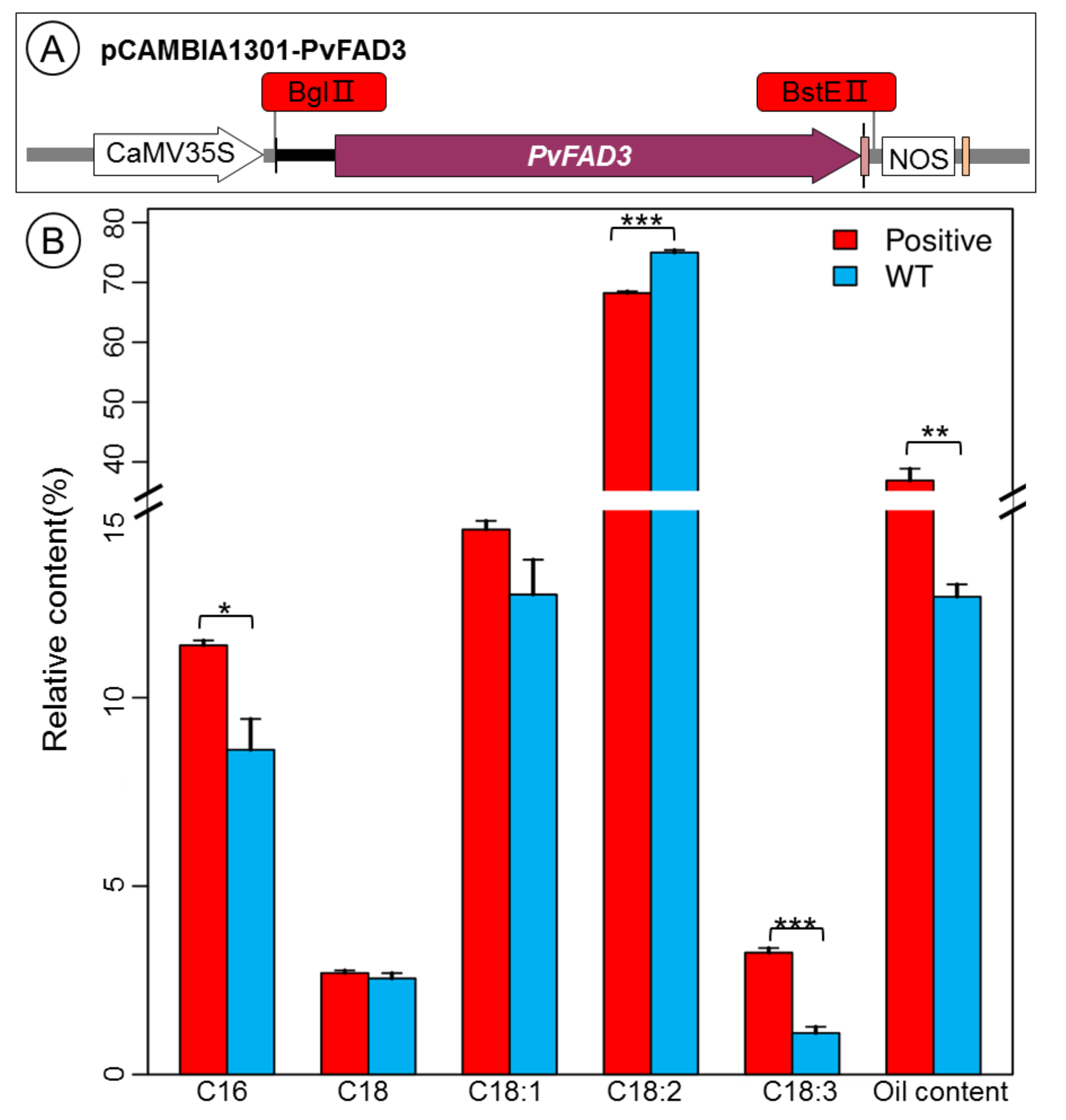
3.4. Fatty Acid Analysis of PvFAD3 Gene-Transformed Tobacco Seeds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bussmann, R.W.; Paniagua Zambrana, N.; Téllez, C. Plukenetia carolis-vegae (Euphorbiaceae)–A new useful species from northern Peru. Econ. Bot. 2013, 67, 387–392. [Google Scholar] [CrossRef]
- Gutiérrez, L.-F.; Rosada, L.M.; Jiménez, Á. Chemical composition of Sacha Inchi (Plukenetia volubilis L.) seeds and characteristics of their lipid fraction. Grasas Aceites 2011, 62, 76–83. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, U.; Liu, A. Stage-specific metabolization of triacylglycerols during seed germination of Sacha Inchi (Plukenetia volubilis L.). J. Sci. Food Agric. 2015, 95, 1764–1766. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, F.; Kakuda, Y. Sacha inchi (Plukenetia volubilis L.): Nutritional composition, biological activity, and uses. Food Chem. 2018, 265, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zhu, Y.; Zhang, T.; Bai, X.; He, H.; Pan, B.; Fu, Q.; Xu, Z. Analysis of nutritional components in tender stems and leaves of the woody oil crop Plukenetia volubilis. Non-Wood For. Res. 2021, 39, 97–103. [Google Scholar]
- Kodahl, N. Sacha inchi (Plukenetia volubilis L.)—From lost crop of the Incas to part of the solution to global challenges? Planta 2020, 251, 80. [Google Scholar] [CrossRef]
- Baker, E.J.; Miles, E.; Burdge, G.; Yaqoob, P.; Calder, P.C. Metabolism and functional effects of plant-derived omega-3 fatty acids in humans. Prog. Lipid Res. 2016, 64, 30–56. [Google Scholar] [CrossRef]
- Yu, S.Y.; Zhang, X.; Huang, L.B.; Lyu, Y.P.; Zhang, Y.; Yao, Z.J.; Zhang, X.X.; Yuan, J.H.; Hu, Y.H. Transcriptomic analysis of α-linolenic acid content and biosynthesis in Paeonia ostii fruits and seeds. BMC Genom. 2021, 22, 297. [Google Scholar] [CrossRef]
- Berestovoy, M.; Pavlenko, O.S.; Goldenkova-Pavlova, I.V. Plant Fatty Acid Desaturases: Role in the Life of Plants and Biotechnological Potential. Biol. Bull. Rev. 2020, 10, 127–139. [Google Scholar] [CrossRef]
- Puttick, D.; Dauk, M.; Lozinsky, S.; Smith, M.A. Overexpression of a FAD3 Desaturase Increases Synthesis of a Polymethylene-Interrupted Dienoic Fatty Acid in Seeds of Arabidopsis thaliana L. Lipids 2009, 44, 753–757. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Fan, C.; Guo, Z.; Qin, J.; Wu, J.; Li, Q.; Fu, T.; Zhou, Y. Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele-specific markers for high oleic and low linolenic acid contents. Theor. Appl. Genet. 2012, 125, 715–729. [Google Scholar] [CrossRef]
- Domínguez, T.; Hernández, M.L.; Pennycooke, J.C.; Jiménez, P.; Martínez-Rivas, J.M.; Sanz, C.; Stockinger, E.J.; Sánchez-Serrano, J.J.; Sanmartín, M. Increasing ω-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 2010, 153, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Pham, A.-T.; Shannon, J.G.; Bilyeu, K.D. Combinations of mutant FAD2 and FAD3 genes to produce high oleic acid and low linolenic acid soybean oil. Theor. Appl. Genet. 2012, 125, 503–515. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, S.; Zhang, L.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Functional characterization of two microsomal fatty acid desaturases from Jatropha curcas L. J. Plant Physiol. 2013, 170, 1360–1366. [Google Scholar] [CrossRef]
- Wu, L.; Li, F.; Wu, X.; Song, D.; Wang, M. Cloning and expression characteristics of one ω-3 cis 15 fatty acid dehydrogenase gene AhFAD3A in Arachis hypogaea L. Chin. Oil Crop Sci. 2015, 37, 41–47. [Google Scholar]
- Lee, K.-R.; Lee, Y.; Kim, E.-H.; Lee, S.-B.; Roh, K.H.; Kim, J.-B.; Kang, H.-C.; Kim, H.U. Functional identification of oleate 12-desaturase and ω-3 fatty acid desaturase genes from Perilla frutescens var. frutescens. Plant Cell Rep. 2016, 35, 2523–2537. [Google Scholar] [CrossRef]
- Huang, X.; Lu, J.; Liao, B.; Bai, H.; Guan, L.; Zhang, T. Cloning and expression analysis of fatty acid desaturase gene FAD3 from oil peony. Sci. Agric. Sin. 2017, 50, 1914–1921. [Google Scholar]
- Wang, P.; Cao, H.; Chen, D.; Chen, D.; Chen, G.; Yang, J.; Ye, N. Cloning and Expression Analysis of Fatty Acid Desaturase Family Genes in Camellia sinensis. Acta Hortic. Sin. 2020, 47, 1141–1152. [Google Scholar]
- Bhunia, R.K.; Chakraborty, A.; Kaur, R.; Gayatri, T.; Bhattacharyya, J.; Basu, A.; Maiti, M.; Sen, S.K. Seed-specific increased expression of 2S albumin promoter of sesame qualifies it as a useful genetic tool for fatty acid metabolic engineering and related transgenic intervention in sesame and other oil seed crops. Plant Mol. Biol. 2014, 86, 351–365. [Google Scholar] [CrossRef]
- Feng, Y.Z. Transcriptome Sequencing and Study on FAD3 Gene Identification and Function in Eucommia Ulmides Oliv; Chinese Academy of Forestry: Beijing, China, 2016. [Google Scholar]
- Liu, G.; Wu, Z.; Peng, Y.; Shang, X.; Xie, Y.; Arnold, R.J. Transcriptome analyses reveals the dynamic nature of oil accumulation during seed development of Plukenetia volubilis L. Sci. Rep. 2020, 10, 20467. [Google Scholar] [CrossRef]
- Wang, X.; Liu, A. Expression of Genes Controlling Unsaturated Fatty Acids Biosynthesis and Oil Deposition in Developing Seeds of Sacha Inchi (Plukenetia volubilis L.). Lipids 2014, 49, 1019–1031. [Google Scholar] [CrossRef]
- Wang, X.; Xu, R.; Wang, R.; Liu, A. Transcriptome analysis of Sacha Inchi (Plukenetia volubilis L.) seeds at two developmental stages. BMC Genom. 2012, 13, 716. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ivak Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Liu, G.; Chen, H.P.; Peng, Y.; Xie, Y.J.; Chen, S.X. Variations in fatty acid composition of Sacha inchi seeds during growth and development. China Oils Fats 2018, 43, 57–62. [Google Scholar]
- Shanklin, J.; Whittle, E.; Fox, B.G. Eight Histidine Residues Are Catalytically Essential in a Membrane-Associated Iron Enzyme, Stearoyl-CoA Desaturase, and Are Conserved in Alkane Hydroxylase and Xylene Monooxygenase. Biochemisty 1994, 33, 12787–12794. [Google Scholar] [CrossRef]
- Guan, L.; Hou, K.; Chen, J.; Xu, Y.; Wu, W. Phylogenetic and functional differentiation of the omega-6 and omega-3 fatty acid dehydrogenase families. Hereditas 2013, 35, 643–654. [Google Scholar]
- Czumaj, A.; Śledziński, T. Biological role of unsaturated fatty acid desaturases in health and disease. Nutrients 2020, 12, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Wang, G.; Pan, X. China Food Composition, 2nd ed.; Peking Univerisity Medical Press: Beijing, China, 2009. [Google Scholar]
- Shen, Q.; Zhu, W.X.; Qin, X.R.; Zhao, Y.; Wang, X.P.; Li, M.; Du, C.F. Oil Content and fatty acid components of 30 Perilla frutescens samples in Guizhou. Guizhou Agric. Sci. 2014, 42, 5–8. [Google Scholar]
- Porras-Loaiza, P.; Jiménez-Munguía, M.T.; Sosa-Morales, M.E.; Palou, E.; López-Malo, A. Physical properties, chemical characterization and fatty acid composition of Mexican chia (Salvia hispanica L.) seeds. Int. J. Food Sci. Technol. 2014, 49, 571–577. [Google Scholar] [CrossRef]
- Xing, L.; Zhao, F.-M.; Cao, Y.-F.; Wang, M.; Mei, S.; Li, S.-P.; Cai, Z.-Y. Principal component analysis of mineral elements and fatty acids composition in flaxseed from ten different regions. Spectrosc. Spectr. Anal. 2014, 34, 2538–2543. [Google Scholar]
- Kodahl, N.; Sørensen, M. Sacha Inchi (Plukenetia volubilis L.) Is an Underutilized Crop with a Great Potential. Agronomy 2021, 11, 1066. [Google Scholar] [CrossRef]
- Li, S.-S.; Wang, L.-S.; Shu, Q.-Y.; Wu, J.; Chen, L.-G.; Shao, S.; Yin, D.-D. Fatty acid composition of developing tree peony (Paeonia section Moutan DC.) seeds and transcriptome analysis during seed development. BMC Genom. 2015, 16, 208. [Google Scholar] [CrossRef] [Green Version]
- McCartney, A.W.; Dyer, J.M.; Dhanoa, P.K.; Kim, P.K.; Andrews, D.W.; McNew, J.A.; Mullen, R.T. Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J. 2004, 37, 156–173. [Google Scholar] [CrossRef] [Green Version]
- Long, M.; Rosenberg, C.; Gilbert, W. Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl. Acad. Sci. USA 1995, 92, 12495–12499. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, O.; Watanabe, C.; Iba, K. Hormonal regulation of tissue-specific ectopic expression of an Arabidopsis endoplasmic reticulum-type ω-3 fatty acid desaturase (FAD3) gene. Planta 2001, 213, 833–840. [Google Scholar] [CrossRef]
- Feng, J.; Dong, Y.; Liu, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Genome-wide identification of membrane-bound fatty acid desaturase genes in Gossypium hirsutum and their expressions during abiotic stress. Sci. Rep. 2017, 7, srep45711. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.L.; Yin, Z.J.; Xiao, L.; Xu, Y.N.; Qu, L.Q. Identification and evaluation of ω-3 fatty acid desaturase genes for hyperfortifying α-linolenic acid in transgenic rice seed. J. Exp. Bot. 2012, 63, 3279–3287. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Chen, B.; Win, A.N.; Fu, C.; Lian, J.; Liu, X.; Wang, R.; Zhang, X.; Chai, Y. Omega-3 fatty acid desaturase gene family from two ω-3 sources, Salvia hispanica and Perilla frutescens: Cloning, characterization and expression. PLoS ONE 2018, 13, e0191432. [Google Scholar] [CrossRef]
- Román, Á.; Andreu, V.; Hernández, M.L.; Lagunas, B.; Picorel, R.; Martínez-Rivas, J.M.; Alfonso, M. Contribution of the different omega-3 fatty acid desaturase genes to the cold response in soybean. J. Exp. Bot. 2012, 63, 4973–4982. [Google Scholar] [CrossRef] [Green Version]
- Duan, W.; Lijun, W.; Shimei, Y.; Shen, Q.; Zhao, D. Advances on formation and regulation mechanism of α-linolenic acid in seeds. J. Plant Genet. Resour. 2020, 21, 49–62. [Google Scholar]
- Wu, D.; Yang, S.M.; Shang, Z.W.; Xu, J.; Zhao, D.G.; Wang, H.B.; Shen, Q. Genome-wide analysis of the fatty acid desaturase gene family reveals the key role of PfFAD3 in α-linolenic acid biosynthesis in Perilla Seeds. Front. Genet. 2021, 12, 735862. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Wu, Z.; Shang, X.; Peng, Y.; Gao, L. Overexpression of PvFAD3 Gene from Plukenetia volubilis Promotes the Biosynthesis of α-Linolenic Acid in Transgenic Tobacco Seeds. Genes 2022, 13, 450. https://doi.org/10.3390/genes13030450
Liu G, Wu Z, Shang X, Peng Y, Gao L. Overexpression of PvFAD3 Gene from Plukenetia volubilis Promotes the Biosynthesis of α-Linolenic Acid in Transgenic Tobacco Seeds. Genes. 2022; 13(3):450. https://doi.org/10.3390/genes13030450
Chicago/Turabian StyleLiu, Guo, Zhihua Wu, Xiuhua Shang, Yan Peng, and Liqiong Gao. 2022. "Overexpression of PvFAD3 Gene from Plukenetia volubilis Promotes the Biosynthesis of α-Linolenic Acid in Transgenic Tobacco Seeds" Genes 13, no. 3: 450. https://doi.org/10.3390/genes13030450
APA StyleLiu, G., Wu, Z., Shang, X., Peng, Y., & Gao, L. (2022). Overexpression of PvFAD3 Gene from Plukenetia volubilis Promotes the Biosynthesis of α-Linolenic Acid in Transgenic Tobacco Seeds. Genes, 13(3), 450. https://doi.org/10.3390/genes13030450




