Differential Gene Expression Analysis of SoCBL Family Calcineurin B-like Proteins: Potential Involvement in Sugarcane Cold Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. cDNA First-Strand Synthesis
2.3. PCR Primer Design and Amplification Conditions
2.4. Sequence Analysis and Prediction
2.5. Real-Time Quantitative PCR for Gene Expression Analysis
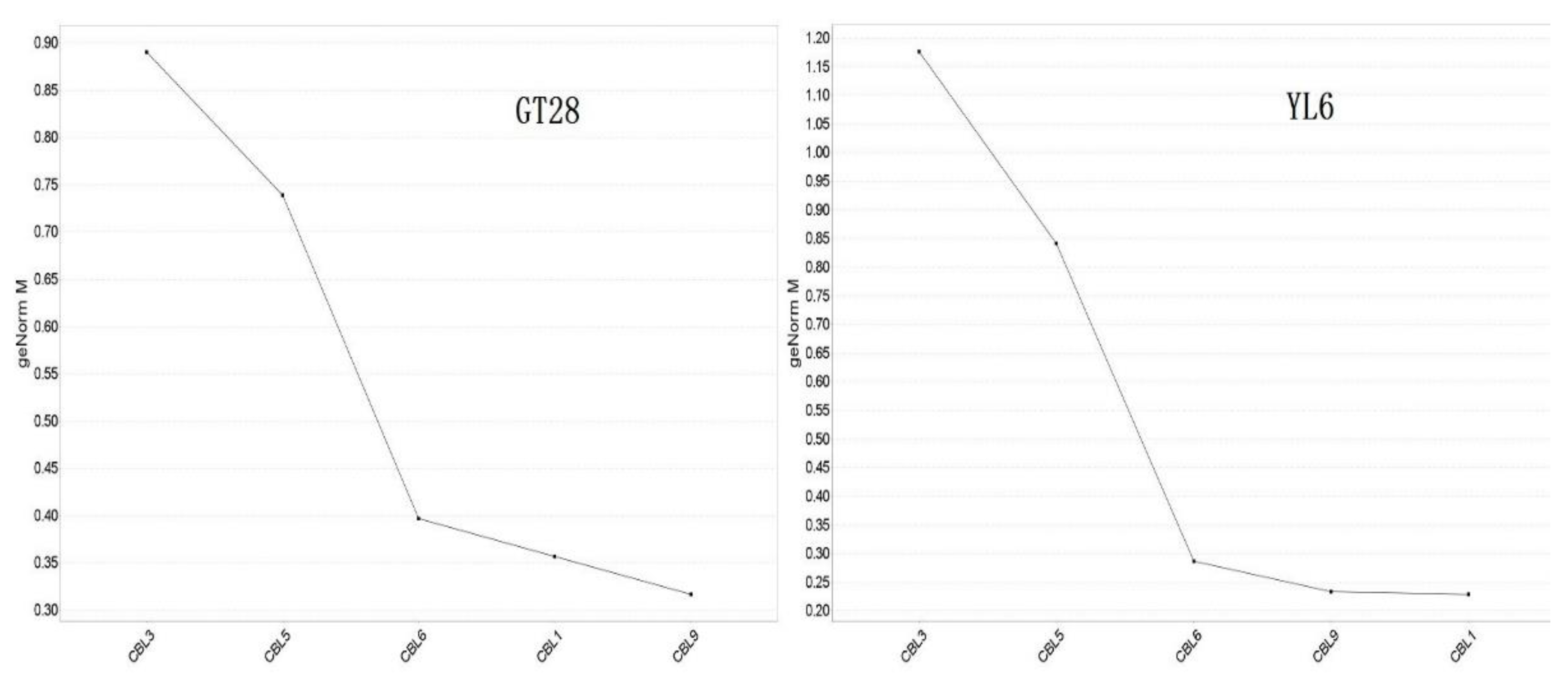
2.6. Statistical Analysis
3. Results
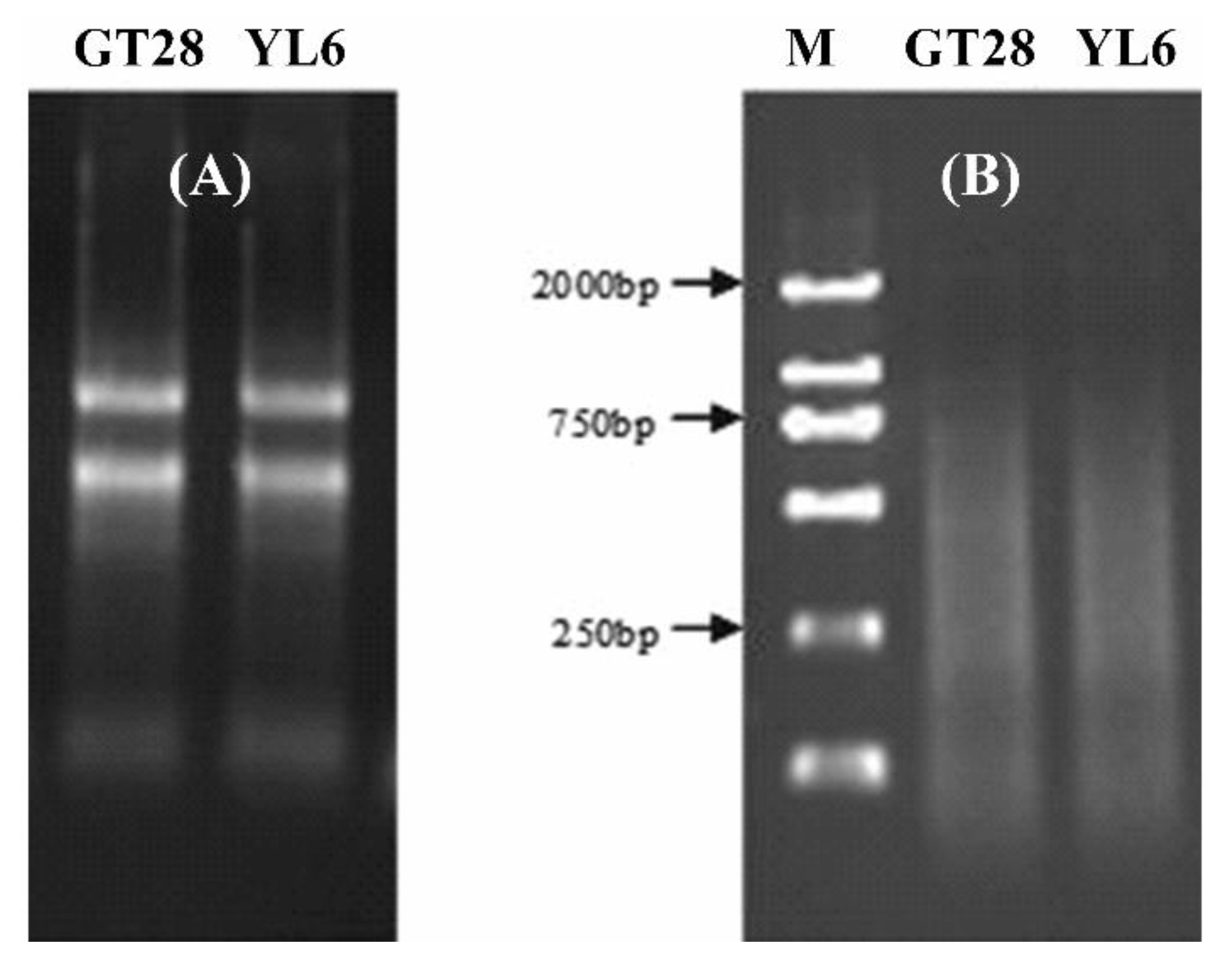
3.1. Total RNA and cDNA Quality of Sugarcane Leaves
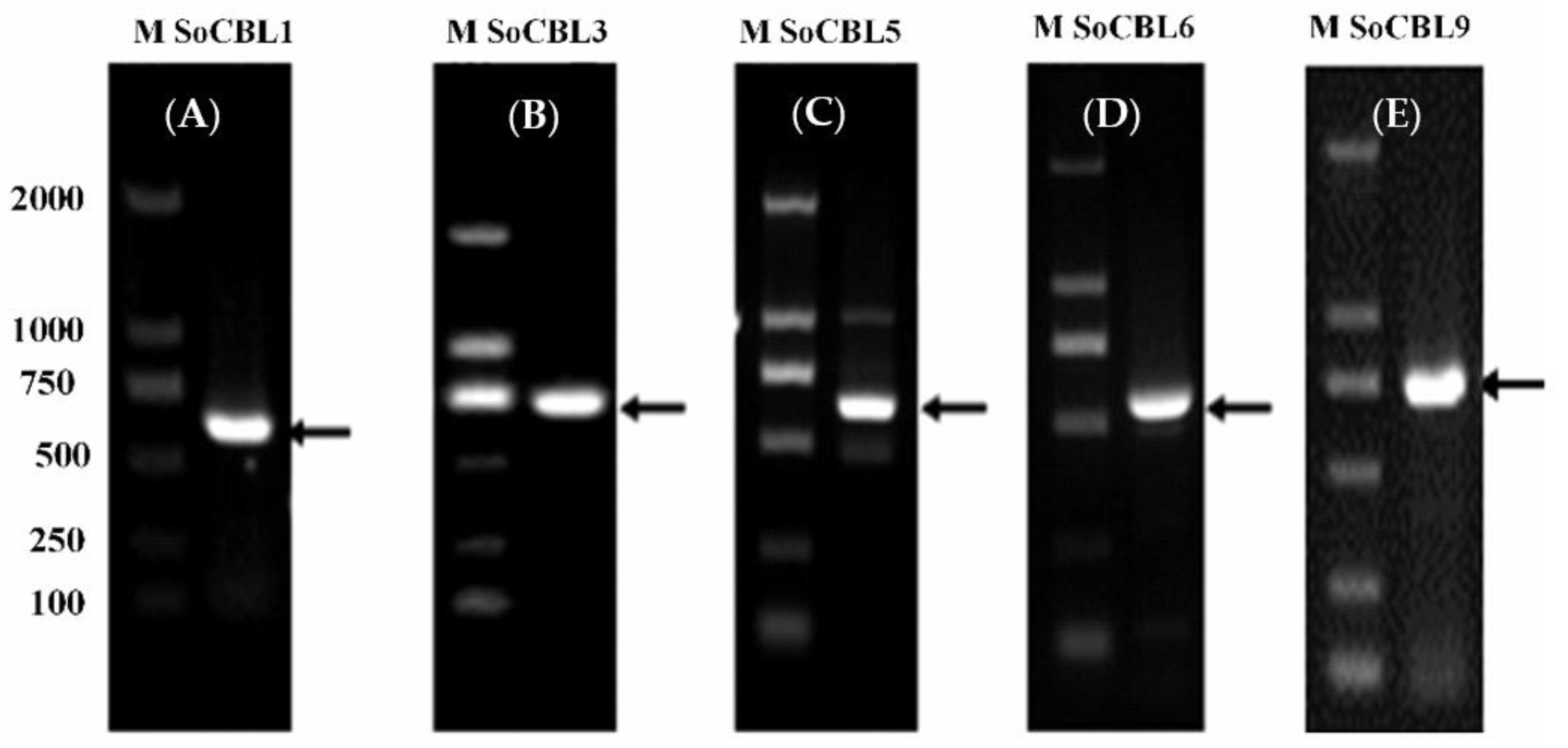
3.2. Cloning and Sequencing of Sugarcane SoCBL Genes
3.3. Bioinformatics Analysis of Sugarcane SoCBL
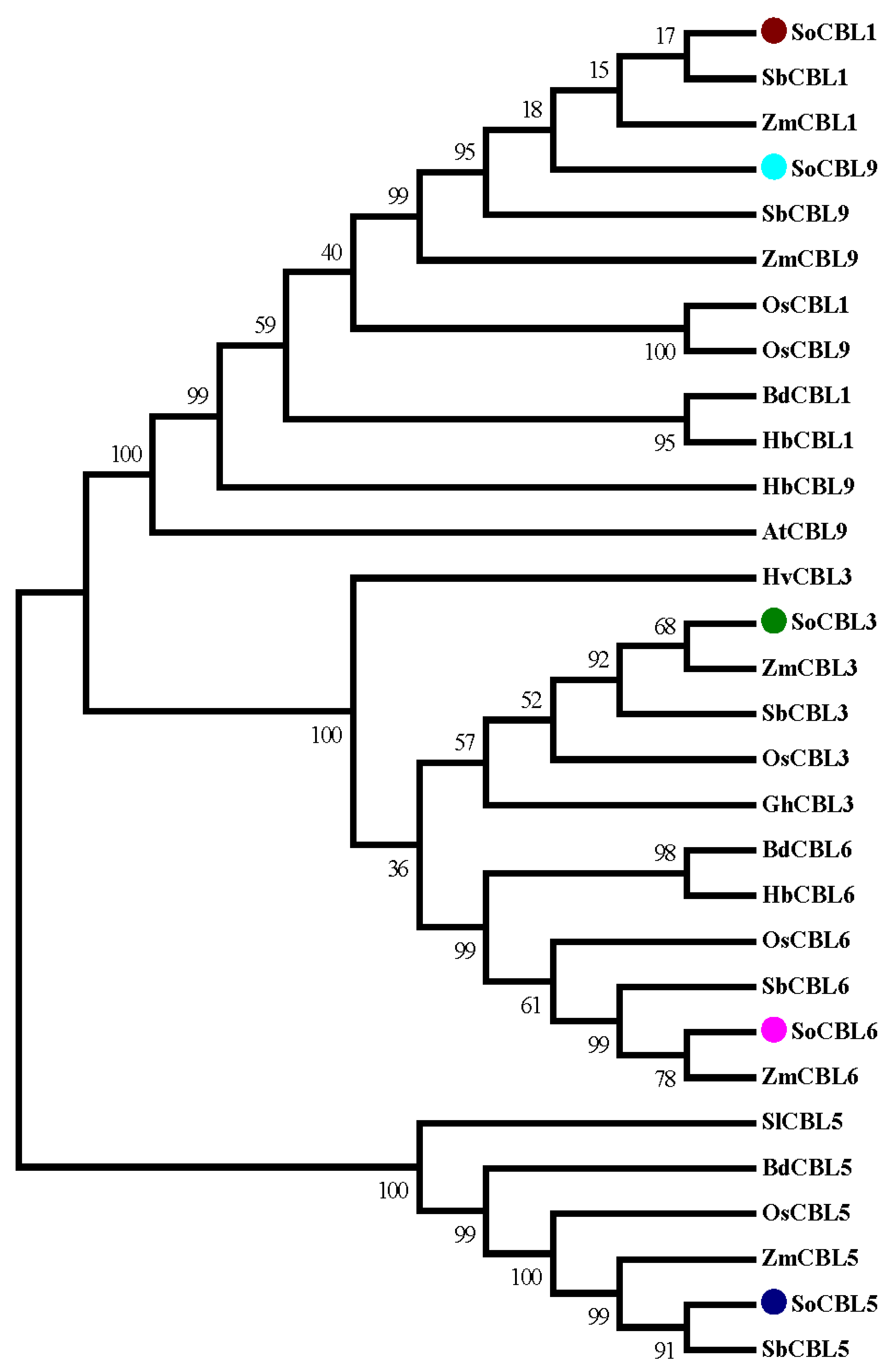
3.4. Phylogenetic Analysis of Sugarcane SoCBL
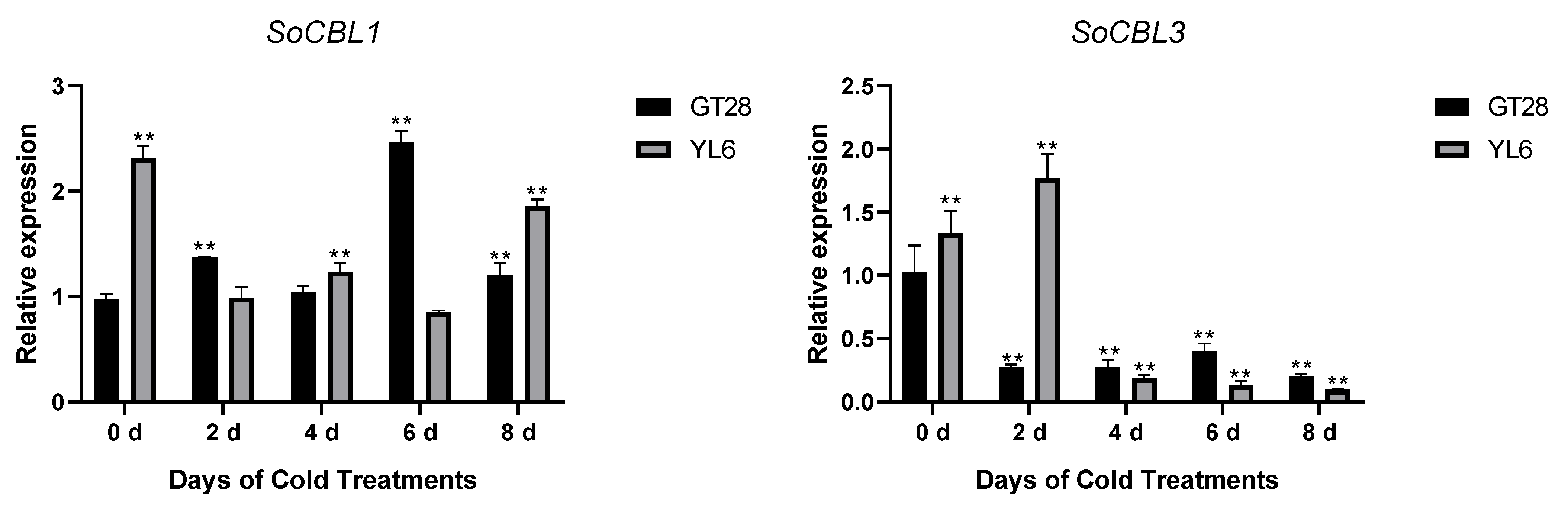
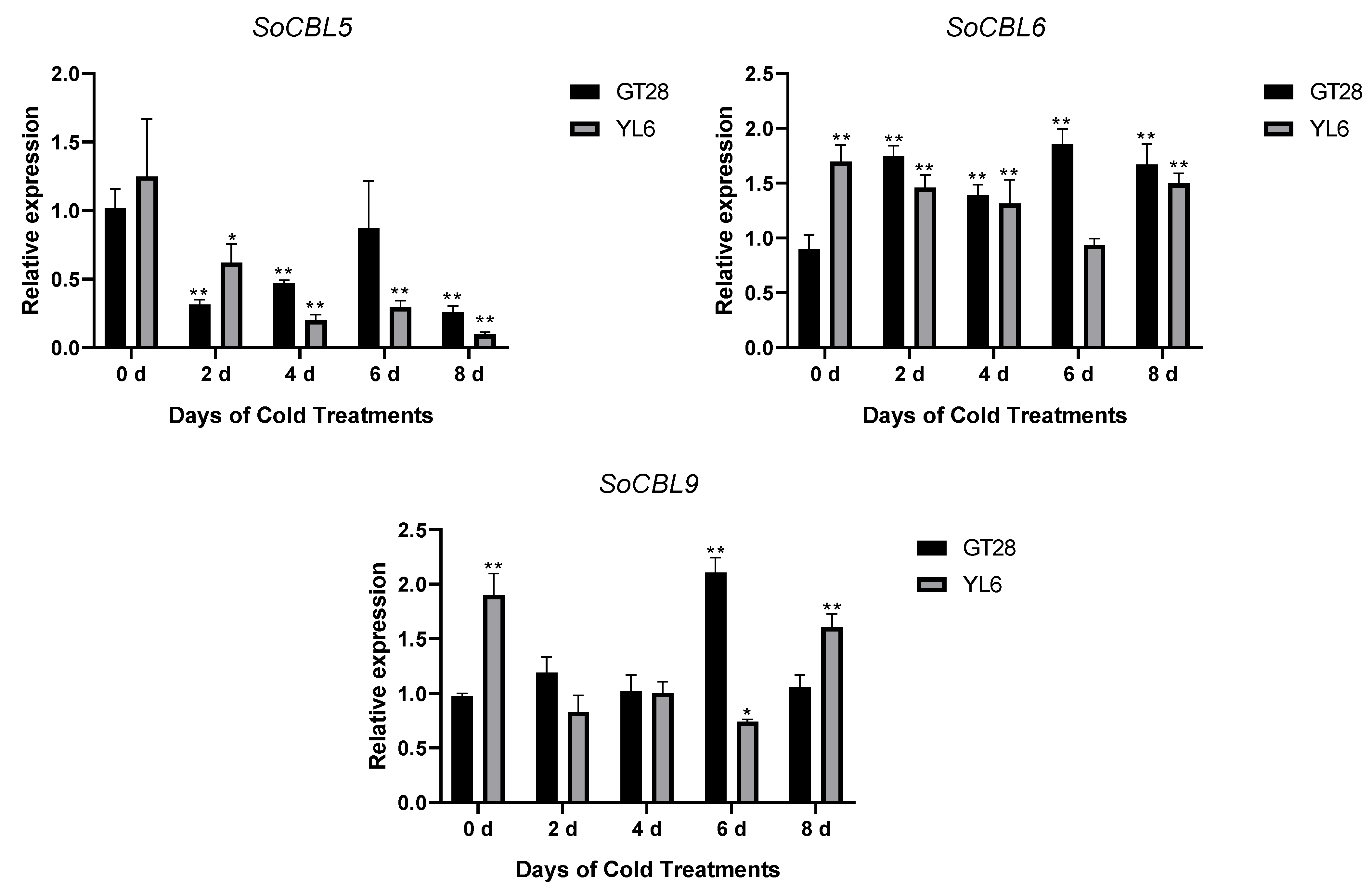
3.5. Change in Expression of SoCBL Genes in Response to Low-Temperature Stress
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14 (Suppl. 1), S165–S183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the Crossroads of Signaling. Plant Cell 2002, 14, S401–S417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, J.-K. A Calcium Sensor Homolog Required for Plant Salt Tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudla, J.; Xu, Q.; Harter, K.; Gruissem, W.; Luan, S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 1999, 96, 4718–4723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmon, A.C.; Gribskov, M.; Harper, J.F. CDPKs—A kinase for every Ca2+ signal? Trends Plant Sci. 2000, 5, 154–159. [Google Scholar] [CrossRef]
- Snedden, W.A.; Fromm, H. Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 2001, 151, 35–66. [Google Scholar] [CrossRef] [Green Version]
- Batistič, O.; Kudla, J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 2009, 1793, 985–992. [Google Scholar] [CrossRef] [Green Version]
- Kolukisaoglu, Ü.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium Sensors and Their Interacting Protein Kinases: Genomics of the Arabidopsis and Rice CBL-CIPK Signaling Networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef] [Green Version]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and Calcineurin B–like Proteins. Plant Cell 2002, 14, S389–S400. [Google Scholar] [CrossRef] [Green Version]
- Luan, S. The CBL–CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.-S.; Shi, W.; Zhu, J.-K. SOS3 Function in Plant Salt Tolerance Requires N-Myristoylation and Calcium Binding. Plant Cell 2000, 12, 1667–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheong, Y.H.; Kim, K.-N.; Pandey, G.K.; Gupta, R.; Grant, J.J.; Luan, S. CBL1, a Calcium Sensor That Differentially Regulates Salt, Drought, and Cold Responses in Arabidopsis. Plant Cell 2003, 15, 1833–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheong, Y.H.; Sung, S.J.; Kim, B.-G.; Pandey, G.K.; Cho, J.-S.; Kim, K.-N.; Luan, S. Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol. Cells 2010, 29, 159–165. [Google Scholar] [CrossRef]
- Pandey, G.K.; Cheong, Y.H.; Kim, K.-N.; Grant, J.J.; Li, L.; Hung, W.; D’Angelo, C.; Weinl, S.; Kudla, J.; Luan, S. The Calcium Sensor Calcineurin B-Like 9 Modulates Abscisic Acid Sensitivity and Biosynthesis in Arabidopsis. Plant Cell 2004, 16, 1912–1924. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.; Park, J.; Ryu, M.; Yoon, I.S.; Kim, K.-N. Calcineurin B-like proteins in rice can bind with calcium ion and associate with the Arabidopsis CIPK family members. Plant Sci. 2009, 177, 577–583. [Google Scholar] [CrossRef]
- Mahajan, S.; Sopory, S.K.; Tuteja, N. Cloning and characterization of CBL-CIPK signalling components from a legume (Pisum sativum). FEBS J. 2006, 273, 907–925. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.F.; Yu, A.L.; Tian, G.; Du, Y.W.; Guo, E.H.; Diao, X.M. Identification of CBL genes from foxtail millet (Setaria italica L. Beauv) and its expression under drought and salt stresses. Acta Agron. Sin. 2013, 39, 360. [Google Scholar] [CrossRef]
- Farani, T.; Gentile, A.; Tavares, R.; Ribeiro, C.; Menossi, M. Characterization of a protein-protein interaction network of the CBL-interacting protein kinase 8 from sugarcane. Genet. Mol. Res. 2015, 14, 483–491. [Google Scholar] [CrossRef]
- Ling, Q.; Zeng, Q.; Wu, J.; Hu, F.; Li, Q.; Qi, Y. Analysis of CBL1 and CBL6 gene expression in sugarcane under abiotic stress. Mol. Plant Breed. 2018, 16, 377–385. [Google Scholar] [CrossRef]
- Ling, Q.P.; Wu, J.Y.; Yang, Z.D.; Zhou, W.L.; Huang, Y.; Li, Q.W.; Zeng, Q.Y. Cloning and Expression Characteristic Analysis of SsCBL4 gene from Sugarcane (Saccharum spp.). J. Agric. Bio. 2019, 27, 1351–1359. [Google Scholar]
- Su, W.; Huang, L.; Ling, H.; Mao, H.; Huang, N.; Su, Y.; Ren, Y.; Wang, D.; Xu, L.; Muhammad, K.; et al. Sugarcane calcineurin B-like (CBL) genes play important but versatile roles in regulation of responses to biotic and abiotic stresses. Sci. Rep. 2020, 10, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, V.; Weinl, S.; Blazevic, D.; D’Angelo, C.; Batistic, O.; Kolukisaoglu, Ü.; Bock, R.; Schulz, B.; Harter, K.; Kudla, J. The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 2003, 36, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Barrena, M.J.; Martinez-Ripoll, M.; Albert, A. Structural Biology of a Major Signaling Network that Regulates Plant Abiotic Stress: The CBL-CIPK Mediated Pathway. Int. J. Mol. Sci. 2013, 14, 5734–5749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, L.; Riggs, W. Nourishing urbanism: A case for a new urban paradigm. Int. J. Agric. Sustain. 2010, 8, 116–126. [Google Scholar] [CrossRef]
- Pang, X.; Zhu, P.; Zhou, Q.; Huang, Q.; Tan, Q.; Chun, L. Research progress on sugarcane response to cold stress. Guizhou Agric. Sci. 2015, 43, 39–43. [Google Scholar]
- Zhu, P.; Pang, X.; Tan, Q.; Liang, C.; Yan, L.; Zhou, Q. Effects of chilling on photosynthesis and photosynthetic pigment contents in leaves of different sugarcane varieties. Chin. J. Trop. Crop. 2019, 40, 51–57. [Google Scholar]
- Ding, C.; Yang, Q.; Li, F.; Weifu, L. Effect of cold stress on the content of free-proline in leaf of Saccharum spontaneum L. and sclerostachya (II). J. Anhui Agric. Sci. 2006, 34, 30–33. [Google Scholar] [CrossRef]
- Wang, H.; Jiao, X.; Kong, X.; Hamera, S.; Wu, Y.; Chen, X.; Fang, R.; Yan, Y. A Signaling Cascade from miR444 to RDR1 in Rice Antiviral RNA Silencing Pathway. Plant Physiol. 2016, 170, 2365–2377. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Liang, Y.; Zhang, B.; Song, X.; Li, Y.; Qin, Z.; Li, D.; Chen, R.; Zhou, Z.; Deng, Y.; et al. Integration of Transcriptional and Post-transcriptional Analysis Revealed the Early Response Mechanism of Sugarcane to Cold Stress. Front. Genet. 2021, 11, 1715. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Su, Y.; Zou, J.; Wang, Z.; Xu, L.; Que, Y. miRNA alteration is an important mechanism in sugarcane response to low-temperature environment. BMC Genom. 2017, 18, 833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Wu, Q.; Teng, L.; Tang, F.; Pi, Z.; Shen, S. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015, 15, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Byrns, B.; Badawi, M.A.; Diallo, A.B.; Danyluk, J.; Sarhan, F.; Laudencia-Chingcuanco, D.; Zou, J.; Fowler, D.B. Transcriptomic Insights into Phenological Development and Cold Tolerance of Wheat Grown in the Field. Plant Physiol. 2017, 176, 2376–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Batistic, O.; Kudla, J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar] [CrossRef]
- Fuglsang, A.T.; Guo, Y.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K.S.; et al. Arabidopsis Protein Kinase PKS5 Inhibits the Plasma Membrane H+-ATPase by Preventing Interaction with 14-3-3 Protein. Plant Cell 2007, 19, 1617–1634. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Li, Q.-H.; Yu, Y.-N.; Qiao, Y.-M.; Haq, S.; Gong, Z.-H. The CBL–CIPK Pathway in Plant Response to Stress Signals. Int. J. Mol. Sci. 2020, 21, 5668. [Google Scholar] [CrossRef]
- Chen, P.; Yang, J.; Mei, Q.; Liu, H.; Cheng, Y.; Ma, F.; Mao, K. Genome-Wide Analysis of the Apple CBL Family Reveals That Mdcbl10.1 Functions Positively in Modulating Apple Salt Tolerance. Int. J. Mol. Sci. 2021, 22, 12430. [Google Scholar] [CrossRef]
- Jiang, X.-S.; Wassif, C.; Backlund, P.S.; Song, L.; Holtzclaw, L.; Li, Z.; Yergey, A.L.; Porter, F.D. Activation of Rho GTPases in Smith–Lemli–Opitz syndrome: Pathophysiological and clinical implications. Hum. Mol. Genet. 2010, 19, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.Q. Study on the Physiological and Biochemical Characteristics and Differentially Expressed Proteome during Late Growth Stage of Sugarcane under Cold Stress. Ph.D. Thesis, Guangxi University, Nanning, China, 2013. [Google Scholar]
- Li, H.; Cong, Y.C.; Chang, Y.; Lin, J.; Sheng, B.L. Isolation of a calcineurin b-like protein gene PbCBL2 from Pyrus betulaefolia and preliminary study of gene function. Acta Hortic. Sin. 2013, 40, 1445–1455. [Google Scholar]

| Primer | Sequence (5′-3′) | Size of Fragment (bp) |
|---|---|---|
| CBL1-F | ATGGGGTGCTTCCATTCCACGGCGA | 642 |
| CBL1-R | TCACGTGACGAGATCGTC(C/G) ACTTC | |
| CBL3-F | ATG(T/G)TGCAGTGCCTGGA(T/C) GG | 678 |
| CBL3-R | TCAG(T/G)TATCATCGAC(T/C)TGAGA | |
| CBL5-F | ATGGGCTGTCTGCAAACAAAGCACG | 657 |
| CBL5-R | TTAGACAGCCATGTCTGTTTC | |
| CBL6-F | ATGGTGGACTT(T/C) GTTCGACGGCT | 672 |
| CBL6-R | TCACGCATCCTCTACTTG(A/C) GAGTTG | |
| CBL9-F | ATGGG(A/G) TGCTTCCATTCCACGGC | 693 |
| CBL9-R | TCACGTGACGAGATC(A/G) TC(G/C) ACTTC |
| Primer | Sequence (5′-3′) | Size of Fragment (bp) |
|---|---|---|
| GAPDH-F | AAGGGTGGTGCCAAGAAGG | 145 |
| GAPDH-R | CAAGGGGAGCAAGGCAGTT | |
| CBL1-F | TATGGATGGCACAGGGTTTATT | 131 |
| CBL1-R | CAGCGTCCGAAAATGTCTTATC | |
| CBL3-F | AGCAAGAAGGAGAGCCTGTTC | 123 |
| CBL3-R | AAGGGGAGCATTAGGATGAAAT | |
| CBL5-F | ACACAAAAGGTGATGGGAAGAT | 98 |
| CBL5-R | CTTGAGGTAGGGAAGGGTCATA | |
| CBL6-F | GTGGTTGATGATGGCTTGATTA | 172 |
| CBL6-R | CACTTGGATGGAACACAGAAAG | |
| CBL9-F | TATGGATGGCACAGGGTTTATT | 131 |
| CBL9-R | CAGCGTCCGAAAATGTCTTATC |
| Protein | Accession Number | MW | pI |
|---|---|---|---|
| SoCBL1 | AGO81718.1 | 24.5 kD | 4.6 |
| SoCBL3 | AGO81719.1 | 25.9 kD | 4.6 |
| SoCBL5 | AGO81720.1 | 25.2 kD | 5.0 |
| SoCBL6 | AGO81721.1 | 25.6 kD | 4.7 |
| SoCBL9 | AGO81722.1 | 26.3 kD | 4.5 |
| Protein | Casein Kinase II Phosphorylation Site | N-Myristoylation Site | Phosphorylation Site | N-Glycosylation Site | CAMP- and cGMP-Dependent Protein Kinase Phosphorylation Site | EF Calcium-Binding Domain |
|---|---|---|---|---|---|---|
| SoCBL1 | 28~31, 45~48, 151~154, 156~159, 171~174, 206~209 | 2~7 | 7~9, 112~114 | 161~173, 67~102, 104~139, | ||
| SoCBL3 | 42~45, 79~82, 150~153, 165~168, 170~173, 176~179, 220~223 | 155~160 | 157~160, 200~203 | 54~57, 80~83 | 79~114, 116~151, 160~195 | |
| SoCBL5 | 31~34, 106~109, 143~146, 165~168, 212~215 | 2~7 | 67~69 | 189~192 | 43~46 | 70~105, 107~142, 151~186 |
| SoCBL6 | 148~151, 168~171, 174~177, 218~221 | 153~158 | 76~78 | 155~158, 198~201 | 52~55, 78~81 | 79~114, 116~151, 160~195 |
| SoCBL9 | 45~48, 62~65, 168~171, 173~176, 188~191, 223~226 | 19~24 | 24~26, 129~131 | 6~9 | 84~119, 121~156, 165~200 |
| Protein | Number of α-Helices | Number of Extended Strands | Number of β-Turns | Number of Random Coils |
|---|---|---|---|---|
| SoCBL1 | 106 (49.8%) | 16 (7.5%) | 14 (6.6%) | 77 (36.2%) |
| SoCBL3 | 126 (56.0%) | 13 (5.8%) | 18 (8.9%) | 68 (30.2%) |
| SoCBL5 | 110 (50.5%) | 23 (10.6%) | 14 (7.2%) | 71 (32.7%) |
| SoCBL6 | 121 (54.3%) | 13 (5.8%) | 16 (7.2%) | 73 (32.7%) |
| SoCBL9 | 118 (51.3%) | 19 (8.3%) | 16 (7.0%) | 77 (33.5%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.-Q.; Song, X.-P.; Zhang, X.-Q.; Huang, Y.-X.; Liang, Y.-J.; Zhou, S.; Yang, C.-F.; Yang, L.-T.; Huang, X.; Li, Y.-R. Differential Gene Expression Analysis of SoCBL Family Calcineurin B-like Proteins: Potential Involvement in Sugarcane Cold Stress. Genes 2022, 13, 246. https://doi.org/10.3390/genes13020246
Zhang B-Q, Song X-P, Zhang X-Q, Huang Y-X, Liang Y-J, Zhou S, Yang C-F, Yang L-T, Huang X, Li Y-R. Differential Gene Expression Analysis of SoCBL Family Calcineurin B-like Proteins: Potential Involvement in Sugarcane Cold Stress. Genes. 2022; 13(2):246. https://doi.org/10.3390/genes13020246
Chicago/Turabian StyleZhang, Bao-Qing, Xiu-Peng Song, Xiao-Qiu Zhang, Yu-Xin Huang, Yong-Jian Liang, Shan Zhou, Cui-Fang Yang, Li-Tao Yang, Xing Huang, and Yang-Rui Li. 2022. "Differential Gene Expression Analysis of SoCBL Family Calcineurin B-like Proteins: Potential Involvement in Sugarcane Cold Stress" Genes 13, no. 2: 246. https://doi.org/10.3390/genes13020246
APA StyleZhang, B.-Q., Song, X.-P., Zhang, X.-Q., Huang, Y.-X., Liang, Y.-J., Zhou, S., Yang, C.-F., Yang, L.-T., Huang, X., & Li, Y.-R. (2022). Differential Gene Expression Analysis of SoCBL Family Calcineurin B-like Proteins: Potential Involvement in Sugarcane Cold Stress. Genes, 13(2), 246. https://doi.org/10.3390/genes13020246






