Marker-Assisted Pyramiding of CRa and CRd Genes to Improve the Clubroot Resistance of Brassica rapa
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant and P. brassicae Materials
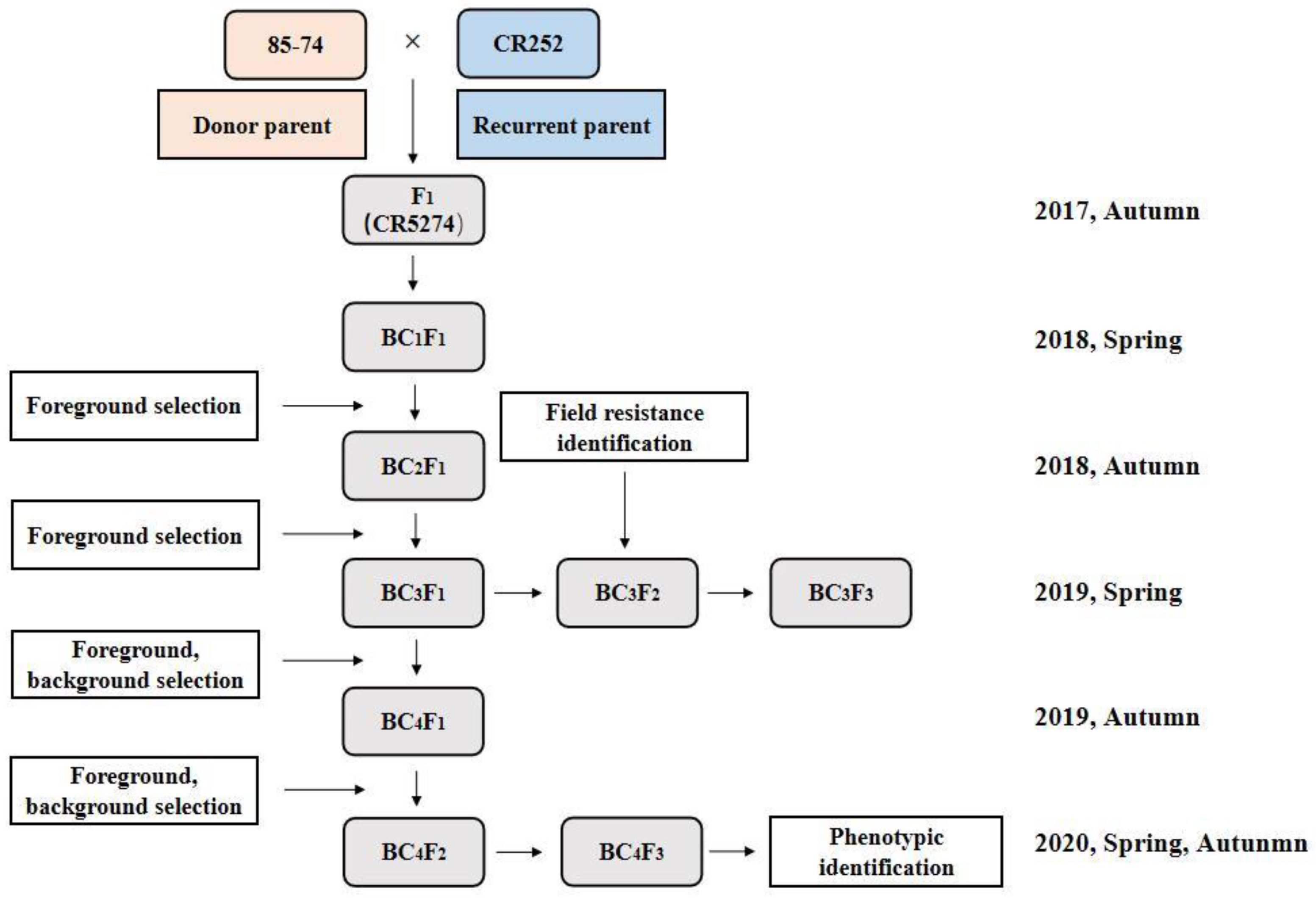
2.2. Breeding Program for Gene Pyramiding
2.3. Development and Validation of Polymorphic Markers between Parents
2.4. Investigation of the Phenotypic Characteristics
3. Results
3.1. Polymorphic-Marker Screening for Foreground Selection
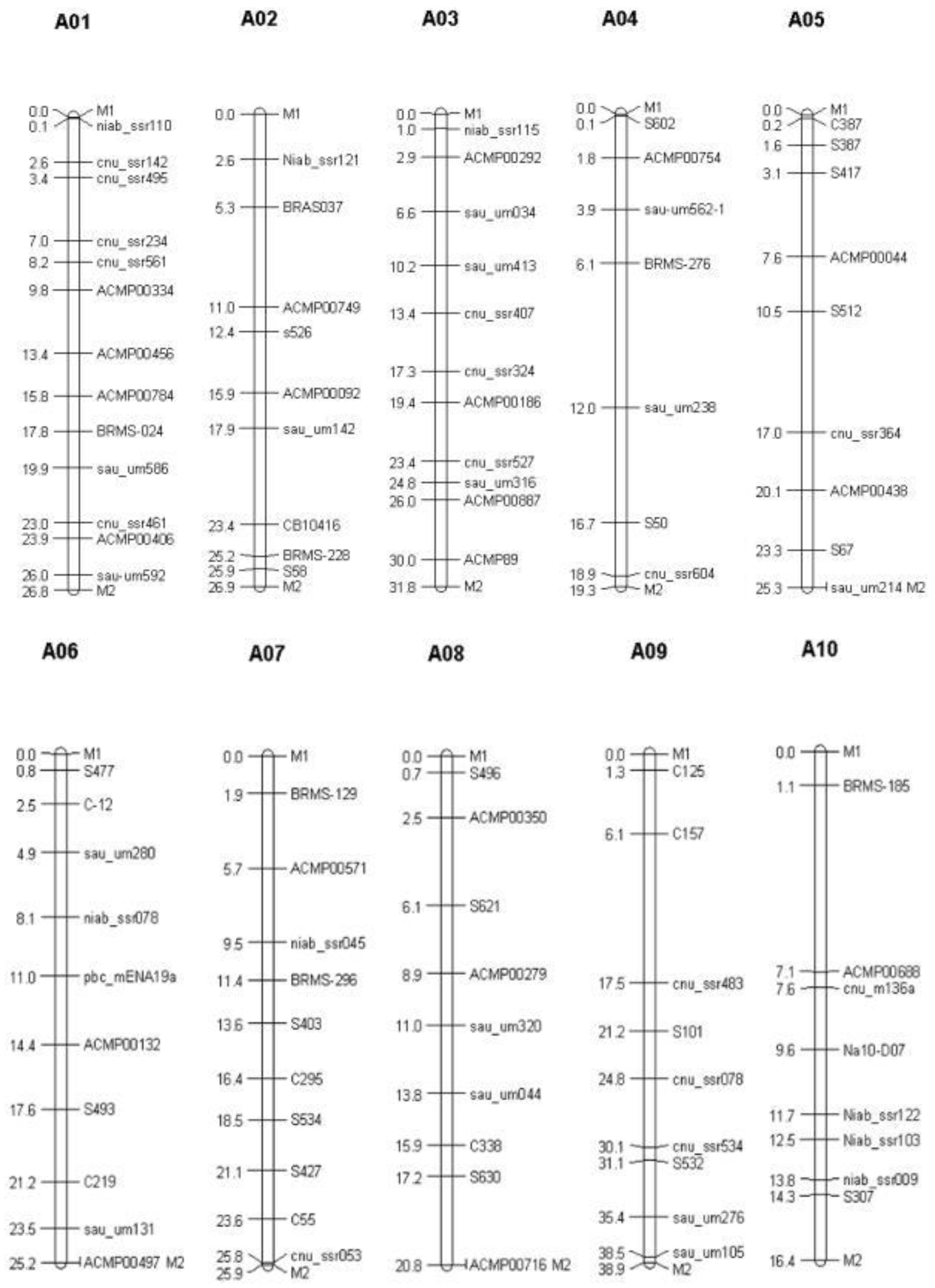
3.2. Polymorphic Markers’ Screening for Background Selection
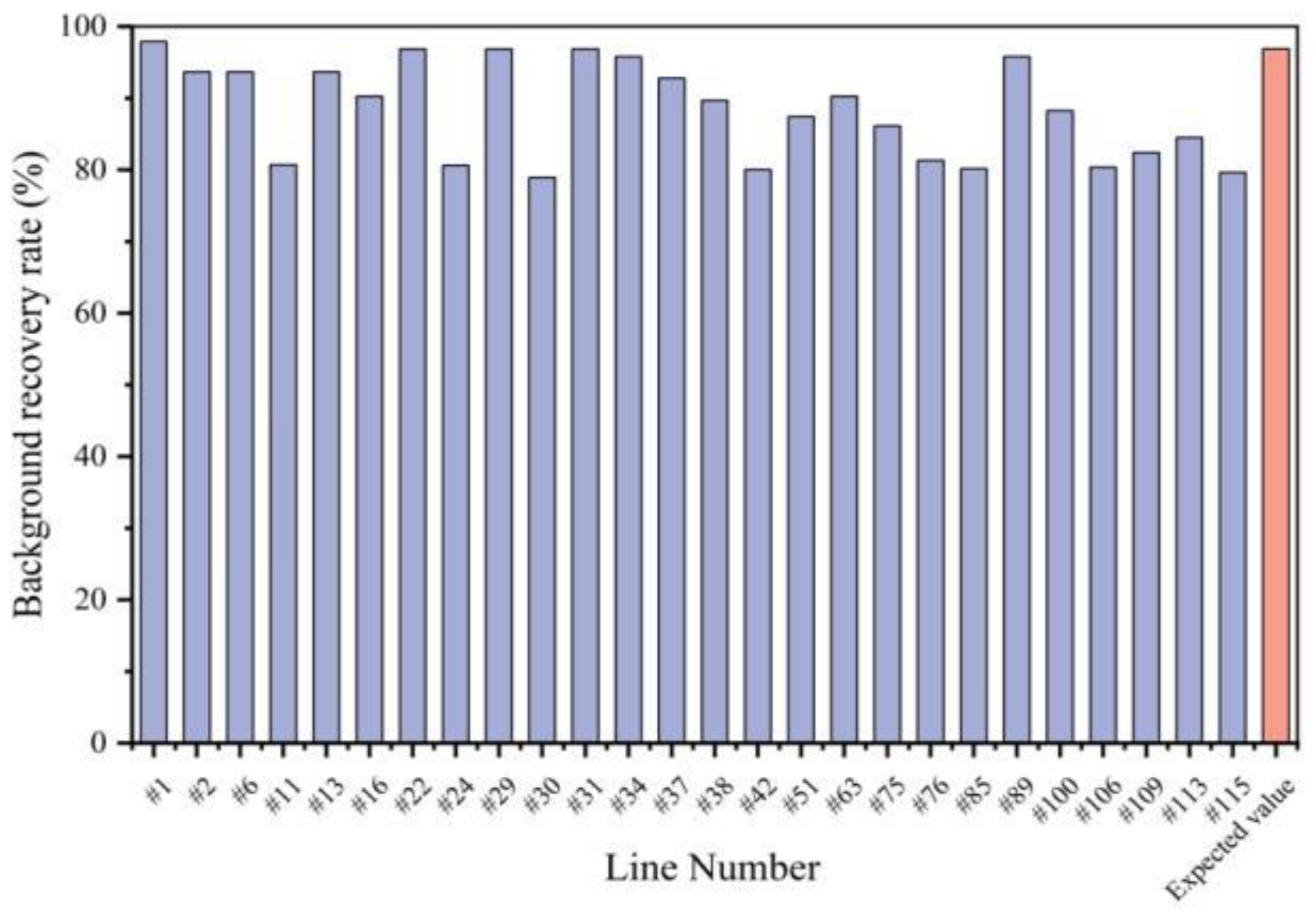
3.3. Foreground and Background Selection for Each Generation
3.4. Clubroot-Resistance Identification and Morphological-Trait Evaluation of Pyramided Line
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crute, I.R.; Gray, A.R.; Crisp, P.; Buczacki, S.T. Variation in Plasmodiophora brassicae and resistance to clubroot disease in brassicas and allied crops—A critical review. Plant Breed Abstr. 1980, 50, 91–104. [Google Scholar] [CrossRef]
- Dixon, G.R. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Donald, C.; Porter, I. Integrated Control of Clubroot. J. Plant Growth Regul. 2009, 28, 289–303. [Google Scholar] [CrossRef]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Matsumoto, S.; Hirai, M. Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 2003, 107, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. Plasmodiophora brassicae: Aspects of pathogenesis and resistance in Brassica oleracea. Euphytica 1995, 83, 139–146. [Google Scholar] [CrossRef]
- Mehraj, H.; Akter, A.; Miyaji, N.; Miyazaki, J.; Shea, D.J.; Fujimoto, R.; Doullah, U. Genetics of clubroot and fusarium wilt disease resistance in Brassica Vegetables: The application of marker assisted breeding for disease resistance. Plants 2020, 9, 726. [Google Scholar] [CrossRef]
- Suwabe, K.; Suzuki, G.; Nunome, T.; Hatakeyama, K.; Mukai, Y.; Fukuoka, H.; Matsumoto, S. Microstructure of a Brassica rapa genome segment homoeologous to the resistance gene cluster on Arabidopsis chromosome 4. Breed. Sci. 2012, 62, 170–177. [Google Scholar] [CrossRef]
- Hirai, M.; Harada, T.; Kubo, N.; Tsukada, M.; Suwabe, K.; Matsumoto, S. A novel locus for CR in Brassica rapa and its linkage markers. Theor. Appl. Genet. 2004, 108, 639–643. [Google Scholar] [CrossRef]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Kondo, M.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Hirai, M.; Matsumoto, S. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: The genetic origin of clubroot resistance. Genetics 2006, 173, 309–319. [Google Scholar] [CrossRef]
- Matsumoto, E.; Yasui, C.; Ohi, M.; Tsukada, M. Linkage analysis of RFLP markers for CR and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). Euphytica 1998, 104, 79–86. [Google Scholar] [CrossRef]
- Piao, Z.Y.; Deng, Y.Q.; Choi, S.R.; Park, Y.J.; Lim, Y.P. SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 2004, 108, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Saito, A.; Hayashida, N.; Taguchi, G.; Matsumoto, E. Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 2008, 117, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Fu, P.; Li, X.; Zhan, Z.; Yu, S.; Piao, Z. Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa ssp. pekinensis). Front. Plant Sci. 2018, 9, 653. [Google Scholar] [CrossRef]
- Chen, J.; Jing, J.; Zhan, Z.; Zhang, T.; Zhang, C.; Piao, Z. Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS ONE 2013, 8, e85307. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Song, T.; Falk, K.C.; Zhang, X.; Liu, X.; Chang, A.; Lahlali, R.; McGregor, L.; Gossen, B.D.; Yu, F.; et al. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genom. 2014, 15, 1166. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.Q.; Zhang, X.G.; Peng, G.; Falk, K.C.; Strelkov, S.E.; Gossen, B.D. Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 2017, 7, 4516. [Google Scholar] [CrossRef] [PubMed]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.O.; Okporie, E.O.; Onyishi, G.; Utobo, E.; Ekwu, L.G.; Swaray, S.; et al. Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.). Biotechnol. Biotechnol. Equip. 2019, 33, 440–455. [Google Scholar] [CrossRef]
- Bharani, M.; Nagarajan, P.; Rabindran, R.; Saraswathi, R.; Balasubramanian, P.; Ramalingam, J. Bacterial leaf blight resistance genes (Xa21, xa13 and xa5) py-ramiding through molecular marker-assisted selection into rice cultivars. Arch. Phytopathol. Plant Prot. 2010, 43, 1032–1043. [Google Scholar] [CrossRef]
- Fu, C.; Wu, T.; Liu, W.; Wang, F.; Li, J.; Zhu, X.; Huang, H.; Liu, Z.; Liao, Y.; Zhu, M.; et al. Genetic improvement of resistance to blast and bacterial blight of the elite maintainer line Rongfeng B in hybrid rice (Oryza sativa L.) by using marker-assisted selection. Afr. J. Biotechnol. 2012, 11, 13104–13124. [Google Scholar] [CrossRef]
- Kloppers, F.J.; Pretorius, Z.A. Effects of combinations amongst genes Lr13, Lr34 and Lr37 on components of resistance in wheat to leaf rust. Plant Pathol. 1997, 46, 737–750. [Google Scholar] [CrossRef]
- Shanti, M.L.; George, M.L.; Cruz, C.M.; Bernardo, M.A.; Nelson, R.J.; Leung, H.; Reddy, J.N.; Sridhar, R. Identification of resistance genes effective against rice bacterial blight pathogen in eastern India. Plant Dis. 2001, 85, 506–512. [Google Scholar] [CrossRef][Green Version]
- Singh, V.K.; Singh, A.P.; Singh, S.P.; Ellur, R.K.; Choudhary, V.; Sarkel, S.; Singh, D.; Krishnan, S.G.; Nagarajan, M.; Vinod, K.K.; et al. Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker-assisted backcross breeding. Field Crop. Res. 2012, 128, 8–16. [Google Scholar] [CrossRef]
- Prasanna, H.C.; Sinha, D.P.; Rai, G.K.; Krishna, R.; Kashyap, S.P.; Singh, N.K.; Singh, M.; Malathi, V.G. Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathol. 2014, 64, 256–264. [Google Scholar] [CrossRef]
- Matsumoto, E.; Ueno, H.; Aruga, D.; Sakamoto, K.; Hayashida, N. Accumulation of three CR genes through marker-assisted selection in Chinese cabbage (Brassica rapa ssp. pekinensis). J. Jpn. Soc. Hortic. Sci. 2012, 81, 184–190. [Google Scholar] [CrossRef]
- Wallenhammar, A.; Arwidsson, O. Detection of plasmodiophora brassicae by PCR in naturally infested soils. Eur. J. Plant Pathol. 2001, 107, 313–321. [Google Scholar] [CrossRef]
- Watson, I.A.; Singh, D. The future for rust resistant wheat in Australia. J. Aust. Inst. Agric. Sci. 1952, 18, 190–197. [Google Scholar]
- Hittalmani, S.; Parco, A.; Mew, T.V.; Zeigler, R.S.; Huang, N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 2000, 100, 1121–1128. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S.; Anandan, A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Sun, J.; Yu, S.; Yang, Z.; Wang, Z.; Huang, F.; Dun, B.C.; Gong, J.; Liu, Y.; Li, Y.; et al. Genetic variation analysis of field isolates of clubroot and their responses to Brassica napus lines containing resistant genes CRb and PbBa8.1 and their combination in homozygous and heterozygous state. Mol. Breed. 2019, 39, 53. [Google Scholar] [CrossRef]
- Tomita, H.; Shimizu, M.; Asad-Ud Doullah, M.; Fujimoto, R.; Okazaki, K. Accumulation of quantitative trait loci conferring broad-spectrum clubroot resistance in Brassica oleracea. Mol. Breed. 2013, 32, 889–900. [Google Scholar] [CrossRef]
- Pang, W.; Liang, Y.; Zhan, Z.; Li, X.; Piao, Z. Development of a Sinitic clubroot differential set for the pathotype classification of Plasmodiophora brassicae. Front. Plant Sci. 2020, 11, 568771. [Google Scholar] [CrossRef]
- Li, X.; Ramchiary, N.; Choi, S.R.; Van Nguyen, D.; Hossain, M.J.; Yang, H.; Lim, Y.P. Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome 2010, 53, 939–947. [Google Scholar] [CrossRef]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Mapping quantitative trait loci for leaf and heading-related traits in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Environ. Biotechnol. 2011, 52, 494–501. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, V.K.; Singh, S.P.; Pandian, R.T.P.; Ellur, R.K.; Singh, D.; Prolay, K.; Bhowmick, S.; Gopala Krishnan, M.; Nagarajan, K.K.; et al. Molecular breeding for the development of multiple disease resistance in Basmati rice. AoB Plants 2012, 2012, pls029. [Google Scholar] [CrossRef]
- Das, G.; Rao, G.J.N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef]
- Ramalingam, J.; Raveendra, C.; Savitha, P.; Vidya, V.; Chaithra, T.L.; Velprabakaran, S.; Saraswathi, R.; Ramanathan, A.; Pillai, M.P.A.; Arumugachamy, S.; et al. Gene pyramiding for achieving enhanced resistance to bacterial blight, blast, and sheath blight diseases in rice. Front. Plant Sci. 2020, 11, 591457. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Chiu, C.H.; Yap, R.; Tseng, Y.C.; Wu, Y.P. Pyramiding bacterial blight resistance genes in Tainung82 for broad spectrum resistance using marker-assisted selection. Int. J. Mol. Sci. 2020, 21, 1281. [Google Scholar] [CrossRef]
- Yamanaka, N.; Hossain, M. Pyramiding three rust-resistance genes confers a high level of resistance in soybean (Glycine max). Plant Breed. 2019, 138, 686–695. [Google Scholar] [CrossRef]
- Shi, A.; Chen, P.; Zheng, C.; Hou, A.; Zhu, S. Gene Pyramiding for Soybean Mosaic Virus resistance using microsatellite markers. Int. Meet. ASA-CSSA-SSSA 2006, 13. [Google Scholar] [CrossRef]
- Hanson, P.; Lu, S.F.; Wang, J.F.; Chen, W.; Kenyon, L.; Tan, C.W.; Tee, K.L.; Wang, Y.Y.; Hsu, Y.C.; Schafleitner, R.; et al. Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Sci. Hortic. 2016, 201, 346–354. [Google Scholar] [CrossRef]
- Arunakumari, K.; Durgarani, C.V.; Satturu, V.; Sarikonda, K.R.; Chittoor, P.D.R.; Vutukuri, B.; Laha, G.S.; Nelli, A.P.K.; Gattu, S.; Jamal, M.; et al. Marker-assisted pyramiding of genes conferring resistance against bacterial blight and blast diseases into Indian rice variety MTU1010. Rice Sci. 2016, 23, 306–316. [Google Scholar] [CrossRef]
- Jamaloddin, M.; Durga Rani, C.V.; Swathi, G.; Anuradha, C.; Vanisri, S.; Rajan, C.P.C.; Krishnam Raju, S.; Bhuvaneshwari, V.; Jagadeeswar, R.; Laha, G.S.; et al. Marker assisted gene pyramiding (MAGP) for bacterial blight and blast resistance into mega rice variety “Tellahamsa”. PLoS ONE 2020, 15, e0234088. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Li, Q.; Xu, Q.; Liu, J.; Huang, F.; Zhan, Z.; Qin, P.; Zhou, X.; Yu, W.; Zhu, L.; et al. CRb and PbBa8.1 synergically increases resistant genes expression upon infection of Plamodiophora brassicae in Brassica napus. Genes 2020, 11, 202. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Pang, W.; Chen, B.; Zhang, C.; Piao, Z. Transcriptome analysis of Brassica rapa near-isogenic lines carrying clubroot-resistant and -susceptible alleles in response to Plasmodiophora brassicae during early infection. Front. Plant Sci. 2016, 6, 1183. [Google Scholar] [CrossRef]
- Cobb, J.N.; Biswas, P.S.; Platten, J.D. Back to the future: Revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 2019, 132, 647–667. [Google Scholar] [CrossRef]
- Chen, S.; Xu, C.; Lin, X.; Zhang, Q. Improving bacterial blight resistance of ’6078’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Plant Breed. 2001, 120, 133–137. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]

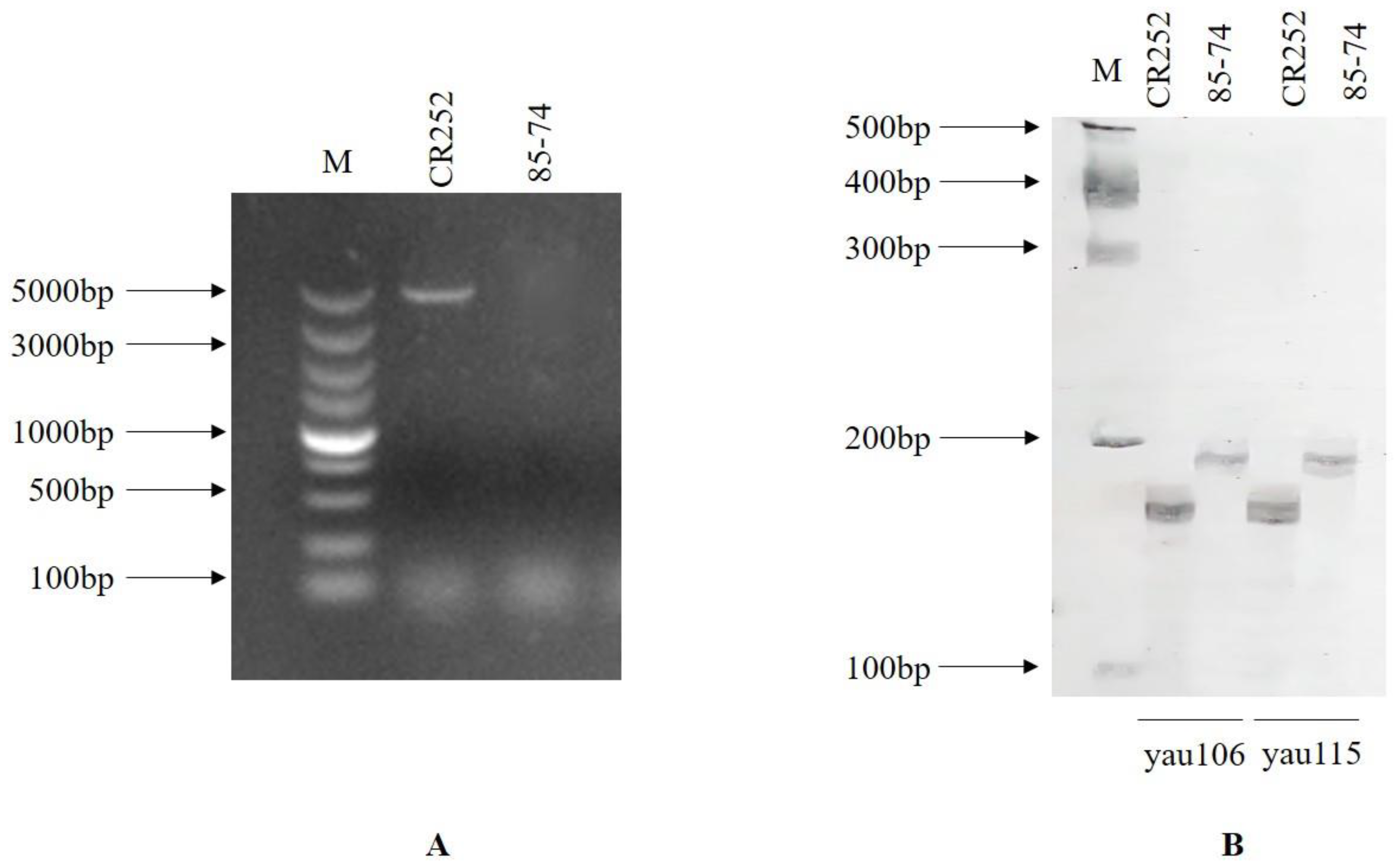
| Markers | CR Gene | Chromosome | Primer Sequence |
|---|---|---|---|
| yau106 | CRd | A3 | F: GGTCACCAATCGAAGCCTTT R: GCATGCGGGTATACACATCT |
| yau115 | CRd | A3 | F: CCGTTTGGTTTCCCTTGCAA R: GTTTAACACAGCAACAACAATGG |
| CRa full-length | CRa | A3 | F: ATGGATTTCTCTCTTTTCC R: TTAACATGAGGGAGTTTCCAG |
| Total | Level 0 | Level 1 | Level 2 | Level 3 | Incidence Index | |
|---|---|---|---|---|---|---|
| 85–74 | 34 | 34 | 0 | |||
| CR252 | 30 | 30 | 100 | |||
| F1 (CR5274) | 60 | 60 | 0 |
| Total | Level 0 | Level 1 | Level 2 | Level 3 | Resistance: Susceptibility | Theoretical Ratio | χ2 | |
|---|---|---|---|---|---|---|---|---|
| CR5274BC1F1 | 60 | 28 | 6 | 26 | 28:32 | 1:1 | 0.27 | |
| CR5274F(BC4F2) | 165 | 116 | 4 | 8 | 37 | 116:49 | 3:1 | 0.49 |
| Total | Level 0 | Level 1 | Level 2 | Level 3 | Resistance: Susceptibility | Theoretical Ratio | χ2 | |
|---|---|---|---|---|---|---|---|---|
| CR252 | 8 | 8 | - | - | - | |||
| 85-74 | 8 | 8 | ||||||
| CR5274F(BC4F2) * | 67 | 39 | 10 | 3 | 15 | 49:18 | 3:1 | 0.12 |
| CR5274①-4 | 15 | 15 | ||||||
| CR5274⑥-2 | 15 | 15 | ||||||
| CR5274⑥-4 | 14 | 14 | ||||||
| CR5274⑥-8 | 15 | 15 | ||||||
| CR5274⑩-4 | 15 | 15 |
| Materials | Level | 85–74 | CR252 | CR5274⑥-4 | CR5274⑥-8 |
|---|---|---|---|---|---|
| LNND-2 (Pb3) | 0 | 28 | 30 | 29 | |
| (Pb3) | 1 | ||||
| 2 | |||||
| 3 | 23 | ||||
| CQPL-99 (Pb4) | 0 | 18 | 29 | 30 | |
| (Pb12) | 1 | ||||
| 2 | 3 | ||||
| 3 | 20 | ||||
| SCXC-60 (Pb5) | 0 | 19 | 27 | 22 | |
| (Pb5) | 1 | ||||
| 2 | |||||
| 3 | 16 | ||||
| YNTC-75 (Pb8) | 0 | 15 | 28 | 26 | |
| (Pb8) | 1 | ||||
| 2 | |||||
| 3 | 22 | ||||
| LAB-5 (Pb11) | 0 | 20 | 26 | 30 | |
| (Pb9) | 1 | ||||
| 2 | |||||
| 3 | 17 |
| Agronomic Traits | CR252 | CR5274⑥-4 | CR5274⑥-8 |
|---|---|---|---|
| Plant height (cm) | 34.00 ± 2.64 | 33.33 ± 2.08 | 31.00 ± 1.73 |
| Plant width (cm) | 49.33 ± 2.30 | 48.00 ± 2.64 | 44.33 ± 2.04 |
| Plant weight (g) | 2.07 ± 0.18 | 2.35 ± 0.17 | 2.07 ± 0.18 |
| Number of outer leaves | 9.00 ± 1.00 | 9.00 ± 3.00 | 9.66 ± 1.52 |
| Leaf length (cm) | 32.83 ± 2.51 | 33.66 ± 2.84 | 32.00 ± 2.64 |
| Petiole length (cm) | 18.50 ± 1.50 | 19.83 ± 1.25 | 19.50 ± 3.50 |
| Leaf width (cm) | 19.33 ± 2.88 | 19.33 ± 1.52 | 20.00 ± 2.00 |
| Petiole width (cm) | 5.30 ± 0.57 | 5.83 ± 0.28 | 6.00 ± 0.50 |
| Petiole color | Green | Green | Green |
| Head weight (g) | 1.12 ± 0.08 | 1.18 ± 0.01 | 1.14 ± 0.141 |
| Head shape | Folded | Folded | Folded |
| Head solidity | Compaction | Compaction | Compaction |
| Head length (cm) | 23.30 ± 1.13 | 23.33 ± 1.52 | 23.66 ± 1.52 |
| Head width (cm) | 11.50 ± 1.32 | 12.00 ± 1.00 | 12.66 ± 0.57 |
| Stem height (cm) | 3.50 ± 0.50 | 4.5 ± 1.32 | 3.00 ± 0.00 |
| Stem width (cm) | 2.93 ± 0.11 | 3.06 ± 0.11 | 2.76 ± 0.40 |
| Head color | Light yellow | Light yellow | Light yellow |
| Growth period (day) | 80d ± 0.00 | 80d ± 0.00 | 80d ± 0.00 |
| Pod Length (cm) | Pod Width (cm) | Number of Seeds per Pod | Hundred-Grain Weight (g) | |
|---|---|---|---|---|
| CR5274-1 | 5.20 ± 0.26 | 0.50 ± 0.00 | 12.00 ± 2.00 | 0.34 ± 0.00 |
| CR5274-2 | 5.10 ± 0.20 | 0.53 ± 0.05 | 11.33 ± 1.52 | 0.34 ± 0.11 |
| CR5274-3 | 5.13 ± 0.11 | 0.60 ± 0.00 | 12.66 ± 0.57 | 0.35 ± 0.11 |
| CR5274-6 | 5.10 ± 0.26 | 0.53 ± 0.05 | 11.66 ± 2.30 | 0.35 ± 0.15 |
| CR5274-22 | 5.00 ± 0.20 | 0.53 ± 0.05 | 12.33 ± 2.51 | 0.35 ± 0.26 |
| CR5274-29 | 5.03 ± 0.15 | 0.50 ± 0.00 | 11.00 ± 1.00 | 0.34 ± 0.10 |
| CR5274-31 | 5.03 ± 0.05 | 0.60 ± 0.00 | 12.33 ± 1.52 | 0.35 ± 0.10 |
| CR5274-34 | 5.13 ± 0.15 | 0.53 ± 0.05 | 12.00 ± 2.00 | 0.35 ± 0.15 |
| CR252 | 5.10 ± 0.30 | 0.5 ± 0.05 | 12.00 ± 2.00 | 0.34 ± 0.20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wei, Y.; Ma, Y.; Cao, G.; Ma, S.; Zhang, T.; Zhan, Z.; Piao, Z. Marker-Assisted Pyramiding of CRa and CRd Genes to Improve the Clubroot Resistance of Brassica rapa. Genes 2022, 13, 2414. https://doi.org/10.3390/genes13122414
Li X, Wei Y, Ma Y, Cao G, Ma S, Zhang T, Zhan Z, Piao Z. Marker-Assisted Pyramiding of CRa and CRd Genes to Improve the Clubroot Resistance of Brassica rapa. Genes. 2022; 13(12):2414. https://doi.org/10.3390/genes13122414
Chicago/Turabian StyleLi, Xiaonan, Yingxia Wei, Yingmei Ma, Guizhu Cao, Siwen Ma, Tianyu Zhang, Zongxiang Zhan, and Zhongyun Piao. 2022. "Marker-Assisted Pyramiding of CRa and CRd Genes to Improve the Clubroot Resistance of Brassica rapa" Genes 13, no. 12: 2414. https://doi.org/10.3390/genes13122414
APA StyleLi, X., Wei, Y., Ma, Y., Cao, G., Ma, S., Zhang, T., Zhan, Z., & Piao, Z. (2022). Marker-Assisted Pyramiding of CRa and CRd Genes to Improve the Clubroot Resistance of Brassica rapa. Genes, 13(12), 2414. https://doi.org/10.3390/genes13122414





