Abstract
Structural variations such as copy number variants (CNVs) have been associated with multiple autoimmune diseases. In this study, we explored the association of the Fc gamma receptor 3B gene (FCGR3B) copy number variation (CNV) with rheumatoid arthritis (RA) susceptibility and related serological traits in the Pakistani population. We also performed a meta-analysis of four published FCGR3B CNV studies along with the current study. A total of 927 subjects (597 RA cases, 330 healthy controls) were recruited from three rheumatology centers in Pakistan. Anti-cyclic citrullinated peptide (anti-CCP) antibodies and rheumatoid factor (RF) were measured in RA patients. FCGR3B copy number was assayed using the TaqMan® CN assay (Hs04211858_cn, Applied Biosystems, Foster City, CA, USA) and the copy number was estimated by using CopyCaller® software (version 2.1; Applied Biosystems, USA). Logistic regression was applied to calculate the odds ratio (OR) of RA risk associated with FCGR3B CNV using sex and age as covariates in R. Meta-analysis on four previously published studies and the current study was performed using the random-effect model. We observed a significant association between FCGR3B copy number < 2 and RA susceptibility (OR = 1.53; 95% CI: 1.05 to 2.22; p = 0.0259) and anti-CCP seropositivity (OR 2.56; 95% CI: 1.34 to 4.89; p = 0.0045). A non-significant association of FCGR3B copy number < 2 was also observed between increased rheumatoid factor (RF) seropositivity (OR = 1.74; 95% CI:0.93 to 3.26; p = 0.0816). Meta-analysis on 13,915 subjects (7005 RA cases and 6907 controls) also showed significant association of copy number < 2 with the increased risk of RA (OR = 1.30; 95% CI: 1.07 to 1.56; p = 0.00671). FCGR3B copy number < 2 is associated with increased RA risk and anti-CCP seropositivity.
1. Introduction
Rheumatoid arthritis (RA) is a chronic multisystem autoimmune disorder that shows a disparity in clinical presentation [1]. Hyperplasia of the synovial membrane due to the increased intrusion of inflammatory cells is the hallmark of RA [2]. It mainly affects the joints, leading to restricted mobility, deformity, and in the worst cases, early death [3]. The epidemiological data suggest that RA affects ~1% of the general population globally and ~0.5% in Pakistan [4]. The interplay of genetic and environmental factors is an important part of RA expression. Over the last two decades, many small-scale candidate gene association studies [5] and large-scale genome-wide association studies (GWAS) have identified more than 100 RA susceptibility loci [6]. In addition to single-nucleotide polymorphisms (SNPs), several studies have also reported a significant association of copy number variation (CNV) with RA susceptibility [7]. CNV has been defined as the structural genetic variation in DNA sequence (~1 kilobases or larger) present in an altered CN as compared to a reference genome. Copy number variants (CNVs) can be duplications or deletions [8]. Copy number variable regions may cover almost 12% (~360 mega-bases) of the human genome [9]. Many of these CNVs are present at high frequency (>5%) in the general population [10]. CNVs are understudied structural polymorphisms and an important potential source of variation in gene expression associated with altered phenotypes. Recent data suggest a key role of CNVs in numerous immune response-related genes in multiple autoimmune diseases, including RA [11,12].
The Fc receptor protein is present on the surface of immune cells and mediates numerous imperative functions, including the eradication of antibody-bound foreign pathogens [13]. The Fc gamma receptor (FcγR) is specific for the Fc portion of immunoglobulin G (IgG) [14]. FcγRs have different levels of affinity for different IgG subclasses. FcγRIIIb belongs to the family of low-affinity FcγRs and is encoded by the FCGR3B gene, which is present in a complex cluster containing five genes (FCGR2A, FCGR3A, FCGR2C, FCGR3B, FCGR2B) on chromosome 1q23.3 [15]. FCGR3B encodes a stimulatory receptor, and the neutrophil surface contains its highest expression [16]. FcγRIIIb is a glycosyl-phosphatidylinositol (GPI) associated receptor with no cytoplasmic domain. The exact function of FCGR3B in the immune system is not fully understood but recent studies suggest a potential role in neutrophil extracellular traps (NETs) formation [17]. The interaction of FcγRIIIb with multiple immune complexes and activation of neutrophils has suggested its significant role in multiple autoimmune diseases, including RA [18]. A low copy number (<2) of FCGR3B has been implicated in multiple autoimmune diseases, including systemic lupus erythematosus (SLE) [19], Sjogren’s syndrome [20], and RA [11].
Association studies of FCGR3B CNV with RA susceptibility have reported conflicting results [10,11,20,21,22,23,24,25]. Most of the previously published studies [11,21,22,23,24] reported significant association, and a few [10,20,25] reported no significant association of <2 FCGR3B CNV with RA susceptibility. The present study explores the FCGR3B CNV in RA case-control subjects recruited from the Pakistani population and then performs a meta-analysis with published studies.
2. Materials and Methods
2.1. Study Subjects
The study population comprised 597 RA cases and 330 healthy controls (927 subjects in total). Blood samples and related clinical data were collected after obtaining written informed consent from each subject at three rheumatology centers (Rehmat Noor Clinic, Military Hospital, and Pakistan Institute of Medical Sciences) in Pakistan. The study was also approved by the University of Pittsburgh IRB (IRB no. PRO12110472) in Pittsburgh, USA, where the samples were processed for genetic analysis. All the RA cases (mean age ± SD = 42.09 ± 12.06, 77.3% women) included in this study were diagnosed by rheumatologists following the American College of Rheumatology (ACR) 1987 classification criteria for RA [26]. All controls (mean age ± SD = 35.41 ± 12.62, 41.2% women) were recruited from the general population and were free from any autoimmune disease at the time of recruitment. All cases and controls were enrolled during the same period from September 2015 to May 2017. Of RA patients, 84% were positive for anti-cyclic citrullinated peptide (anti-CCP) antibodies and 83% were positive for rheumatoid factor (RF).
2.2. Genomic DNA Extraction
Whole blood was used to extract the genomic DNA by either the GeneJET Whole Blood Genomic DNA Purification kit (Thermo Scientific, Waltham, MA, USA) or standard phenol chloroform method, followed by DNA quantification using NanoDrop (Thermo Scientific, Waltham, MA, USA).
2.3. Measurement of FCGR3B CNV
The 384-well-dried DNA plates were used to run quantitative polymerase chain reaction (qPCR) for the measurement of FCGR3B CNV using commercially available TaqMan® CNV assay (Hs04211858_cn, FAM-MGB dual-labeled probe) together with RNaseP reference assay (4403326, VIC-TAMRA dual-labeled probe) in a duplex reaction, according to manufacturer’s protocol (Applied Biosystems, Thermo Scientific, Waltham, MA, USA). The location of the target site for the primer-probe set was within the fifth intron of FCGR3B (https://www.thermofisher.com/order/genome-database/details/cnv/Hs04211858_cn?CID=&ICID=&subtype=; accessed on 12 September 2022). The padded amplicon (target sequence plus extra nucleotides on both sides) is provided in the Supplementary info, where the context sequence surrounding the TaqMan® probe is shown in brackets: [AGGAGAACTAACTCAATGTAAACAT]. The qPCR was performed on a QuantStudio™ 12K Flex system (Applied Biosystems, Thermo Scientific, Waltham, MA, USA) and fluorescence signals were normalized to ROX reference dye. All samples were tested in quadruplicate. Each 384-well DNA plate contained an equal proportion of case and control samples with two additional reference samples. The qPCR data were analyzed using the 0.2 cycle threshold and Ct auto-baseline as recommended by the manufacturer. CNV assignments were made using the CopyCaller® software (version 2.1; Applied Biosystems, Thermo Scientific, Waltham, MA, USA). CopyCaller® software provides calculated and predicted (discrete) copy number assignments using qPCR data along with some quality control (QC) measures, like confidence estimate and Z-score, for each copy number call. Each plate was analyzed individually to minimize the effect of experimental plate-to-plate variations. Confidence estimate ≥ 90%, Z-score < 1.75, zero-copy ΔCT threshold, and exclusion of samples where reference assay (VIC-labeled RNaseP) had CT greater than 32.0 were used as QC measures during analyses. The CNVs were classified into three commonly observed groups: CNV = 2, CNV < 2, and CNV > 2. Rare instances of CNVs of 4 and 5 were also observed. CNVs were detected in a range of 0 to 5 copies of FCGR3B while having 2 copies being the common/normal occurrence. Those carrying duplications or deletions were assessed as compared to those carrying 2 copies of FCGR3B.
2.4. Statistical Analysis
To test the association between RA susceptibility and FCGR3B copy number, we performed a logistic regression with copy number represented as a 3-category (non-ordinal) variable. CNV = 2 was the reference group and was compared separately to the CNV < 2 and CNV > 2 groups. Age and sex were used as covariates. Odds ratios (ORs), 95% confidence intervals (CI), and the corresponding p values were calculated. The association of CNV with anti-cyclic citrullinated peptide (anti-CCP) and rheumatoid factor (RF) seropositivity was also assessed using logistic regression with age and sex included as covariates in a case-only analysis. All statistical analyses were performed using R version 4.0.2 (http://www.r-project.org; accessed on 1 October 2021). R code used for the analyses can be found online (https://github.com/MuhammadMuaazAslam/Copy-Number-Variation; accessed on 7 November 2022).
2.5. Meta-Analysis
We used four terms (Copy Number Variation, Rheumatoid Arthritis, Polymorphism, and FCGR3B) in different combinations to search for previously reported studies in PubMed (https://www.ncbi.nlm.nih.gov/pubmed/: accessed on 1 October 2021) and Google Scholar (https://scholar.google.com/: accessed on 1 October 2021). We also examined the references of the selected studies to identify additional reported data not indexed in the above-mentioned two online databases. While we did not apply any filters for geographical location, race/ethnicity, or linguistic group during our initial search, we focused on studies of European-descent subjects for our meta-analysis. Our search resulted in four studies in populations of European ancestry [10,23,24,27], showing either significant or no significant association of FCGR3B copy number < 2 versus = 2 with RA susceptibility. To ensure comparability, we focused on studies that reported data for <2 FCGR3B CN in comparison to 2 CN and excluded studies that reported < 2 CN in comparison to ≥2 CN. Information about population type, number of cases and controls, ORs, and p-values were collected for meta-analysis. Meta-analysis was performed using R version 4.0.2 (http://www.r-project.org: accessed on 1 October 2021) and ‘metafor’ version 2.1.0 package [28].
3. Results
For every sample, the CNV was classified into one of the three groups: less than two (CNV < 2), equal to two (CNV = 2), or greater than two (CNV > 2). No variation was found in three reference samples across all tested plates, implying no bias in the CNV assignment. Copy numbers of 0, 1, 2, 3, 4, and 5 were observed in 4.3%, 20.5%, 57.60%, 13.4%, 3.3%, and 0.76% of subjects, respectively. The distribution in the combined case-control sample was 57.60% (n = 534/927) for 2 copies, 24.81% (n = 230/927) for <2 copies, and 17.58% (n = 163/927) for >2 copies. CNV distribution in cases and controls is illustrated in Figure 1 and unrounded CNV distribution is presented in Supplementary Figures S1 and S2.
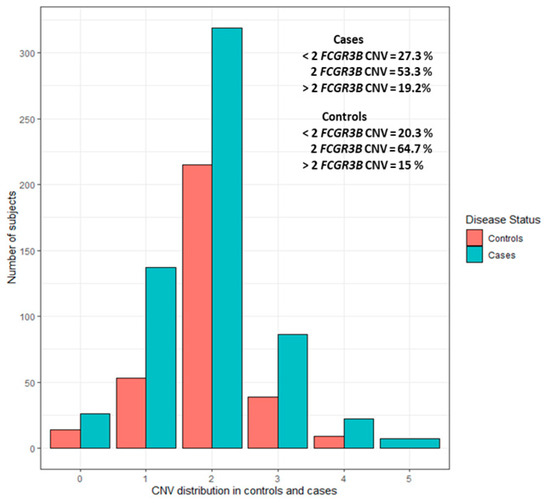
Figure 1.
CNV distribution in cases and controls separately.
Association results and distribution of three CNV groups (<2, 2, >2) between cases and controls are summarized in Table 1. The frequency of CNV < 2 was significantly higher in RA cases (27.30%; n = 163/597) as compared to controls (20.30%; n = 67/330) with an OR of 1.53 (95% CI: 1.05 to 2.22; p = 0.02589). CNV < 2 also showed a significant association with increased anti-CCP seropositivity in RA cases with an OR of 2.56 (95% CI: 1.34 to 4.89, p = 0.0045). However, only a trend of association was observed between CNV < 2 and RF seropositivity (OR = 1.74; 95% CI: 0.93 to 3.26; p = 0.0816) (Table 2). CNV > 2 showed no significant association with RA susceptibility (OR = 1.33; 95% CI: 0.87 to 2.03; p = 0.1851), anti-CCP (OR = 1.8 CI: 0.86 to 3.77; p = 0.117) or RF seropositivity (OR = 1.19; CI: 0.6 to 2.34; p = 0.6216).

Table 1.
Association results of FCGR3B CNV with RA susceptibility.

Table 2.
Association results of FCGR3B CNV with anti-CCP and RF seropositivity in RA patients.
We performed a meta-analysis with a restricted maximum-likelihood estimator in the random-effects model on 13,915 subjects (7005 RA cases and 6907 controls) from five studies including our sample (Figure 2). The model showed an I2 value of 44.2%, indicating sufficient homogeneity of the studies included in the meta-analysis. The meta-analysis confirmed the association of FCGR3B CNV < 2 and increased risk of RA (OR = 1.30; 95% CI: 1.07 to 1.56; p = 0.00671).
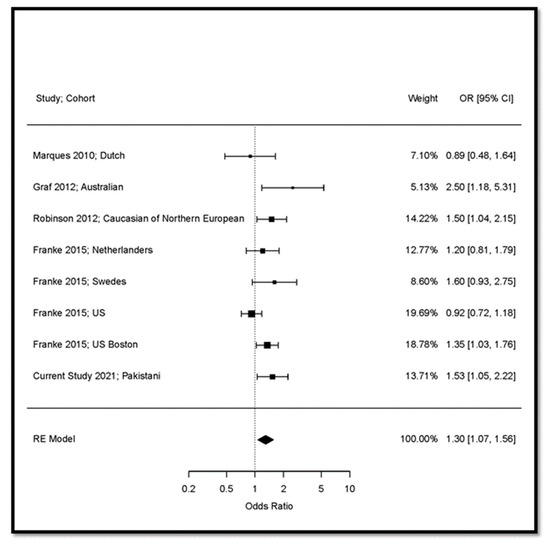
Figure 2.
Random effects meta-analysis of the association of FCGR3B CNV (<2 versus 2 copy numbers) with RA. US: United States of America; UK: United Kingdom; OR: Odds Ratio; RE: Random Effects; CI: Confidence Interval.
4. Discussion
Lengths of DNA segments vary due to structural variations, such as CNV between different individuals. CNVs are a key source of human genetic diversity and are well established as the heritable cause of numerous complex genetic diseases. The expression of a gene can be affected by CNVs. Gene dosage changes due to CNVs can lead to pathological conditions [29]. CNVs in various genomic regions have been linked with numerous diseases, including RA. CNVs in C4B [30], FCGR3B [23], CCL3L1 [31]), VPREB1 [12], and LCE3B [32] have been associated with RA susceptibility and related conditions. FCGR3B is located in chromosomal region 1q21–23, which is associated with autoimmune diseases. Low (<2) and high (>2) CNVs contributing to the altered gene expression of FCGR3B have also been associated with autoimmune phenotypes, including RA [22].
In our study, we investigated the association of FCGR3B CNV with RA susceptibility in 927 case-control subjects from the Pakistani population. To our knowledge, no such study has been carried out on this population. We found a significant association of RA risk with <2 copies of the FCGR3B CNV, while no significant association was observed with >2 copies. Previous studies did not find a significant association between >2 CN and RA susceptibility either. For CN < 2, our results are consistent with some previously published datasets, in which a significant association of CNV < 2 with RA susceptibility was also detected [11,21,22,23,24]. However, there has been an inconsistency in reports of FCGR3B CNV association with RA susceptibility, where some reported no associations [10,25]. FCGR3B lies in a complex genomic region, which is difficult to study. The discrepancies in earlier reported studies from multiple populations may be due to the complexity of the FCGR3B region or a lack of adequate study power. To address the power issues, we combined the results of four reported studies with the current study in a meta-analysis comprising 7005 RA cases and 6907 controls. The meta-analysis confirmed the association of CNV < 2 with RA risk (OR = 1.30; p = 0.00671).
To investigate a possible underlying mechanism of the association of CNV < 2 with RA, we also examined the association of CNV < 2 with the occurrence of anti-CCP antibodies, which is an important diagnostic marker for RA along with RF positivity. Interestingly, CNV < 2 was strongly associated with anti-CCP (OR = 2.56; p = 0.00453), while only a trend of association was observed with RF (OR = 1.74; p = 0.0816). While there is no reported significant association of CNV < 2 with anti-CCP antibodies, one study presented no association between low FCGR3B CNV and anti-CCP status in RA [23]. The possible explanation for this association could be the potentially impaired interaction of low FCGR3B with autoantigen-autoantibody immune complexes, ultimately contributing to autoimmunity. Alternatively, this may be a consequence of the CNVs’ extended effects on the neighboring gene(s), e.g., FCGR2C (involved in phagocytosis and immune complex clearance), rather than being a direct effect of the loss of FCGR3B [21]. Our findings with RF are consistent with previously reported studies that also found a trend for association (p = 0.08) or a nominally significant association (p = 0.02) [11]. Our results are also consistent with another previously published study by Franke et.al. [24] that reported a significant association of low FCGR3B CNV with an increased risk of antibody-positive RA in Caucasians.
As a potential limitation of our study, a small number of samples might have been misclassified regarding the FCGR3B CNV status using the qPCR approach due to the complex FCGR locus structure; however, considering the large number of samples analyzed, these putative misclassified samples are not expected to significantly affect our study results.
5. Conclusions
In summary, we investigated the association of FCGR3B CNV and RA susceptibility in Pakistanis and observed a significant association of CNV < 2 with an increased risk of RA risk. Our meta-analysis of 5 studies (4 published and the current one) confirmed this significant association. It appears that this genetic association with RA risk may be mediated, at least in part, by the association of CNV < 2 with anti-CCP antibodies, as CNV < 2 was also associated with increased levels of those antibodies. Additional studies on larger samples and comprehensive analyses of the entire FCGR locus are warranted to further explore and understand the association of FCGR3B CNV with RA and related clinical phenotypes. Such an understanding can advance our knowledge of underlying biological mechanisms, which in turn may lead to preventive measures and/or new treatments for RA patients.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes13122238/s1, Supplementary Text: Padded amplicon; Figure S1: Scatter plot of unrounded CNV. Cases and controls are color coded differently; Figure S2: Histogram of unrounded CNV.
Author Contributions
Conceptualization, M.M.A., F.Y.D. and M.I.K.; data curation, M.M.A., F.Y.D. and J.M.M.; formal analysis, M.M.A., F.Y.D., K.-H.F. and E.F.; funding acquisition, P.J. and M.I.K.; investigation, M.M.A., F.Y.D., M.I.K. and P.J.; methodology: M.I.K., F.Y.D., P.J. and E.F.; project administration, M.I.K. and F.Y.D.; resources, M.I.K., P.J. and J.M.M.; software, M.M.A., K.-H.F. and E.F.; supervision, M.I.K. and P.J.; validation, M.M.A., K.-H.F., M.I.K. and E.F.; visualization, K.-H.F. and M.M.A.; writing—original draft preparation, M.M.A.; writing—review and editing, M.M.A., F.Y.D., J.M.M., K.-H.F., E.F. and M.I.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of University of Pittsburgh, PA, USA (IRB Title = Genetic Studies of Chronic Diseases in Pakistan; Approval code = PRO12110472; Approval Date = 27 November 2012).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All the data reported in this study are presented here. The R code used to analyze the data can be found at an online resource (https://github.com/MuhammadMuaazAslam/Copy-Number-Variation).
Acknowledgments
We would like to acknowledge all the rheumatologists and their staff for their support for the study subjects’ recruitment. Further, we would like to acknowledge all the study participants for their willingness to contribute to this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Karami, J.; Aslani, S.; Jamshidi, A.; Garshasbi, M.; Mahmoudi, M. Genetic implications in the pathogenesis of rheumatoid arthritis; an updated review. Gene 2019, 702, 8–16. [Google Scholar] [CrossRef] [PubMed]
- McInnes, I.B.; Schett, G. The Pathogenesis of Rheumatoid Arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Alam, J.; Jantan, I.; Bukhari, S.N.A. Rheumatoid arthritis: Recent advances on its etiology, role of cytokines and pharmacotherapy. Biomed. Pharmacother. 2017, 92, 615–633. [Google Scholar] [CrossRef] [PubMed]
- Ullah, Z.; Ullah, M.I.; Hussain, S.; Kaul, H.; Lone, K.P. Determination of Serum Trace Elements (Zn, Cu, and Fe) in Pakistani Patients with Rheumatoid Arthritis. Biol. Trace Elem. Res. 2017, 175, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.M.; John, P.; Fan, K.-H.; Bhatti, A.; Aziz, W.; Ahmed, B.; Feingold, E.; Demirci, F.Y.; Kamboh, M.I. Investigating the GWAS-Implicated Loci for Rheumatoid Arthritis in the Pakistani Population. Dis. Markers 2020, 2020, 1910215. [Google Scholar] [CrossRef]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef]
- Kim, K.; Bang, S.-Y.; Lee, H.-S.; Bae, S.-C. Update on the genetic architecture of rheumatoid arthritis. Nat. Rev. Rheumatol. 2017, 13, 13–24. [Google Scholar] [CrossRef]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef]
- Marques, R.B.; Thabet, M.M.; White, S.J.; Houwing-Duistermaat, J.J.; Bakker, A.M.; Hendriks, G.-J.; Zhernakova, A.; Huizinga, T.W.; Van Der Helm-Van Mil, A.H.; Toes, R.E. Genetic Variation of the Fc Gamma Receptor 3B Gene and Association with Rheumatoid Arthritis. PLoS ONE 2010, 5, e13173. [Google Scholar] [CrossRef]
- McKinney, C.; Fanciulli, M.; Merriman, M.E.; Phipps-Green, A.; Alizadeh, B.Z.; Koeleman, B.P.C.; Dalbeth, N.; Gow, P.J.; Harrison, A.A.; Highton, J.; et al. Association of variation in Fc receptor 3B gene copy number with rheumatoid arthritis in Caucasian samples. Ann. Rheum. Dis. 2010, 69, 1711–1716. [Google Scholar] [CrossRef]
- Aslam, M.M.; John, P.; Fan, K.H.; Bhatti, A.; Feingold, E.; Demirci, F.Y.; Kamboh, M.I. Association of VPREB1 Gene Copy Number Variation and Rheumatoid Arthritis Susceptibility. Dis. Markers 2020, 2020, 7189626. [Google Scholar] [CrossRef]
- Daëron, M. Fc Receptor Biology. Annu Rev. Immunol. 1997, 15, 203–234. [Google Scholar] [CrossRef]
- Willcocks, L.C.; Lyons, P.A.; Clatworthy, M.R.; Robinson, J.I.; Yang, W.; Newland, S.A.; Plagnol, V.; McGovern, N.N.; Condliffe, A.M.; Chilvers, E.R.; et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J. Exp. Med. 2008, 205, 1573–1582. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Ravetch, J.V. Fcγ Receptors: Old Friends and New Family Members. Immunity 2006, 24, 19–28. [Google Scholar] [CrossRef]
- Ravetch, J.V. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J. Exp. Med. 1989, 170, 481–497. [Google Scholar] [CrossRef]
- Fetz, A.E.; Radic, M.Z.; Bowlin, G.L. Human neutrophil FcγRIIIb regulates neutrophil extracellular trap release in response to electrospun polydioxanone biomaterials. Acta Biomater. 2021, 130, 281–290. [Google Scholar] [CrossRef]
- Salmon, J.E.; Pricop, L. Human receptors for immunoglobulin G: Key elements in the pathogenesis of rheumatic disease. Arthritis Rheum. 2001, 44, 739–750. [Google Scholar] [CrossRef]
- Fanciulli, M.; Norsworthy, P.J.; Petretto, E.; Dong, R.; Harper, L.; Kamesh, L.; Heward, J.M.; Gough, S.C.L.; De Smith, A.; Blakemore, A.I.F.; et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat. Genet. 2007, 39, 721–723. [Google Scholar] [CrossRef]
- Mamtani, M.; Anaya, J.M.; He, W.; Ahuja, S.K. Association of copy number variation in the FCGR3B gene with risk of autoimmune diseases. Genes Immun. 2010, 11, 155–160. [Google Scholar] [CrossRef]
- Rahbari, R.; Zuccherato, L.W.; Tischler, G.; Chihota, B.; Ozturk, H.; Saleem, S.; Tarazona-Santos, E.; Machado, L.R.; Hollox, E.J. Understanding the Genomic Structure of Copy-Number Variation of the Low-Affinity Fcγ Receptor Region Allows Confirmation of the Association of FCGR3B Deletion with Rheumatoid Arthritis. Hum. Mutat. 2017, 38, 390–399. [Google Scholar] [CrossRef]
- Lee, Y.H.; Bae, S.C.; Seo, Y.H.; Kim, J.H.; Choi, S.J.; Ji, J.D.; Song, G.G. Association between FCGR3B copy number variations and susceptibility to autoimmune diseases: A meta-analysis. Inflamm. Res. 2015, 64, 983–991. [Google Scholar] [CrossRef]
- Graf, S.W.; Lester, S.; Nossent, J.C.; Hill, C.L.; Proudman, S.M.; Lee, A.; Rischmueller, M. Low copy number of the FCGR3B gene and rheumatoid arthritis: A case-control study and meta-analysis. Arthritis Res. Ther. 2012, 14, R28. [Google Scholar] [CrossRef]
- Franke, L.; el Bannoudi, H.; Jansen, D.T.S.L.; Kok, K.; Trynka, G.; Diogo, D.; Swertz, M.; Fransen, K.; Knevel, R.; Gutierrez-Achury, J.; et al. Association analysis of copy numbers of FC-γ receptor genes for rheumatoid arthritis and other immune-mediated phenotypes. Eur. J. Hum. Genet. 2016, 24, 263–270. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Wang, C.-M.; Chang, S.-W.; Cheng, C.-H.; Wu, Y.-J.J.; Lin, J.-C.; Yang, B.; Ho, H.-H.; Wu, J. Association of FCGR3A and FCGR3B copy number variations with systemic lupus erythematosus and rheumatoid arthritis in Taiwanese patients. Arthritis Rheumatol. 2014, 66, 3113–3121. [Google Scholar] [CrossRef]
- Arnett, F.C.; Edworthy, S.M.; Bloch, D.A.; McShane, D.J.; Fries, J.F.; Cooper, N.S.; Healey, L.A.; Kaplan, S.R.; Liang, M.H.; Luthra, H.S.; et al. The american rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988, 31, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.I.; Carr, I.M.; Cooper, D.L.; Rashid, L.H.; Martin, S.G.; Emery, P.; Isaacs, J.D.; Barton, A.; Wilson, A.G.; Barrett, J.H.; et al. Confirmation of association of FCGR3B but not FCGR3A copy number with susceptibility to autoantibody positive rheumatoid arthritis. Hum. Mutat. 2012, 33, 741–749. [Google Scholar] [CrossRef]
- Viechtbauer, W. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef]
- McCarroll, S.A.; Altshuler, D.M. Copy-number variation and association studies of human disease. Nat Genet. 2007, 39 (Suppl. 7), S37–S42. [Google Scholar] [CrossRef]
- Rigby, W.F.C.; Wu, Y.L.; Zan, M.; Zhou, B.; Rosengren, S.; Carlson, C.; Hilton, W.; Yu, C.Y. Increased frequency of complement C4B deficiency in rheumatoid arthritis. Arthritis Rheum. 2012, 64, 1338–1344. [Google Scholar] [CrossRef]
- Ben Kilani, M.S.; Achour, Y.; Perea, J.; Cornelis, F.; Bardin, T.; Chaudru, V.; Maalej, A.; Petit-Teixeira, E. Characterization of copy number variants for CCL3L1 gene in rheumatoid arthritis for French trio families and Tunisian cases and controls. Clin. Rheumatol. 2016, 35, 1917–1922. [Google Scholar] [CrossRef] [PubMed]
- Bergboer, J.G.M.; Umićević-Mirkov, M.; Fransen, J.; den Heijer, M.; Franke, B.; van Riel, P.L.C.M.; Schalkwijk, J.; Coenen, M.J.H.; on behalf of the Nijmegen Biomedical Study principal. A Replication Study of the Association between Rheumatoid Arthritis and Deletion of the Late Cornified Envelope Genes LCE3B and LCE3C. PLoS ONE 2012, 7, e32045. [Google Scholar] [CrossRef] [PubMed][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).