The Identification of Large Rearrangements Involving Intron 2 of the CDH1 Gene in BRCA1/2 Negative and Breast Cancer Susceptibility
Abstract
1. Introduction
2. Material and Methods
2.1. Patients and Data Collection
2.2. DNA and RNA Preparations
2.3. Zoom-in CGH-Array
2.4. Breakpoint Sequencing
2.5. qPCR-HRM
2.6. RNA Analysis
2.7. Immunochemistry
2.8. Nomenclature
2.9. Statical Analysis
3. Results
4. Discussion
| N | Rearrangements | Rearrangement Coordinates (hg18/GRCh36) | Gain/Loss | Reported Frequency | Size (bp) | Start# | End# | Classification | Reference |
|---|---|---|---|---|---|---|---|---|---|
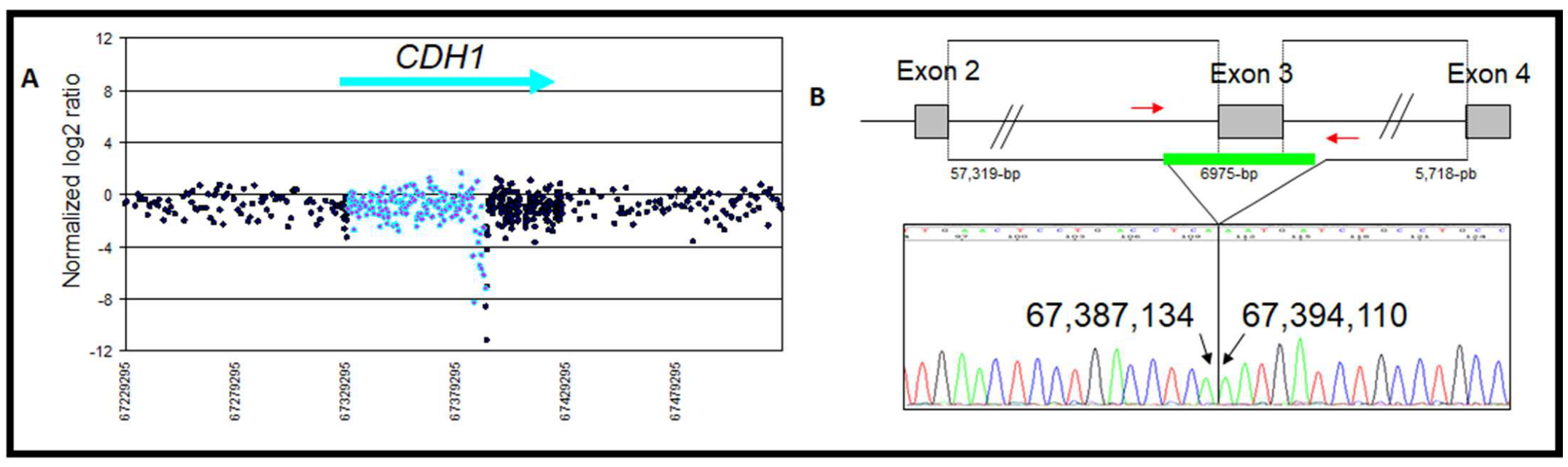
| 1 | Exon 3 deletion | chr16: 67,387,135–67,394,109 | Loss | 1/148 | 6975 | Intron 2 AluSx | Intron 3 AluSx | Deleterious c.164-5939_387+812del, p.Val55GlyfsX38 | This study and [29] |
| 2 | Full CDH3 sequence and CDH1 exon 1–2 deletion | chr16: 67,193,822–67,387,415 | Loss | 2/93 | 193,594 | 5′ region AluSp | Intron 2 AluSg | Deleterious c.-124-u134874_164-5659del, p. | [23] |
| 3 | Exon 1–2 deletion | chr16: 67,324,886–67,330,557 | Loss | 1/93 | 5672 | 5′ region AluSx | Intron 2 AluSg | Deleterious c.-124-u3810_163+742del, p. | [23,37] |
| 4 | Intron 2 deletion | chr16: 67,358,862–67,362,674 | Loss | 1/148 | 3811 | Intron 2 AluSx | Intron 2 AluJo | CNV | This study |
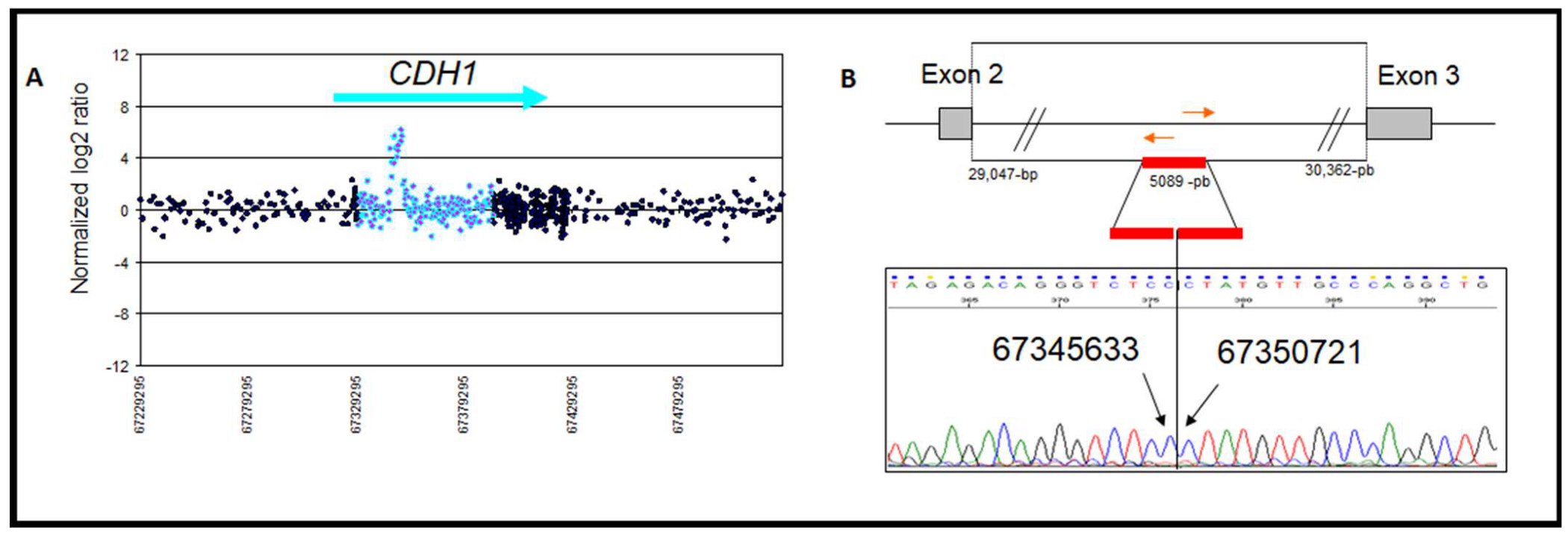
| 5 | Intron 2 duplication | chr16: 67,345,633–67,350,721 | Gain | 1/148 | 5089 | Intron 2 AluJo | Intron 2 FLAM_C | CNV | This study |
| 6 | Intron 2 CNV 67021 | chr16: 67,345,074–67,351,437 | Gain | 24/450 | 6364 | Intron 2 | Intron 2 | CNV | [38] |
| 7 | Intron 2 CNV 77387 | chr16: 67,345,117–67,348,065 | Gain | 9/90 | 2949 | Intron 2 MER-53 | Intron 2 | CNV | [39] |
| 8 | Intron 2 CNV 88182 | chr16: 67,347,663–67,348,065 | Loss | 1/90 | 403 | Intron 2 | Intron 2 | CNV | [39] |
| 9 | Intron 2 CNV 5831 | chr16: 67,330,369–67,331,573 | Loss | 1/36 | 1205 | Intron 2 AluJo | Intron 2 | CNV | [40] |
| 10 | Exon 1 deletion | chr16: 67,328,695–67,328,844 | Loss | 1/93 | 150 | AluJo | Intron 1 AluJo | Deleterious | [23] |
| 11 | Exon 14–16 deletion | chr16: 67,416,845–67,424,923 | Loss | 1/93 | 8078 | FLAM_C Intron 13 | 3′ region AluJb | Deleterious | [23] |
| 12 | Exon 16 deletion | chr16: 67,424,298–67,425,126 | Loss | 1/93 | 828 | AluSq Intron 15 | 3′ region AluJb | Deleterious | [23,37] |
| 13 | Exon 4–16 duplication CNV 77388 | chr16: 67,397,988–67,426,849 | Gain | 1/90 | 28,862 | Intron 3 MIR | 3′ region | Deleterious c.388-1840_*1946del, p.Ala130MetfsX155 | [39] |
| 14 | Exon 13–14 duplication CNV 9761 | chr16: 67,414,790–67,420,815 | Gain | 1/112 | 6026 | Intron 13 | Intron 14 | Deleterious c.1937-13_2296-243del p.Gln647ValfsX10 | [41] |
| 15 | Intron 15 CNV 67022 | chr16: 67,421,302–67,424,310 | Loss | 5/450 | 3009 | Intron 15 | Intron 15 | CNV | [38] |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rostami, P.; Zendehdel, K.; Shirkoohi, R.; Ebrahimi, E.; Ataei, M.; Imanian, H.; Najmabadi, H.; Akbari, M.R.; Sanati, M.H. Gene Panel Testing in Hereditary Breast Cancer. Arch. Iran. Med. Acad. Med. Sci. IR Iran 2020, 23, 155–162. [Google Scholar]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Am. Assoc. Adv. Sci. 1994, 266, 66–71. [Google Scholar] [CrossRef]
- Wooster, R.; Bignell, G.; Lancaster, J.; Swift, S.; Seal, S.; Mangion, J.; Collins, N.; Gregory, S.; Gumbs, C.; Micklem, G. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995, 378, 789–792. [Google Scholar] [CrossRef]
- Ford, D.; Easton, D.F.; Stratton, M.; Narod, S.; Goldgar, D.; Devilee, P.; Bishop, D.T.; Weber, B.; Lenoir, G.; Chang-Claude, J.; et al. Genetic Heterogeneity and Penetrance Analysis of the BRCA1 and BRCA2 Genes in Breast Cancer Families. Am. J. Hum. Genet. 1998, 62, 676–689. [Google Scholar] [CrossRef]
- Han, M.-R.; Long, J.; Choi, J.-Y.; Low, S.-K.; Kweon, S.-S.; Zheng, Y.; Cai, Q.; Shi, J.; Guo, X.; Matsuo, K.; et al. Genome-wide association study in East Asians identifies two novel breast cancer susceptibility loci. Hum. Mol Genet. 2016, 25, 3361–3371. [Google Scholar] [CrossRef] [PubMed]
- van der Post, R.S.; Vogelaar, I.P.; Carneiro, F.; Guilford, P.; Huntsman, D.; Hoogerbrugge, N.; Caldas, C.; Schreibei, K.E.C.; Hardwick, R.H.; Ausems, M.G.E.M. Hereditary diffuse gastric cancer: Updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J. Med. Genet. 2015, 52, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Hansford, S.; Kaurah, P.; Li-Chang, H.; Woo, M.; Senz, J.; Pinheiro, H.; Schrader, K.A.; Schaeffer, D.F.; Shumansky, K.; Zogopoulos, G. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol. 2015, 1, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Seevaratnam, R.; Coburn, N.; Cardoso, R.; Dixon, M.; Bocicariu, A.; Helyer, L. A systematic review of the indications for genetic testing and prophylactic gastrectomy among patients with hereditary diffuse gastric cancer. Gastric Cancer. 2012, 15 (Suppl. 1), S153–S163. [Google Scholar] [CrossRef] [PubMed]
- Ben Aissa-Haj, J.; Kabbage, M.; Othmen, H.; Saulnier, P.; Kettiti, H.T.; Jaballah-Gabten, A.; Ferah, A.L.; Medhioub, M.; Khsiba, A.; Mahmoudi, M. CDH1 Germline Variants in a Tunisian Cohort with Hereditary Diffuse Gastric Carcinoma. Genes 2022, 13, 400. [Google Scholar] [CrossRef]
- Stemmler, M.P. Cadherins in development and cancer. Mol. Biosyst. 2008, 4, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Sheng, Q.; Nakanishi, K.; Ohashi, A.; Wu, J.; Christ, N.; Liu, X.; Jasin, M.; Couch, F.J.; Livingston, D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 2006, 22, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Benusiglio, P.R.; Malka, D.; Rouleau, E.; Pauw, A.D.; Buecher, B.; Noguès, C.; Fourme, E.; Colas, C.; Coulet, F.L.; Warcoin, M. CDH1 germline mutations and the hereditary diffuse gastric and lobular breast cancer syndrome: A multicentre study. J. Med. Genet. 2013, 50, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Wendt, C.; Margolin, S. Identifying breast cancer susceptibility genes—A review of the genetic background in familial breast cancer. Acta Oncol. 2019, 58, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Balmaña, J.; Díez, O.; Castiglione, M. BRCA in breast cancer: ESMO Clinical Recommendations. Ann. Oncol. 2009, 20, iv19–iv20. [Google Scholar] [CrossRef]
- European Institute of Oncology. Understanding How CDH1 Germline Mutations Affect Hereditary Lobular Breast Cancer. Clinical Trial Registration. 2019. Available online: https://clinicaltrials.gov/ct2/show/NCT04206891 (accessed on 1 October 2022).
- Corso, G.; Montagna, G.; Figueiredo, J.; La Vecchia, C.; Fumagalli Romario, U.; Fernandes, M.S.; Seixas, S.L.; Roviello, F.L.; Travato, C.; Guerini-Rocco, E.; et al. Hereditary Gastric and Breast Cancer Syndromes Related to CDH1 Germline Mutation: A Multidisciplinary Clinical Review. Cancers 2020, 12, 1598. [Google Scholar] [CrossRef]
- Rahman, N.; Stone, J.G.; Coleman, G.; Gusterson, B.; Seal, S.; Marossy, A.; Lakhani, S.R.; Ward, A.; Nash, A.; McKinna, A. Lobular carcinoma in situ of the breast is not caused by constitutional mutations in the E-cadherin gene. Br. J. Cancer 2000, 82, 568–570. [Google Scholar] [CrossRef]
- Lei, H.; Sjöberg-Margolin, S.; Salahshor, S.; Werelius, B.; Jandáková, E.; Hemminki, K.; Lindblom, A.; Vorechovsky, I. CDH1 mutations are present in both ductal and lobular breast cancer, but promoter allelic variants show no detectable breast cancer risk. Int. J. Cancer 2002, 98, 199–204. [Google Scholar] [CrossRef]
- Salahshor, S.; Haixin, L.; Huo, H.; Kristensen, V.N.; Loman, N.; Sjöberg-Margolin, S.; Borg, A.; Borresen-Dale, A.-L.; Vorechovsky, I.; Lindblom, A. Low frequency of E-cadherinalterations in familial breast cancer. Breast Cancer Res 2001, 3, 199. [Google Scholar] [CrossRef]
- Schrader, K.A.; Masciari, S.; Boyd, N.; Salamanca, C.; Senz, J.; Saunders, D.N.; Yorida, E.; Maines-Bandiera, S.; Kaurah, P.; Tung, N.; et al. Germline mutations in CDH1 are infrequent in women with early-onset or familial lobular breast cancers. J. Med. 2011, 48, 64–68. [Google Scholar] [CrossRef]
- Suriano, G. Characterization of a Recurrent Germ Line Mutation of the E-Cadherin Gene: Implications for Genetic Testing and Clinical Management. Clin. Cancer Res. 2005, 11, 5401–5409. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Senz, J.; Kaurah, P.; Pinheiro, H.; Sanges, R.; Haegert, A.; Corso, G.; Schouten, J.; Fitzgerals, R.; Vogelsang, H.; et al. Germline CDH1 deletions in hereditary diffuse gastric cancer families. Hum. Mol. Genet. 2009, 18, 1545–1555. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, E.; Lefol, C.; Moncoutier, V.; Castera, L.; Houdayer, C.; Caputo, S.; Bieche, I.; Buisson, M.; Mazoyer, S.; Stoppa-Lyonnet, D.; et al. A missense variant within BRCA1 exon 23 causing exon skipping. Cancer Genet. Cytogenet. 2010, 202, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, E.; Lefol, C.; Tozlu, S.; Andrieu, C.; Guy, C.; Copigny, F.; Nogues, C.; Bieche, I.; Lidereau, R. High-resolution oligonucleotide array-CGH applied to the detection and characterization of large rearrangements in the hereditary breast cancer gene.BRCA1. Clin. Genet. 2007, 72, 199–207. [Google Scholar] [CrossRef]
- Rouleau, E.; Lefol, C.; Bourdon, V.; Coulet, F.; Noguchi, T.; Soubrier, F.; Bieche, I.; Olschwang, S.; Sobol, H.; Lidereau, R. Quantitative PCR high-resolution melting (qPCR-HRM) curve analysis, a new approach to simultaneously screen point mutations and large rearrangements: Application to MLH1 germline mutations in Lynch syndrome. Hum. Mutat. 2009, 30, 867–875. [Google Scholar] [CrossRef]
- Sebai, M.; Tang, R.; Le Formal, A.; Nashvi, M.; Leary, A.; Rouleau, E. RNAseq splicing profile of CDH1 gene: A description of physiological and pathogenic splicing patterns. In preparation.
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 2020, 22, 245–257. [Google Scholar] [CrossRef]
- Yamada, H.; Shinmura, K.; Ito, H.; Kasami, M.; Sasaki, N.; Shima, H.; Ikeda, M.; Tao, H.; Goto, M.; Ozawa, T.; et al. Germline alterations in the CDH1 gene in familial gastric cancer in the Japanese population. Cancer Sci. 2011, 102, 1782–1788. [Google Scholar] [CrossRef]
- Stemmler, M.P.; Hecht, A.; Kemler, R. E-cadherin intron 2 contains cis-regulatory elements essential for gene expression. Development 2005, 132, 965–976. [Google Scholar] [CrossRef][Green Version]
- Nasri, S.; More, H.; Graziano, F.; Ruzzo, A.; Wilson, E.; Dunbier, A.; McKinney, C.; Merriman, T.; Guilford, P.; Magnani, M.; et al. A novel diffuse gastric cancer susceptibility variant in E-cadherin (CDH1) intron 2: A case control study in an Italian population. BMC Cancer 2008, 8, 138. [Google Scholar] [CrossRef]
- Suzuki, Y.; Yamashita, R.; Shirota, M.; Sakakibara, Y.; Chiba, J.; Mizushima-Sugano, J.; Nakai, K.; Sugano, S. Sequence Comparison of Human and Mouse Genes Reveals a Homologous Block Structure in the Promoter Regions. Genome Res. 2004, 14, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, H.; Bordeira-Carriço, R.; Seixas, S.; Carvalho, J.; Senz, J.; Oliveira, P.; Inacio, P.; Gusmao, L.; Rocha, J.; Huntsman, D.; et al. Allele-specific CDH1 downregulation and hereditary diffuse gastric cancer. Hum. Molec. Genet. 2010, 19, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, S.; Yamada, H.; Takahashi, M.; Morohoshi, Y.; Yamaguchi, N.; Tsunoda, Y.; Hayashi, H.; Sugimura, H.; Komatsu, H. Early-onset diffuse gastric cancer associated with a de novo large genomic deletion of CDH1 gene. Gastric Cancer 2014, 17, 745–749. [Google Scholar] [CrossRef] [PubMed]
- Sanford-Kobayashi, E.; Batalov, S.; Wenger, A.M.; Lambert, C.; Dhillon, H.; Hall, R.J.; Baybayan, P.; Ding, Y.; Rego, S.; Wigby, K.; et al. Approaches to long-read sequencing in a clinical setting to improve diagnostic rate. Sci. Rep. 2022, 12, 16945. [Google Scholar] [CrossRef] [PubMed]
- Geng, K.; Merino, L.G.; Wedemann, L.; Martens, A.; Sobota, M.; Sanchez, Y.P.; Sondergaard, J.N.; White, R.J.; Kutter, C. Target-nriched nanopore sequencing and de novo assembly reveals cooccurrences of complex on-target genomic rearrangements induced by CRISPR-Cas9 in human cells. Genome Res. 2022, 32, 1876–1891. [Google Scholar]
- Carneiro, F.; Oliveira, C.; Seruca, R. Hereditary Diffuse Gastric Cancer and Other Gastric Cancers Associated with Hereditary Predisposition Syndromes. In Molecular Pathology of Neoplastic Gastrointestinal Diseases; Sepulveda, A.R., Lynch, J.P., Eds.; Springer: Boston, MA, USA, 2013; pp. 83–107. [Google Scholar] [CrossRef]
- Conrad, D.F.; Pinto, D.; Redon, R.; Feuk, L.; Gokcumen, O.; Zhang, Y.; Aerts, J.; Andrews, D.T.; Barnes, C.; Campbell, P.; et al. Origins and functional impact of copy number variation in the human genome. Nature 2010, 464, 704–712. [Google Scholar] [CrossRef]
- Matsuzaki, H.; Wang, P.-H.; Hu, J.; Rava, R.; Fu, G.K. High resolution discovery and confirmation of copy number variants in 90 Yoruba Nigerians. Genome Biol. 2009, 10, R125. [Google Scholar] [CrossRef]
- Mills, R.E.; Luttig, C.T.; Larkins, C.E.; Beauchamp, A.; Tsui, C.; Pittard, W.S.; Devine, S.E. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006, 16, 1182–1190. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.F.A.; Hakonarson, H.; Bucan, M. PennCNV: An integrated hidden arkov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef]


| Gene | CDH1 | |||
|---|---|---|---|---|
| Exon/Intron | Exon n°3 | Intron n°2 | ||
| Zoom in gene region (hg18/GRCh36) | chr16: 67,387,135–67,394,109 | chr16: 67,358,862–67,362,674 | chr16: 67,345,633–67,350,721 | |
| Type of rearrangement | Heterozygous deletion of 6975 pb | Heterozygous deletion of 3812 bp | Heterozygous duplication of 5089 bp | |
| c. position | c.164-5939_387+812del | c.163+29048_164-30362del | c.163+15818_163+20906dup | |
| Clinicopathogical characteristicsof the patient | Age at diagnosis | 32 | 58 | 49 |
| Personal history | Bilateral lobular BC and metachronous diffuse gastric carcinoma. | Ductal Invasive BC | A mix of lobular and ductal In situ carcinomas. | |
| Familial history | No history | BC/CRC | BC | |
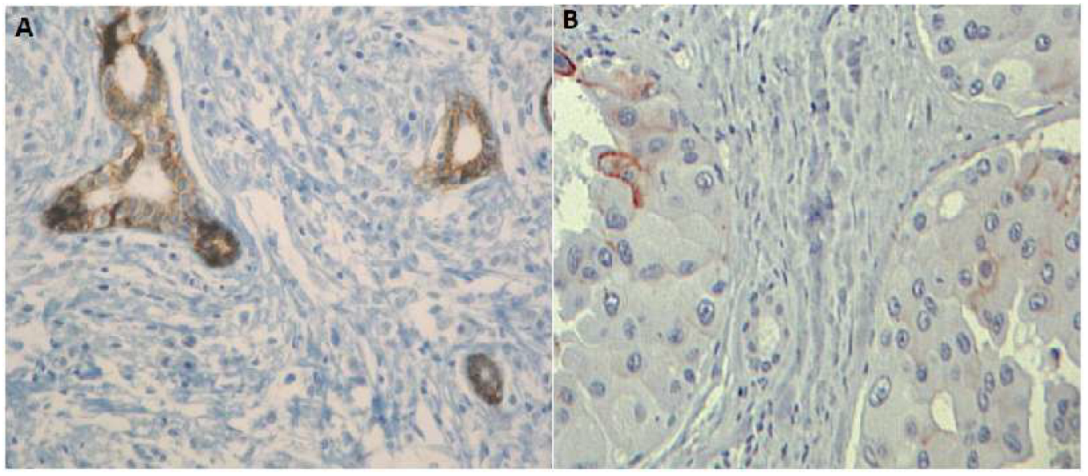
| E-Cadherin expression | Homogeneous loss (Figure 5A) | Heterogeneous loss | Heterogeneous loss (Figure 5B) | |
| Protein change | p.Val55Glyfs*38 | - | - | |
| Classification | Deleterious | CNV | CNV | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Aissa-Haj, J.; Pinheiro, H.; Cornelis, F.; Sebai, M.; Meseure, D.; Briaux, A.; Berteaux, P.; Lefol, C.; Des Guetz, G.; Trassard, M.; et al. The Identification of Large Rearrangements Involving Intron 2 of the CDH1 Gene in BRCA1/2 Negative and Breast Cancer Susceptibility. Genes 2022, 13, 2213. https://doi.org/10.3390/genes13122213
Ben Aissa-Haj J, Pinheiro H, Cornelis F, Sebai M, Meseure D, Briaux A, Berteaux P, Lefol C, Des Guetz G, Trassard M, et al. The Identification of Large Rearrangements Involving Intron 2 of the CDH1 Gene in BRCA1/2 Negative and Breast Cancer Susceptibility. Genes. 2022; 13(12):2213. https://doi.org/10.3390/genes13122213
Chicago/Turabian StyleBen Aissa-Haj, Jihenne, Hugo Pinheiro, François Cornelis, Molka Sebai, Didier Meseure, Adrien Briaux, Philippe Berteaux, Cedric Lefol, Gaëtan Des Guetz, Martine Trassard, and et al. 2022. "The Identification of Large Rearrangements Involving Intron 2 of the CDH1 Gene in BRCA1/2 Negative and Breast Cancer Susceptibility" Genes 13, no. 12: 2213. https://doi.org/10.3390/genes13122213
APA StyleBen Aissa-Haj, J., Pinheiro, H., Cornelis, F., Sebai, M., Meseure, D., Briaux, A., Berteaux, P., Lefol, C., Des Guetz, G., Trassard, M., Stevens, D., Vialard, F., Bieche, I., Noguès, C., Tang, R., Oliveira, C., Stoppat-Lyonnet, D., Lidereau, R., & Rouleau, E. (2022). The Identification of Large Rearrangements Involving Intron 2 of the CDH1 Gene in BRCA1/2 Negative and Breast Cancer Susceptibility. Genes, 13(12), 2213. https://doi.org/10.3390/genes13122213







