Genome-Wide DNA Methylation Profiling Solves Uncertainty in Classifying NSD1 Variants
Abstract
1. Introduction
2. Methods
2.1. Study Cohort
2.2. Molecular Genetics
2.3. Methylation Analysis
3. Results
3.1. DNAm profiling allows the classification of rare/private NSD1 variants, confirming or ruling out the diagnosis of Sotos syndrome
3.2. Clinical and Molecular Re-Evaluation of Subjects Carrying Reclassified NSD1 Variants
3.3. Robustness of the SVM Classifier and Sample Size Requirement
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Approval and Consent to Participate
References
- Tatton-Brown, K.; Weksberg, R. Molecular mechanisms of childhood overgrowth. Am. J. Med. Genet. C Semin. Med. Genet. 2013, 163, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Manor, J.; Lalani, S.R. Overgrowth Syndromes-Evaluation, Diagnosis, and Management. Front. Pediatr. 2020, 8, 574857. [Google Scholar] [CrossRef] [PubMed]
- Kurotaki, N.; Imaizumi, K.; Harada, N.; Masuno, M.; Kondoh, T.; Nagai, T.; Ohashi, H.; Naritomi, K.; Tsukahara, M.; Makita, Y.; et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 2002, 30, 365–366. [Google Scholar] [CrossRef] [PubMed]
- Tatton-Brown, K.; Douglas, J.; Coleman, K.; Baujat, G.; Cole, T.R.; Das, S.; Horn, D.; Hughes, H.E.; Temple, I.K.; Faravelli, F.; et al. Genotype-phenotype associations in Sotos syndrome: An analysis of 266 individuals with NSD1 aberrations. Am. J. Hum. Genet. 2005, 77, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Douglas, J.; Hanks, S.; Temple, I.K.; Davies, S.; Murray, A.; Upadhyaya, M.; Tomkins, S.; Hughes, H.E.; Cole, T.R.; Rahman, N. NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am. J. Hum. Genet. 2003, 72, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Tatton-Brown, K.; Rahman, N. Sotos syndrome. Eur. J. Hum. Genet. 2007, 15, 264–271. [Google Scholar] [CrossRef]
- Tatton-Brown, K.; Cole, T.R.P.; Rahman, N. Sotos Syndrome. 2004 [Updated 2019 Aug 1]. In GeneReviews® [Internet]; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993–2022; Available online: https://www.ncbi.nlm.nih.gov/books/ (accessed on 1 August 2019).
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Quinodoz, M.; Peter, V.G.; Cisarova, K.; Royer-Bertrand, B.; Stenson, P.D.; Cooper, D.N.; Unger, S.; Superti-Furga, A.; Rivolta, C. Analysis of missense variants in the human genome reveals widespread gene-specific clustering and improves prediction of pathogenicity. Am. J. Hum. Genet. 2022, 109, 457–470. [Google Scholar] [CrossRef]
- Bernier, R.; Golzio, C.; Xiong, B.; Stessman, H.A.; Coe, B.P.; Penn, O.; Witherspoon, K.; Gerdts, J.; Baker, C.; Vulto-van Silfhout, A.T.; et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 2014, 158, 263–276. [Google Scholar] [CrossRef]
- Tatton-Brown, K.; Loveday, C.; Yost, S.; Clarke, M.; Ramsay, E.; Zachariou, A.; Elliott, A.; Wylie, H.; Ardissone, A.; Rittinger, O.; et al. Mutations in Epigenetic Regulation Genes Are a Major Cause of Overgrowth with Intellectual Disability. Am. J. Hum. Genet. 2017, 100, 725–736. [Google Scholar] [CrossRef]
- Cytrynbaum, C.; Choufani, S.; Weksberg, R. Epigenetic signatures in overgrowth syndromes: Translational opportunities. Am. J. Med. Genet. Part C 2019, 81, 491–501. [Google Scholar] [CrossRef]
- Chater-Diehl, E.; Goodman, S.J.; Cytrynbaum, C.; Turinsky, A.L.; Choufani, S.; Weksberg, R. Anatomy of DNA methylation signatures: Emerging insights and applications. Am. J. Hum. Genet. 2021, 108, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell. 2015, 14, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Aref-Eshghi, E.; Schenkel, L.C.; Ainsworth, P.; Lin, H.; Rodenhiser, D.I.; Cutz, J.C.; Sadikovic, B. Genomic DNA methylation-derived algorithm enables accurate detection of malignant prostate tissues. Front. Oncol. 2018, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Martin-Herranz, D.E.; Aref-Eshghi, E.; Bonder, M.J.; Stubbs, T.M.; Choufani, S.; Weksberg, R.; Stegle, O.; Sadikovic, B.; Reik, W.; Thornton, J.M. Screening for genes that accelerate the epigenetic aging clock in humans reveals a role for the H3K36 methyltransferase NSD1. Genome Biol. 2019, 20, 146–166. [Google Scholar] [CrossRef] [PubMed]
- Aref-Eshghi, E.; Kerkhof, J.; Pedro, V.P.; France, G.D.; Barat-Houari, M.; Ruiz-Pallares, N.; Andrau, J.C.; Lacombe, D.; Van-Gils, J.; Fergelot, P.; et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020, 106, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Sadikovic, B.; Levy, M.A.; Kerkhof, J.; Aref-Eshghi, E.; Schenkel, L.; Stuart, A.; McConkey, H.; Henneman, P.; Venema, A.; Schwartz, C.E.; et al. Clinical epigenomics: Genome-wide DNA methylation analysis for the diagnosis of Mendelian disorders. Genet. Med. 2021, 23, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Ciolfi, A.; Aref-Eshghi, E.; Pizzi, S.; Pedace, L.; Miele, E.; Kerkhof, J.; Flex, E.; Martinelli, S.; Radio, F.C.; Ruivenkamp, C.A.L.; et al. Frameshift mutations at the C-terminus of HIST1H1E result in a specific DNA hypomethylation signature. Clin. Epigenetics 2020, 12, 7. [Google Scholar] [CrossRef]
- Radio, F.C.; Pang, K.; Ciolfi, A.; Levy, M.A.; Hernández-García, A.; Pedace, L.; Pantaleoni, F.; Liu, Z.; de Boer, E.; Jackson, A.; et al. SPEN haploinsufficiency causes a neurodevelopmental disorder overlapping proximal 1p36 deletion syndrome with an episignature of X chromosomes in females. Am. J. Hum. Genet. 2021, 108, 502–516. [Google Scholar] [CrossRef]
- Ciolfi, A.; Foroutan, A.; Capuano, A.; Pedace, L.; Travaglini, L.; Pizzi, S.; Andreani, M.; Miele, E.; Invernizzi, F.; Reale, C.; et al. Childhood-onset dystonia-causing KMT2B variants result in a distinctive genomic hypermethylation profile. Clin. Epigenetics 2021, 13, 157. [Google Scholar] [CrossRef]
- Choufani, S.; Cytrynbaum, C.; Chung, B.H.; Turinsky, A.L.; Grafodatskaya, D.; Chen, Y.A.; Cohen, A.S.; Dupuis, L.; Butcher, D.T.; Siu, M.T.; et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015, 6, 10207. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.A.; Relator, R.; McConkey, H.; Pranckeviciene, E.; Kerkhof, J.; Barat-Houari, M.; Bargiacchi, S.; Biamino, E.; PalomaresBralo, M.; Cappuccio, G.; et al. Functional correlation of genome-wide DNA methylation profiles in genetic neurodevelopmental disorders. Hum. Mutat. 2022, 43, 1609–1628. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Morris, T.J.; Webster, A.P.; Yang, Z.; Beck, S.; Feber, A.; Teschendorff, A.E. ChAMP: Updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017, 33, 3982–3984. [Google Scholar] [CrossRef] [PubMed]
- Fortin, J.P.; Triche, T.J.; Hansen, K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017, 33, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Salas, L.A.; Koestler, D.C.; Butler, R.A.; Hansen, H.M.; Wiencke, J.K.; Kelsey, K.T.; Christensen, B.C. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina Human Methylation EPIC Bead Array. Genome Biol. 2018, 19, 64. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
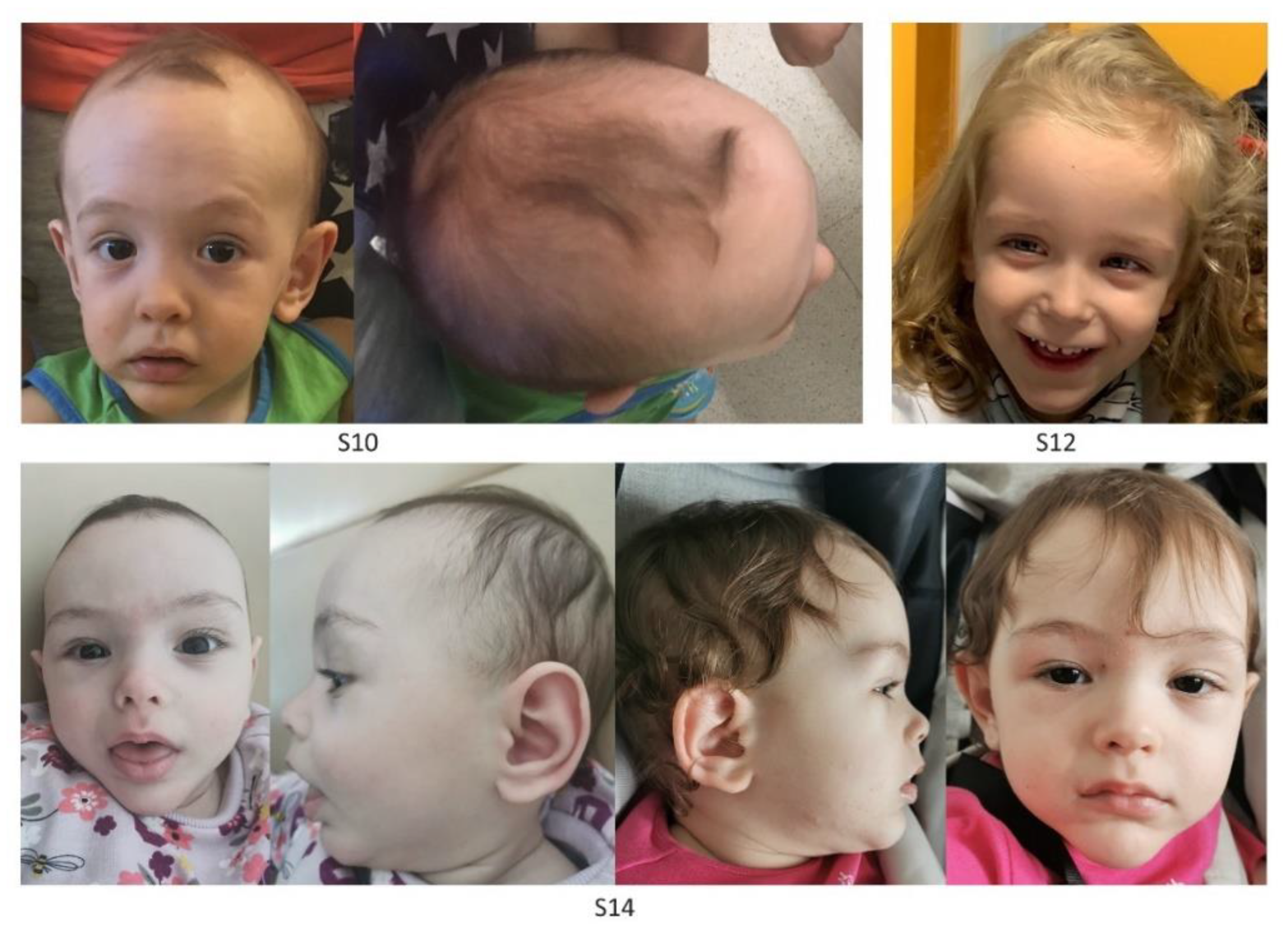

| ID (Age at Last Examination, Years) | S1 (2.9) | S2 (23) | S3 (3) | S4 (2) | S5 (1) | S6 (2) | Tot | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Pathogenic NSD1 Variant | Deletion of Exons 14 and 15 | c.5147-2A > G | c.4638T > A (p.Cys1546*) | c.6010-2A > G | c.6070C > T (p.Gln2024*) | c.2370delA (p.Lys791fs*15) | ||||
| Growth | Birth weight (cent) | 95° | >97° | >97° | 50° | 14° | >97° | 3/6 | ||
| Birth length (cent) | >97° | >97° | 97° | 90° | 10° | >97° | 4/6 | |||
| Birth OFC (cent) | >97° | >97° | >>97° | 50-75° | 64° | >97° | 4/6 | |||
| Postnatal weight (cent) | >97° | >97° | 97° | >97° | >97° | 75°-90° | 5/6 | |||
| Postnatal height (cent) | >97° | 75° | 97° | 90-97° | 75-97° | >97° | 4/6 | |||
| Postnatal OFC (cent) | >97° | >97° | >>97° | >97° | >97° | 75°-90° | 5/6 | |||
| Development | DD/ID | moderate | moderate | mild | moderate | mild | moderate | 6/6 | ||
| Neurology | Hypotonia (Ho) | yes | yes | yes | yes | yes | yes | 6/6 | ||
| Seizures/EEG anomalies | no | no | no | no | yes, febbrile | no | 1/6 | |||
| Brain anomalies | no | mild ectasia of trigone of lateral ventricles; arachnoid cyst | reduced white matter thickness and hyperintensity in the subcortical white matter; wide aspect of the periencephalic spaces; squared aspect of the frontal horns; thin corpus callosum; multicystic aspect of the pineal gland | right posterior fronto-opercular polymicrogiria; thin corpus callosum; thin optic nerves | NA | no | 3/5 | |||
| Craniofacial | Dolichocephaly | yes | yes | yes | yes | yes | yes | 6/6 | ||
| Prominent forehead | yes | yes | yes | yes | yes | yes | 6/6 | |||
| Long/triangular face | yes | yes | yes | yes | yes | yes | 6/6 | |||
| Downslanted palpebral fissures | yes | yes | yes | yes | yes | yes | 6/6 | |||
| Deep set eyes | no | yes | yes | yes | yes | yes | 5/6 | |||
| Depressed nasal bridge | no | yes | yes | yes | yes | yes | 5/6 | |||
| Everted lips | yes | yes | yes | yes | yes | yes | 6/6 | |||
| Pointed/prominent chin | yes | yes | yes | yes | yes | yes | 6/6 | |||
| Musculo-skeletal | Advanced bone age | yes | yes | yes | NA | NA | NA | 3/3 | ||
| Long hands | yes | yes | no | yes | yes | yes | 5/6 | |||
| Joint laxity | yes | yes | no | yes | yes | yes | 5/6 | |||
| Cardiac | ASD OS | no | mild aortic insufficiency | no | transient neonatal bradycardia; ASD OS | moderate tricuspid insufficiency; ASD OS | 4/6 | |||
| Other | no | club feet scoliosis | no | unilateral cryptorchidism; hypoglycemia at birth | prolonged neonatal jaundice | OSAS, mild hepatomegaly | ||||
| ID (Age at Last Examination, Years) | S7 (11) | S8 (1) | S9 (2) | S10 (1) | S11 (0.9) | S12 (2) | S13 (4) | S14 (0.6) | Tot | |
| Identified NSD1 Variant | c.5418G > T (p.Trp1806Cys) | c.4857T > G (p.Cys1619Trp) | c.6454C > G (p.Arg2152Gly) | c.4786T > C (p.Cys1596Arg) | c.6128T > C (p.Phe2043Ser) | c.4917_4925delCTGTATAAC (p.Cys1640_Thr1642del) | c.6371G > C (p.Cys2124 Ser) | c.6215G > C (p.Cys2072Ser) | ||
| Growth | Birth weight (cent) | 25° | 25° | 50° | 90° | 86° | 97° | >97°cc | 66° | 2/8 |
| Birth length (cent) | 25° | 50° | 75° | 97° | 76° | NA | 97° | 71° | 2/7 | |
| Birth OFC (cent) | 25° | 75° | 75° | 90° | 90° | >97° | NA | 75° | 1/7 | |
| Postnatal weight (cent) | 75° | >97° | 97° | >97° | >97° | >97° | 90°-97° | 71° | 6/8 | |
| Postnatal height(cent) | 97° | >97° | >97° | >97° | >97° | >97° | >97° | 99° | 8/8 | |
| Postnatal OFC (cent) | >97° | 97° | >97° | NA | 97° | >97° | >97° | 88° | 6/7 | |
| Development | DD/ID | borderline | mild | mild | moderate | moderate | mild | mild | mild | 8/8 |
| Neurology | Hypotonia | yes | no | yes | yes | yes | yes | yes | yes | 7/8 |
| Seizures/EEG anomalies | no | no | no | no | yes | no | no | no | 1/8 | |
| Brain anomalies # | yes | no | no | yes | yes | yes | NA | yes | 5/7 | |
| Craniofacial | Dolichocephaly | yes | yes | yes | yes | yes | no | yes | yes | 7/8 |
| Prominent forehead | yes | yes | yes | yes | yes | yes | yes | yes | 8/8 | |
| Long/triangular face | yes | yes | yes | yes | yes | yes | yes | yes | 8/8 | |
| Downslanted palpebral fissures | yes | yes | yes | yes | yes | yes | yes | yes | 8/8 | |
| Deep set eyes | yes | yes | yes | no | yes | yes | yes | yes | 7/8 | |
| Depressed nasal bridge | no | no | no | yes | no | no | yes | yes | 3/8 | |
| Everted lips | no | no | no | yes | yes | no | no | yes | 3/8 | |
| Pointed/prominent chin | yes | yes | yes | yes | yes | yes | yes | yes | 8/8 | |
| Musculo-skeletal | Advanced bone age | yes | yes | yes | NA | yes | yes | yes | NA | 6/6 |
| Long hands | yes | yes | yes | no | yes | no | no | yes | 5/8 | |
| Joint laxity | yes | no | no | yes | yes | yes | yes | yes | 6/8 | |
| Cardiac | no | VSD (muscular); DPV | no | no | ASD | no | mitral valve prolapse | 3/8 | ||
| Other | scoliosis | born after ART | scoliosis | feet oedema; cutis laxa; GER; scoliosis | recurrent otitis | mild renal asymmetry | scoliosis | |||
| ID (Age at Last Examination, Years) | S15 (7) | S16 (0.8) | S17 (18.7) | Tot | ||||||
| Identified NSD1 Variant | c.1096G > A (p.Val366Met) | c.7393C > T (p.Gln2465*) | c.1782T > C (p.Pro594=) | |||||||
| Growth | Birth weight (cent) | 75° | 63° | >97° | 1/3 | |||||
| Birth length (cent) | 75° | NA | >97° | 1/2 | ||||||
| Birth OFC (cent) | 97° | 54° | >97° | 2/3 | ||||||
| Postnatal weight (cent) | 75° | 67° | >97° | 1/3 | ||||||
| Postnatal height(cent) | 75° | 16° | >97° | 1/3 | ||||||
| Postnatal OFC cent) | >97° | relative macrocrania | >97° | 2/3 | ||||||
| Development | DD/ID | no | NA | yes | 1/2 | |||||
| Neurology | Hypotonia (Ho) | no | no | yes | 1/3 | |||||
| Seizures/EEG anomalies | yes ^ | no | yes | 2/3 | ||||||
| Brain anomalies | no | no | no | 0 | ||||||
| Craniofacial Craniofacial | Dolichocephaly | no | no | yes | 1/3 | |||||
| Prominent forehead | yes | yes | yes | 2/3 | ||||||
| Long/triangular face | yes | no | yes | 2/3 | ||||||
| Downslanted palpebral fissures | no | no | yes | 1/3 | ||||||
| Deep set eyes | yes | no | no | 1/3 | ||||||
| Depressed nasal bridge | yes | yes | no | 2/3 | ||||||
| Everted lips | yes | no | yes | 2/3 | ||||||
| Pointed/prominent chin | no | no | yes | 1/3 | ||||||
| Musculo-skeletal | Advanced bone age | NA | NA | yes | 1 | |||||
| Long hands | no | no | yes | 1/3 | ||||||
| Joint laxity | no | no | no | 0 | ||||||
| Cardiac | no | no | no | 0 | ||||||
| other | fetal pads | father with macrocephaly; homozygosity for F508; short limbs; dup. Xq21.1 inherited from the mother | scoliosis | |||||||
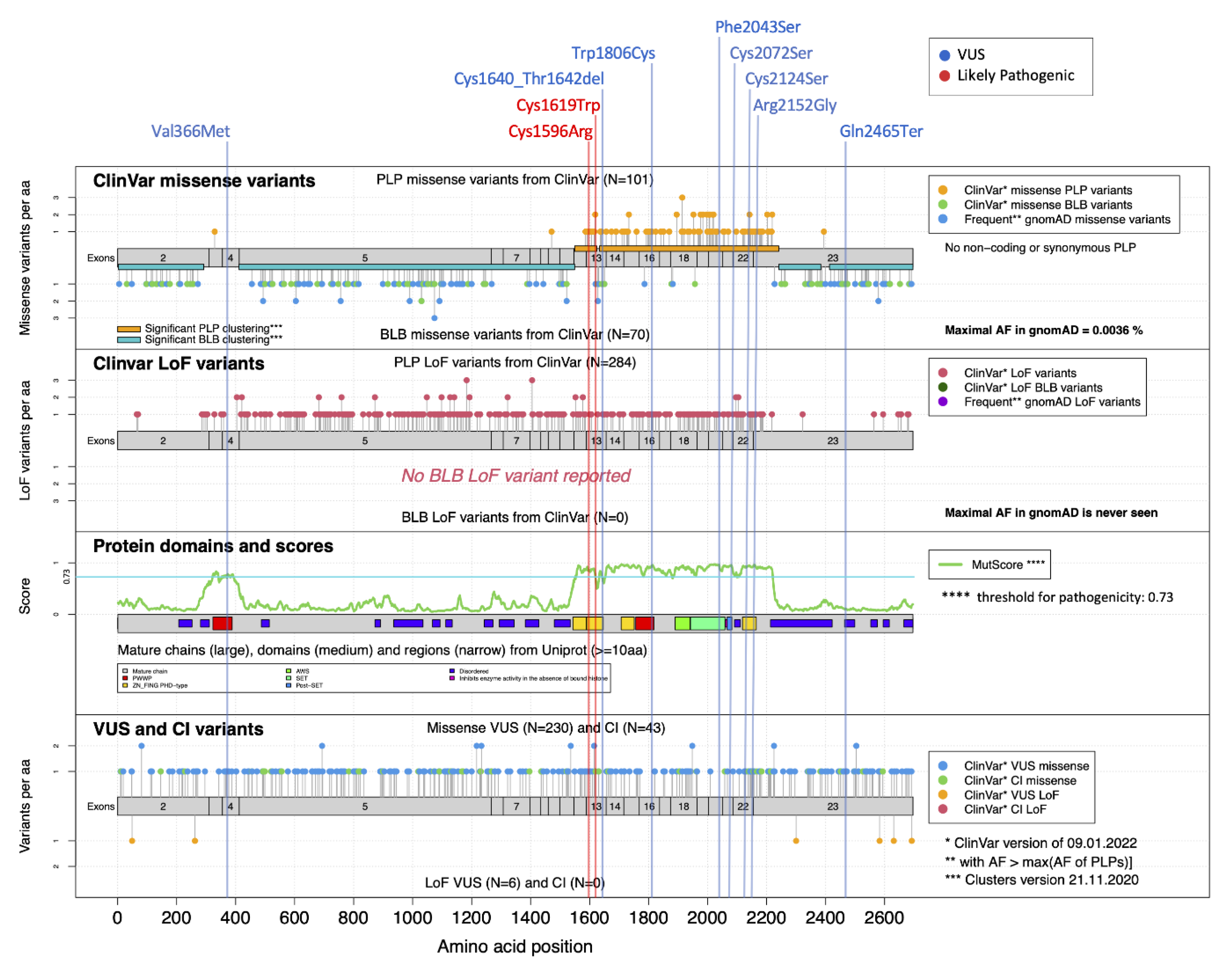
| Patient ID | NSD1 Variant (NM_022455.5) | Type of Variant a | Exon | Inheritance | rsID (maxAF) | ACMG Classification b | Domain c | CADD Score (PHRED) | Sample Set | SVM Score |
|---|---|---|---|---|---|---|---|---|---|---|
| S1 | 5q35.3 microdeletion | SV—multi-exon deletion | 14-15 | NA | NR | 5—P | - | - | training | 0.99 |
| S2 | c.5147-2A > G | SNV—splice site | 15 | de novo | NR | 4—LP | - | 35.0 | training | 0.98 |
| S3 | c.4638T > A, p.(Cys1546 *) | SNV—stop gain | 11 | de novo | NR | 4—LP | - | 35.0 | validation | 0.99 |
| S4 | c.6010-2A > G | SNV—splice site | 20 | de novo | NR | 4—LP | - | 34.0 | training | 0.97 |
| S5 | c.6070C > T; p.(Gln2024 *) | SNV—stop gain | 20 | de novo | NR | 5—P | - | 43.0 | training | 0.99 |
| S6 | c.2370delA; p.(Lys791Serfs*16) | SNV—frameshift | 5 | NA | NR | 4—LP | - | 32.0 | training | 0.99 |
| S7 | c.5418G > T; p.(Trp1806Cys) | SNV—nonsynonymous | 16 | de novo d | NR | 3—VoUS | PWWP2 | 32.0 | testing | 0.99 |
| S8 | c.4857T > G; p.(Cys1619Trp) | SNV—nonsynonymous | 13 | de novo d | NR | 4—LP | PHD-type1 | 26.5 | testing | 0.98 |
| S9 | c.6454C > G; p.(Arg2152Gly) | SNV—nonsynonymous | 22 | de novo d | NR | 3—VoUS | PHD-type4 | 24.9 | testing | 0.84 |
| S10 | c.4786T > C; p.(Cys1596Arg) | SNV—nonsynonymous | 13 | de novo d | NR | 4—LP | PHD-type1 | 28.6 | testing | 0.98 |
| S11 | c.6128T > C; p.(Phe2043Ser) | SNV—nonsynonymous | 20 | NA | NR | 3—VoUS | SET | 30 | testing | 0.99 |
| S12 | c.4917_4925delCTGTATAAC; p.(Cys1640_Thr1642del) | in-frame deletion | 13 | de novo d | NR | 3—VoUS | PHD-type2 | 22.3 | testing | 0.98 |
| S13 | c.6371G > C; p.(Cys2124 Ser) | SNV—nonsynonymous | 22 | de novo d | rs757818289 (no frequency) | 3—VoUS | PHD-type4 | 27.7 | testing | 0.95 |
| S14 | c.6215G > C p.(Cys2072Ser) | SNV—nonsynonymous | 21 | de novo d | NR | 3—VoUS | Post-SET | 28.5 | testing | 0.99 |
| S15 | c.1096G > A; p.(Val366Met) | SNV—nonsynonymous | 4 | de novo | rs1172667661 (0.000003976) | 3—VoUS | PWWP1 | 26.0 | testing | 0.19 |
| S16 | c.7393C > T; p.(Gln2465*) | SNV—stop gain | 23 | paternal | NR | 3—VoUS | - | 39.0 | testing | 0.09 |
| S17 | c.1782T > C; p.(Pro594 = ) | SNV—synonymous | 5 | maternal | rs200002555 (0.00001171) | 1—B | - | 7.5 | testing | 0.08 |
| Sample ID | SMOTE-Balanced Dataset | Unbalanced Dataset | ||||
|---|---|---|---|---|---|---|
| Dataset 1 | Dataset 2 | Dataset 3 | Dataset 1 | Dataset 2 | Dataset 3 | |
| S7 | P (0.982) | P (0.987) | P (0.994) | P (0.812) | P (0.842) | P (0.870) |
| S8 | P (0.971) | P (0.979) | P (0.982) | P (0.727) | P (0.773) | P (0.778) |
| S9 | P (0.830) | P (0.840) | P (0.849) | I (0.262) | I (0.306) | I (0.311) |
| S10 | P (0.977) | P (0.980) | P (0.989) | P (0.748) | P (0.783) | P (0.788) |
| S11 | P (0.989) | P (0.991) | P (0.993) | P (0.791) | P (0.813) | P (0.820) |
| S12 | P (0.975) | P (0.991) | P (0.988) | P (0.716) | P (0.766) | P (0.768) |
| S13 | P (0.952) | P (0.968) | P (0.969) | I (0.482) | P (0.550) | P (0.557) |
| S14 | P (0.974) | P (0.992) | P (0.997) | P (0.838) | P (0.870) | P (0.870) |
| S15 | C (0.172) | C (0.179) | C (0.192) | I (0.473) | P (0.522) | P (0.522) |
| S16 | C (0.119) | C (0.106) | C (0.100) | C (0.029) | C (0.031) | C (0.033) |
| S17 | C (0.019) | C (0.018) | C (0.007) | C (0.006) | C (0.007) | C (0.007) |
| S3 | P (0.986) | P (0.990) | P (0.993) | P (0.809) | P (0.843) | P (0.845) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferilli, M.; Ciolfi, A.; Pedace, L.; Niceta, M.; Radio, F.C.; Pizzi, S.; Miele, E.; Cappelletti, C.; Mancini, C.; Galluccio, T.; et al. Genome-Wide DNA Methylation Profiling Solves Uncertainty in Classifying NSD1 Variants. Genes 2022, 13, 2163. https://doi.org/10.3390/genes13112163
Ferilli M, Ciolfi A, Pedace L, Niceta M, Radio FC, Pizzi S, Miele E, Cappelletti C, Mancini C, Galluccio T, et al. Genome-Wide DNA Methylation Profiling Solves Uncertainty in Classifying NSD1 Variants. Genes. 2022; 13(11):2163. https://doi.org/10.3390/genes13112163
Chicago/Turabian StyleFerilli, Marco, Andrea Ciolfi, Lucia Pedace, Marcello Niceta, Francesca Clementina Radio, Simone Pizzi, Evelina Miele, Camilla Cappelletti, Cecilia Mancini, Tiziana Galluccio, and et al. 2022. "Genome-Wide DNA Methylation Profiling Solves Uncertainty in Classifying NSD1 Variants" Genes 13, no. 11: 2163. https://doi.org/10.3390/genes13112163
APA StyleFerilli, M., Ciolfi, A., Pedace, L., Niceta, M., Radio, F. C., Pizzi, S., Miele, E., Cappelletti, C., Mancini, C., Galluccio, T., Andreani, M., Iascone, M., Chiriatti, L., Novelli, A., Micalizzi, A., Matraxia, M., Menale, L., Faletra, F., Prontera, P., ... Tartaglia, M. (2022). Genome-Wide DNA Methylation Profiling Solves Uncertainty in Classifying NSD1 Variants. Genes, 13(11), 2163. https://doi.org/10.3390/genes13112163











