Comprehensive Analysis of Codon Usage in Quercus Chloroplast Genome and Focus on psbA Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Data
2.2. Nucleotide Composition and Codon Bias Indices
2.3. Relative Synonymous Codon Usage
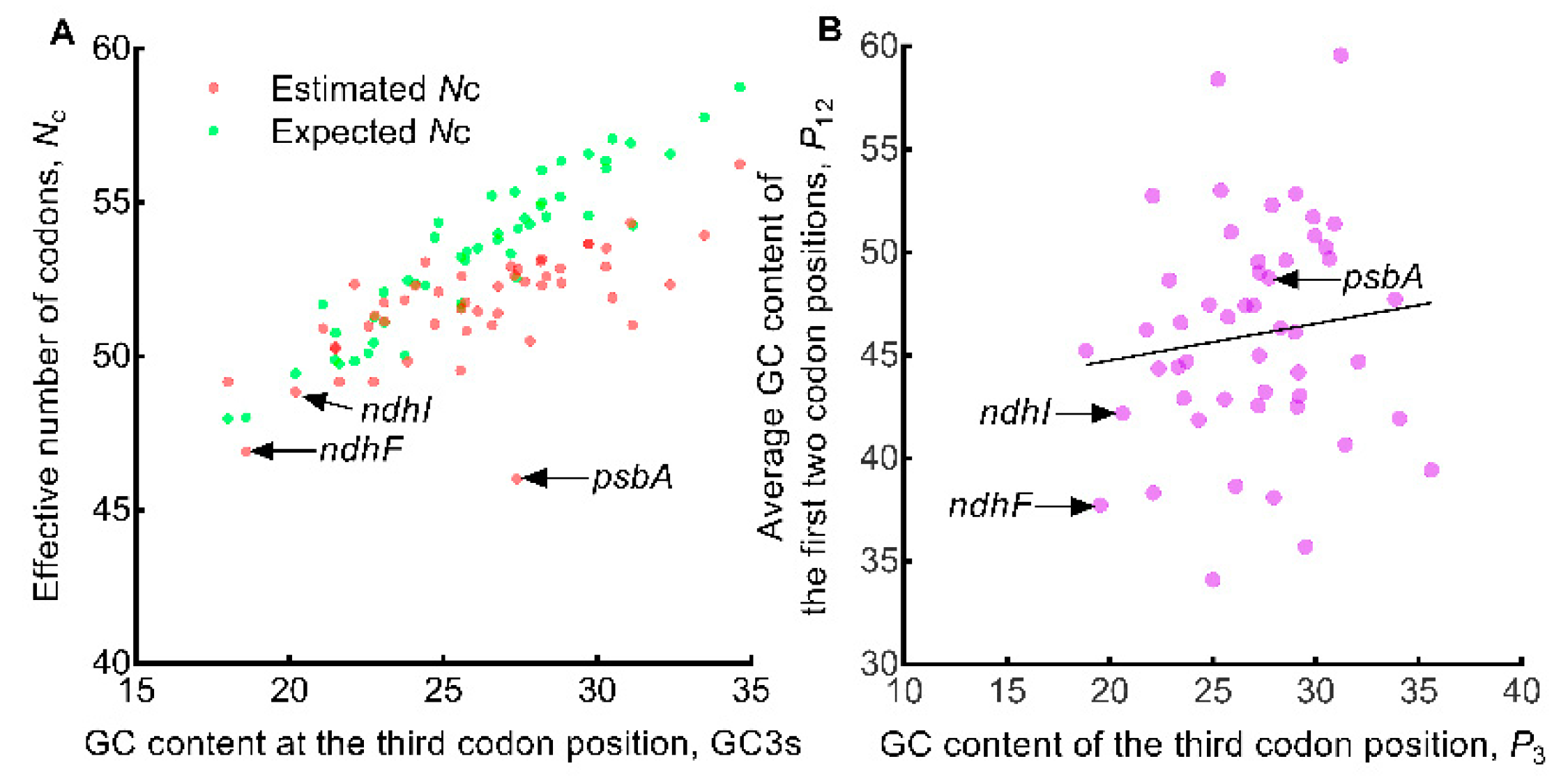
2.4. Estimated Nc versus Expected Nc Plot and P12 versus P3 Plot
2.5. Tests for Context-Dependent Mutation
2.6. Software and Calculation
3. Results
3.1. Base Composition of Synonymous Codons
3.2. Degree of Codon Usage Bias
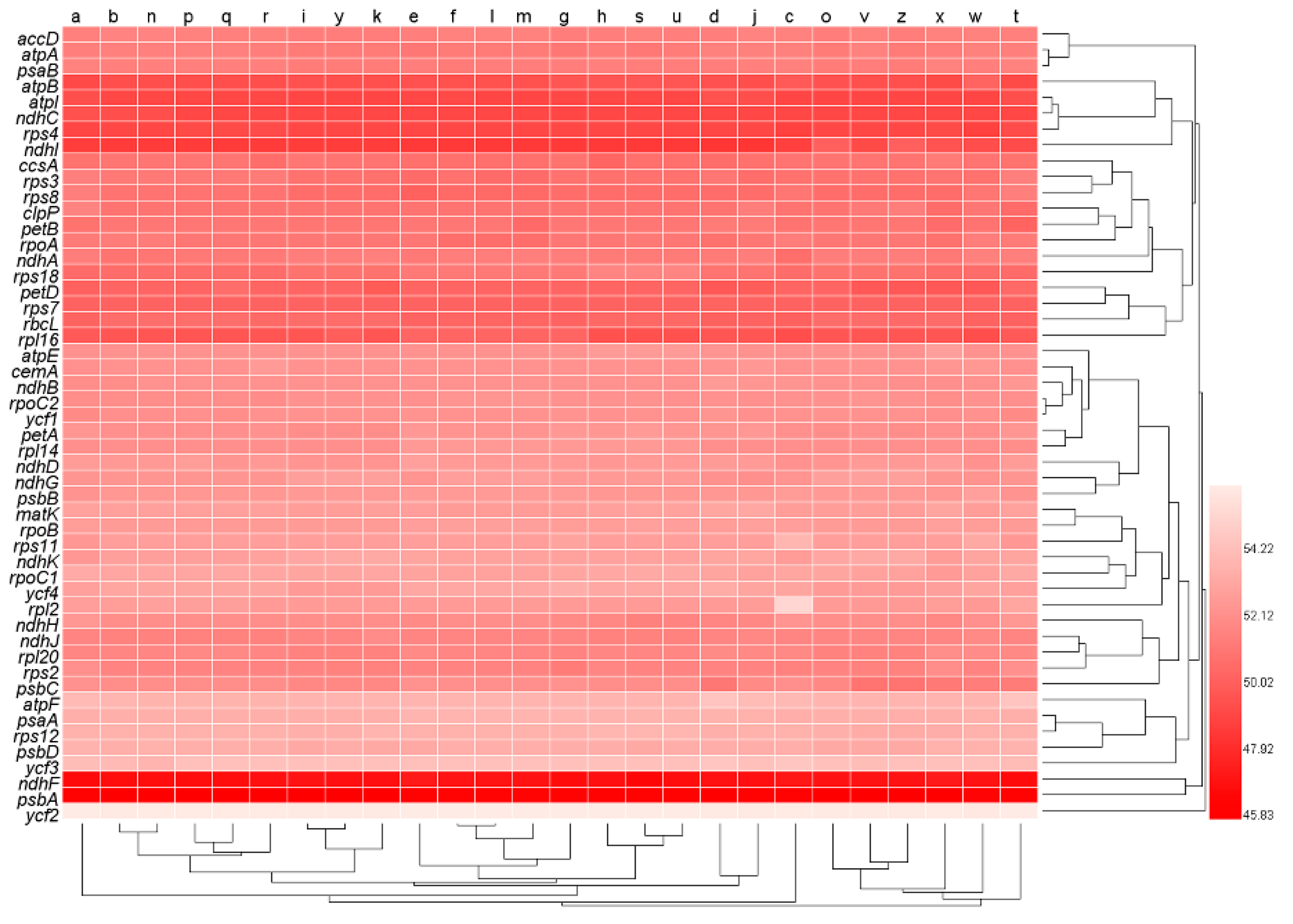
3.3. Pattern of Codon Usage Bias
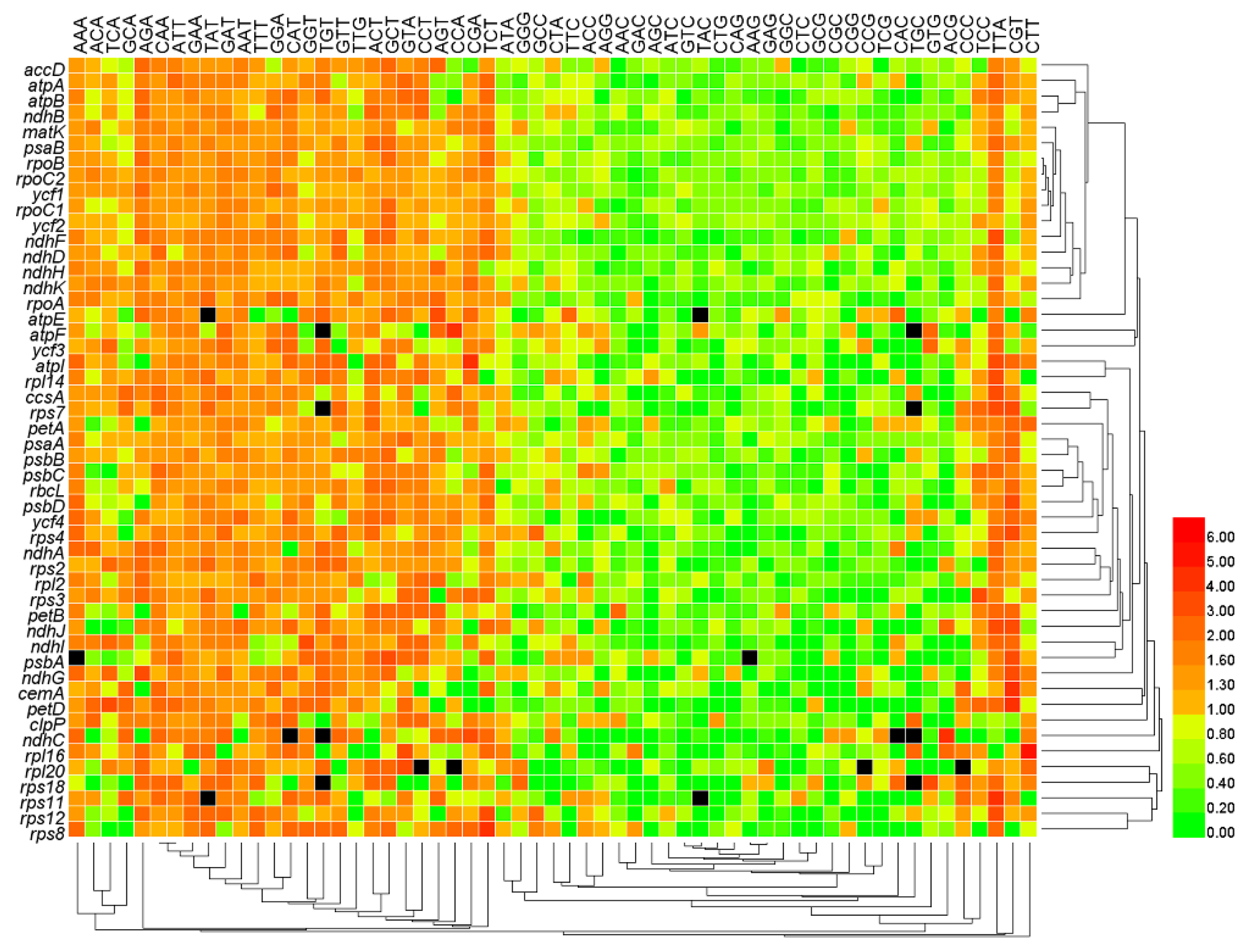
3.4. The Third Base of Codon in Use
3.5. Factors Accounting for Codon Usage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, W.; Zhang, Z.; Lin, L.; Terenius, O. Antheraea pernyi (Lepidoptera: Saturniidae) and Its Importance in Sericulture, Food Consumption, and Traditional Chinese Medicine. J. Econ. Entomol. 2017, 110, 1404–1411. [Google Scholar] [CrossRef] [PubMed]
- Hinsinger, D.D.; Strijk, J.S. Plastome of Quercus xanthoclada and comparison of genomic diversity amongst selected Quercus species using genome skimming. PhytoKeys 2019, 132, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Cavender-Bares, J. Diversification, adaptation, and community assembly of the American oaks (Quercus), a model clade for integrating ecology and evolution. New Phytol. 2018, 221, 669–692. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.W.; Woeste, K.E. Pyrosequencing of the northern red oak (Quercus rubra L.) chloroplast genome reveals high quality polymorphisms for population management. Tree Genet. Genomes 2014, 10, 803–812. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, W.; Wang, G.; Zhou, X.; Qin, L. Research advances in germplasm resource and utilization of Quercus. Sci. Seric. 2019, 45, 577–585. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, L. Investigation of tree species for raising Chinese oak silkworm in Liaoning. Sci. Seric. 1982, 8, 145–150. [Google Scholar]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Daniell, H.; Jin, S.; Zhu, X.G.; Gitzendanner, M.A.; Soltis, D.E.; Soltis, P.S. Green giant-a tiny chloroplast genome with mighty power to produce high-value proteins: History and phylogeny. Plant Biotechnol. J. 2021, 19, 430–447. [Google Scholar] [CrossRef]
- Sowden, R.G.; Watson, S.J.; Jarvis, P. The role of chloroplasts in plant pathology. Essays Biochem. 2017, 62, 21–39. [Google Scholar] [CrossRef]
- Serrano, I.; Audran, C.; Rivas, S. Chloroplasts at work during plant innate immunity. J. Exp. Bot. 2016, 67, 3845–3854. [Google Scholar] [CrossRef]
- Zeng, C.; Jia, T.; Gu, T.; Su, J.; Hu, X. Progress in Research on the Mechanisms Underlying Chloroplast-Involved Heat Tolerance in Plants. Genes 2021, 12, 1343. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Jia, T.; Jiao, Q.; Hu, X. Research Progress in J-Proteins in the Chloroplast. Genes 2022, 13, 1469. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, T.R.; Kozlowski, G.; Liu, M.H.; Yi, L.T.; Song, Y.G. Complete Chloroplast Genome of an Endangered Species Quercus litseoides, and Its Comparative, Evolutionary, and Phylogenetic Study with Other Quercus Section Cyclobalanopsis Species. Genes 2022, 13, 1184. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-j.; Su, N.; Zhang, L.; Tong, R.-c.; Zhang, X.-h.; Wang, J.-r.; Chang, Z.-y.; Zhao, L.; Potter, D. Chloroplast genomes elucidate diversity, phylogeny, and taxonomy of Pulsatilla (Ranunculaceae). Sci. Rep. 2020, 10, 19781. [Google Scholar] [CrossRef]
- Chen, J.; Xie, D.; He, X.; Yang, Y.; Li, X. Comparative Analysis of the Complete Chloroplast Genomes in Allium Section Bromatorrhiza Species (Amaryllidaceae): Phylogenetic Relationship and Adaptive Evolution. Genes 2022, 13, 1279. [Google Scholar] [CrossRef]
- Shi, S.-L.; Jiang, Y.-R.; Yang, R.-S.; Wang, Y.; Qin, L. Codon usage in Alphabaculovirus and Betabaculovirus hosted by the same insect species is weak, selection dominated and exhibits no more similar patterns than expected. Infect. Genet. Evol. 2016, 44, 412–417. [Google Scholar] [CrossRef]
- Iriarte, A.; Lamolle, G.; Musto, H. Codon Usage Bias: An Endless Tale. J. Mol. Evol. 2021, 89, 589–593. [Google Scholar] [CrossRef]
- Kong, W.Q.; Yang, J.H. The complete chloroplast genome sequence of Morus cathayana and Morus multicaulis, and comparative analysis within genus Morus L. PeerJ 2017, 5, e3037. [Google Scholar] [CrossRef]
- Yengkhom, S.; Uddin, A.; Chakraborty, S. Deciphering codon usage patterns and evolutionary forces in chloroplast genes of Camellia sinensis var. assamica and Camellia sinensis var. sinensis in comparison to Camellia pubicosta. J. Integr. Agric. 2019, 18, 2771–2785. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, B.; Li, B.; Zhou, Q.; Wang, G.; Jiang, X.; Wang, C.; Xu, Z. Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ 2020, 8, e8251. [Google Scholar] [CrossRef]
- Morton, B.R. Context-Dependent Mutation Dynamics, Not Selection, Explains the Codon Usage Bias of Most Angiosperm Chloroplast Genes. J. Mol. Evol 2022, 90, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G. Comparative Analysis of the Complete Chloroplast Genomes of Five Quercus Species. Front. Plant Sci. 2016, 07, 959. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-S.; Yang, J.; Hu, H.-L.; Xia, R.-X.; Li, Y.-P.; Su, J.-F.; Li, Q.; Liu, Y.-Q.; Qin, L. A high level of chloroplast genome sequence variability in the Sawtooth Oak Quercus acutissima. Int. J. Biol. Macromol. 2020, 152, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chang, E.-M.; Liu, J.-F.; Huang, Y.-N.; Wang, Y.; Yao, N.; Jiang, Z.-P. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus bawanglingensis Huang, Li et Xing, a Vulnerable Oak Tree in China. Forests 2019, 10, 587. [Google Scholar] [CrossRef]
- Somaratne, Y.; Guan, D.-L.; Wang, W.-Q.; Zhao, L.; Xu, S.-Q. The Complete Chloroplast Genomes of Two Lespedeza Species: Insights into Codon Usage Bias, RNA Editing Sites, and Phylogenetic Relationships in Desmodieae (Fabaceae: Papilionoideae). Plants 2019, 9, 51. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Zang, M.; Li, M.; Fang, Y. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus acutissima. Int. J. Mol. Sci. 2018, 19, 2443. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, J.; Feng, L.; Zhou, T.; Bai, G.; Yang, J.; Zhao, G. Plastid Genome Comparative and Phylogenetic Analyses of the Key Genera in Fagaceae: Highlighting the Effect of Codon Composition Bias in Phylogenetic Inference. Front. Plant Sci. 2018, 9, 82. [Google Scholar] [CrossRef]
- Sheng, J.; She, X.; Liu, X.; Wang, J.; Hu, Z. Comparative analysis of codon usage patterns in chloroplast genomes of five Miscanthus species and related species. PeerJ 2021, 9, e12173. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, L.; Wang, W.; Zhang, Z.; Du, H.; Qu, Z.; Li, X.-Q.; Xiang, H. Differences in Codon Usage Bias between Photosynthesis-Related Genes and Genetic System-Related Genes of Chloroplast Genomes in Cultivated and Wild Solanum Species. Int. J. Mol. Sci. 2018, 19, 3142. [Google Scholar] [CrossRef]
- Lu, S.; Hou, M.; Du, F.K.; Li, J.; Yin, K. Complete chloroplast genome of the Oriental white oak: Quercus aliena Blume. Mitochondrial DNA. Part A DNA Mapp. Seq. Anal. 2016, 27, 2802–2804. [Google Scholar] [CrossRef]
- Du, F.K.; Lang, T.; Lu, S.; Wang, Y.; Li, J.; Yin, K. An improved method for chloroplast genome sequencing in non-model forest tree species. Tree Genet. Genomes 2015, 11, 114. [Google Scholar] [CrossRef]
- Yin, K.; Zhang, Y.; Li, Y.; Du, F. Different Natural Selection Pressures on the atpF Gene in Evergreen Sclerophyllous and Deciduous Oak Species: Evidence from Comparative Analysis of the Complete Chloroplast Genome of Quercus aquifolioides with Other Oak Species. Int. J. Mol. Sci. 2018, 19, 1042. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.C.; Zhou, T.; Yang, J.; Meng, X.; Zhu, J.; Zhao, G. The complete chloroplast genome of Quercus baronii (Quercus L.). Mitochondrial DNA. Part A DNA Mapp. Seq. Anal. 2017, 28, 290–291. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, T.; Zhu, J.; Zhao, J.; Zhao, G. Characterization of the complete plastid genome of Quercus tarokoensis. Conserv. Genet. Resour. 2018, 10, 191–193. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, H.; Ren, T.; Zhao, G. Characterization of the complete plastid genome of Quercus tungmaiensis. Conserv. Genet. Resour. 2018, 10, 457–460. [Google Scholar] [CrossRef]
- Su, H.; Yang, Y.; Ju, M.; Li, H.; Zhao, G. Characterization of the complete plastid genome of Quercus sichourensis. Conserv. Genet. Resour. 2019, 11, 129–131. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, Y.; Ren, T.; Sun, J.; Zhao, G. Remarkably conserved plastid genomes of Quercus group Cerris in China: Comparative and phylogenetic analyses. Nord. J. Bot. 2018, 36, e01921. [Google Scholar] [CrossRef]
- Hu, H.-L.; Zhang, J.-Y.; Li, Y.-P.; Xie, L.; Chen, D.-B.; Li, Q.; Liu, Y.-Q.; Hui, S.-R.; Qin, L. The complete chloroplast genome of the daimyo oak, Quercus dentata Thunb. Conserv. Genet. Resour. 2019, 11, 409–411. [Google Scholar] [CrossRef]
- Ju, M.-M.; Zhang, X.; Yang, Y.-C.; Fan, W.-B.; Zhao, G.-F. The complete chloroplast genome of a critically endangered tree species in China, Cyclobalanopsis obovatifolia (Fagaceae). Conserv. Genet. Resour. 2019, 11, 31–33. [Google Scholar] [CrossRef]
- Hu, H.L.; Wang, L.Z.; Yang, J.; Zhang, R.S.; Li, Q.; Liu, Y.Q.; Qin, L. The complete chloroplast genome of Quercus fenchengensis and the phylogenetic implication. Mitochondrial DNA. Part B Resour. 2019, 4, 3066–3067. [Google Scholar] [CrossRef]
- Feng, L.; Yang, X.; Jiao, Q.; Wang, C.; Yin, Y. The complete chloroplast genome of Quercus robur ‘Fastigiata’. Mitochondrial DNA. Part B Resour. 2019, 5, 129–130. [Google Scholar] [CrossRef]
- Yang, X.; Yin, Y.; Feng, L.; Tang, H.; Wang, F. The first complete chloroplast genome of Quercus coccinea (Scarlet Oak) and its phylogenetic position within Fagaceae. Mitochondrial DNA. Part B Resour. 2019, 4, 3634–3635. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Liu, D.; Nan, C.; Fang, Y.; Huang, F. The complete chloroplast genome sequence of Quercus phillyraeoides (Fagaceae). Mitochondrial DNA. Part B Resour. 2020, 5, 904–905. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, Y.; Yuan, X.; Luo, T.; Wang, Y. The complete chloroplast genome sequence of Qercus pannosa. Mitochondrial DNA Part B 2020, 5, 1777–1778. [Google Scholar] [CrossRef]
- Liu, D.; Li, W.Q.; Xie, X.M.; Liu, D.S.; Li, F.; Gao, G.; Zhuang, Z.J.; Lu, Y.Z.; Li, W. Characterization of the complete chloroplast genome of Quercus virginiana Mill. (Fagaceae). Mitochondrial DNA. Part B Resour. 2021, 6, 868–869. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.B.; Han, E.K.; Choi, I.S.; Kwak, M.; Kim, J.H.; Kim, B.Y.; Lee, J.H. The complete plastid genome sequence of Quercus acuta (Fagaceae), an evergreen broad-leaved oak endemic to East Asia. Mitochondrial DNA. Part B Resour. 2021, 6, 320–322. [Google Scholar] [CrossRef]
- Jiang, X.L.; Mou, H.L.; Luo, C.S.; Xu, G.B. The complete chloroplast genome sequence of Quercus chungii (Fagaceae). Mitochondrial DNA. Part B Resour. 2021, 6, 1789–1790. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Sun, X.; Yang, Q.; Xia, X. An Improved Implementation of Effective Number of Codons (Nc). Mol. Biol. Evol. 2012, 30, 191–196. [Google Scholar] [CrossRef]
- Sharp, P.M.; Tuohy, T.M.F.; Mosurski, K.R. Codon usage in yeast: Cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986, 14, 5125–5143. [Google Scholar] [CrossRef]
- Baldanti, F.; Zhou, J.-h.; Zhang, J.; Sun, D.-j.; Ma, Q.; Chen, H.-t.; Ma, L.-n.; Ding, Y.-z.; Liu, Y.-s. The Distribution of Synonymous Codon Choice in the Translation Initiation Region of Dengue Virus. PLoS ONE 2013, 8, e77239. [Google Scholar] [CrossRef]
- Shi, S.-L.; Xia, R.-X. Codon Usage in the Iflaviridae Family Is Not Diverse Though the Family Members Are Isolated from Diverse Host Taxa. Viruses 2019, 11, 1087. [Google Scholar] [CrossRef]
- Wong, E.H.M.; Smith, D.K.; Rabadan, R.; Peiris, M.; Poon, L.L.M. Codon usage bias and the evolution of influenza A viruses. Codon Usage Biases of Influenza Virus. BMC Evol. Biol. 2010, 10, 253. [Google Scholar] [CrossRef]
- Sueoka, N. Near Homogeneity of PR2-Bias Fingerprints in the Human Genome and Their Implications in Phylogenetic Analyses. J. Mol. Evol. 2001, 53, 469–476. [Google Scholar] [CrossRef]
- Sueoka, N. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. USA 1988, 85, 2653–2657. [Google Scholar] [CrossRef]
- Jia, W.; Higgs, P.G. Codon Usage in Mitochondrial Genomes: Distinguishing Context-Dependent Mutation from Translational Selection. Mol. Biol. Evol. 2008, 25, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liu, J.; Jin, L.; Feng, X.-Y.; Chen, J.-Q. Complex Mutation and Weak Selection together Determined the Codon Usage Bias in Bryophyte Mitochondrial Genomes. J. Integr. Plant Biol. 2010, 52, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Kumar, S. DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A Toolkit for Illustrating Heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2017, 19, 20–30. [Google Scholar] [CrossRef]
- Wang, Z.; Cai, Q.; Wang, Y.; Li, M.; Wang, C.; Wang, Z.; Jiao, C.; Xu, C.; Wang, H.; Zhang, Z. Comparative Analysis of Codon Bias in the Chloroplast Genomes of Theaceae Species. Front. Genet. 2022, 13, 824610. [Google Scholar] [CrossRef]
- Morton, B. Chloroplast DNA codon use: Evidence for selection at the psb A locus based on tRNA availability. J. Mol. Evol. 1993, 37, 273–280. [Google Scholar] [CrossRef]
- Morton, B.R.; Levin, J.A. The atypical codon usage of the plant psbA gene may be the remnant of an ancestral bias. Proc. Natl. Acad. Sci. USA 1997, 94, 11434–11438. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Morton, B.R. Codon Adaptation of Plastid Genes. PLoS ONE 2016, 11, e0154306. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Deng, P.; Feng, K.; Liu, P.; Du, X.; You, F.M.; Weining, S. Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family. Plant Mol. Biol. Report. 2013, 32, 828–840. [Google Scholar] [CrossRef]
- Xu, C.; Cai, X.; Chen, Q.; Zhou, H.; Cai, Y.; Ben, A. Factors Affecting Synonymous Codon Usage Bias in Chloroplast Genome of Oncidium Gower Ramsey. Evol. Bioinform. 2011, 7, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.R.; Nandhini, M.B.; Monalisha, E.; Murugan, K.; Sethuraman, T.; Nagarajan, S.; Rao, N.S.P.; Ganesh, D. Synonymous codon usage in chloroplast genome of Coffea arabica. Bioinformation 2012, 8, 1096–1104. [Google Scholar] [CrossRef]
- Kalkus, A.; Barrett, J.; Ashok, T.; Morton, B.R. Evidence from simulation studies for selective constraints on the codon usage of the Angiosperm psbA gene. PLoS Comput. Biol. 2021, 17, e1009535. [Google Scholar] [CrossRef]



| Accession No. | Species [Ref.] | Letter Series | Genome Size (bp) | Genome GC% | Protein Coding Genes |
|---|---|---|---|---|---|
| NC_020152.1 | Q. rubra [4] | a | 161,304 | 36.80 | 89 |
| NC_026790.1 | Q. aliena [30] | b | 160,921 | 36.88 | 82 |
| NC_026907.1 | Q. spinosa [31] | c | 160,825 | 36.87 | 86 |
| NC_026913.1 | Q. aquifolioides [32] | d | 160,415 | 36.96 | 78 |
| NC_029490.1 | Q. baronii [33] | e | 161,072 | 36.81 | 86 |
| NC_031356.1 | Q. variabilis [22] | f | 161,077 | 36.79 | 86 |
| NC_031357.1 | Q. dolicholepis [22] | g | 161,237 | 36.80 | 86 |
| NC_036370.1 | Q. tarokoensis [34] | h | 161,355 | 36.80 | 86 |
| NC_036930.1 | Q. glauca [27] | i | 160,798 | 36.90 | 86 |
| NC_036936.1 | Q. tungmaiensis [35] | j | 160,702 | 36.94 | 86 |
| NC_036941.1 | Q. sichourensis [36] | k | 160,681 | 36.92 | 86 |
| NC_039428.1 | Q. chenii [37] | l | 161,117 | 36.79 | 86 |
| NC_039429.1 | Q. acutissima [37] | m | 161,127 | 36.79 | 86 |
| NC_039725.1 | Q. dentata [38] | n | 161,250 | 36.83 | 86 |
| NC_039972.1 | Q. obovatifolia [39] | o | 160,817 | 36.90 | 86 |
| NC_043857.1 | Q. wutaishanica [40] * | p | 161,296 | 36.81 | 85 |
| NC_043858.1 | Q. mongolica | q | 161,194 | 36.83 | 86 |
| NC_046388.1 | Q. robur [41] | r | 161,172 | 36.83 | 89 |
| NC_046583.1 | Q. bawanglingensis [24] | s | 161,394 | 36.79 | 86 |
| NC_047481.1 | Q. coccinea [42] | t | 161,298 | 36.80 | 88 |
| NC_048488.1 | Q. phillyraeoides [43] | u | 161,384 | 36.79 | 86 |
| NC_049876.1 | Q. gilva | v | 160,742 | 36.91 | 84 |
| NC_050963.1 | Q. pannosa [44] | w | 161,222 | 36.85 | 85 |
| NC_050972.1 | Q. virginiana [45] | x | 161,221 | 36.81 | 86 |
| NC_054352.1 | Q. acuta [46] | y | 160,533 | 36.93 | 85 |
| NC_057248.1 | Q. chungii [47] | z | 160,731 | 36.91 | 85 |
| Codon Bias | Ranges | A Ending * | C Ending * | G Ending * | T Ending * |
|---|---|---|---|---|---|
| Positive bias | RSCU > 1.6 | 32.95/23.08 | 3.28/0.00 | 2.47/0.00 | 40.15/56.25 |
| 1 < RSCU ≤ 1.6 | 38.11/23.08 | 8.08/25.00 | 10.80/8.33 | 41.41/31.25 | |
| Subtotal | 71.06/46.16 | 11.36/25.00 | 13.27/8.33 | 81.56/87.50 | |
| Negative bias | 0.6 ≤RSCU < 1 | 15.90/15.38 | 27.65/25.00 | 23.61/0.00 | 9.47/0.00 |
| 0 < RSCU < 0.6 | 7.88/38.46 | 42.55/31.25 | 45.22/58.33 | 3.54/6.25 | |
| Subtotal | 27.50/53.84 | 86.49/68.75 | 85.65/91.66 | 16.04/6.25 | |
| No bias | RSCU = 1 | 1.43/0.00 | 2.15/6.25 | 1.08/0.00 | 2.40/6.25 |
| Not in use | RSCU = 0 | 3.72/0.00 | 16.29/12.50 | 16.82/33.33 | 3.03/0.00 |
| Amino acids | Codons | p Value * | |
|---|---|---|---|
| Genome | psbA | ||
| Leu/Pro/Arg | CTN/CCN/CGN | p < 0.001 | p = 0.062 |
| Val/Ala/Gly | GTN/GCN/GGN | p < 0.001 | p = 0.026 |
| Leu + Val/Ser + Pro + Thr + Ala/Arg + Gly | CTN + GTN/NCN/CGN + GGN | p < 0.001 | p < 0.001 |
| Leu/Val | CTN/GTN | p < 0.001 | p = 1.000 |
| Arg/Gly | CGN/GGN | p = 0.021 | p = 0.597 |
| Ser/Pro/Thr/Ala | TCN/CCN/ACN/GCN | p < 0.001 | p = 0.125 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, S.-L.; Liu, Y.-Q.; Xia, R.-X.; Qin, L. Comprehensive Analysis of Codon Usage in Quercus Chloroplast Genome and Focus on psbA Gene. Genes 2022, 13, 2156. https://doi.org/10.3390/genes13112156
Shi S-L, Liu Y-Q, Xia R-X, Qin L. Comprehensive Analysis of Codon Usage in Quercus Chloroplast Genome and Focus on psbA Gene. Genes. 2022; 13(11):2156. https://doi.org/10.3390/genes13112156
Chicago/Turabian StyleShi, Sheng-Lin, Yan-Qun Liu, Run-Xi Xia, and Li Qin. 2022. "Comprehensive Analysis of Codon Usage in Quercus Chloroplast Genome and Focus on psbA Gene" Genes 13, no. 11: 2156. https://doi.org/10.3390/genes13112156
APA StyleShi, S.-L., Liu, Y.-Q., Xia, R.-X., & Qin, L. (2022). Comprehensive Analysis of Codon Usage in Quercus Chloroplast Genome and Focus on psbA Gene. Genes, 13(11), 2156. https://doi.org/10.3390/genes13112156





