Transcriptome Sequencing Reveals Tgf-β-Mediated Noncoding RNA Regulatory Mechanisms Involved in DNA Damage in the 661W Photoreceptor Cell Line
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. 661W Cells Culture and Treatment
2.3. Primary Retinal Neurons Culture and Treatment
2.4. Immunofluorescence Assay
2.5. Comet Assay
2.6. RNA-seq Analysis and Differential Expression Analysis
2.7. Functional Enrichment and Protein-Protein Interaction Analysis
2.8. Construction of the ceRNA Regulatory Network
2.9. Reverse Transcription-Polymerase Chain Reaction (RT-qPCR)
2.10. Cell Viability Assayed by Cell Counting Kit-8 (CCK8) Assay
2.11. siRNAs and miRNA Mimics
2.12. Transfection
2.13. Statistical Analysis
3. Results
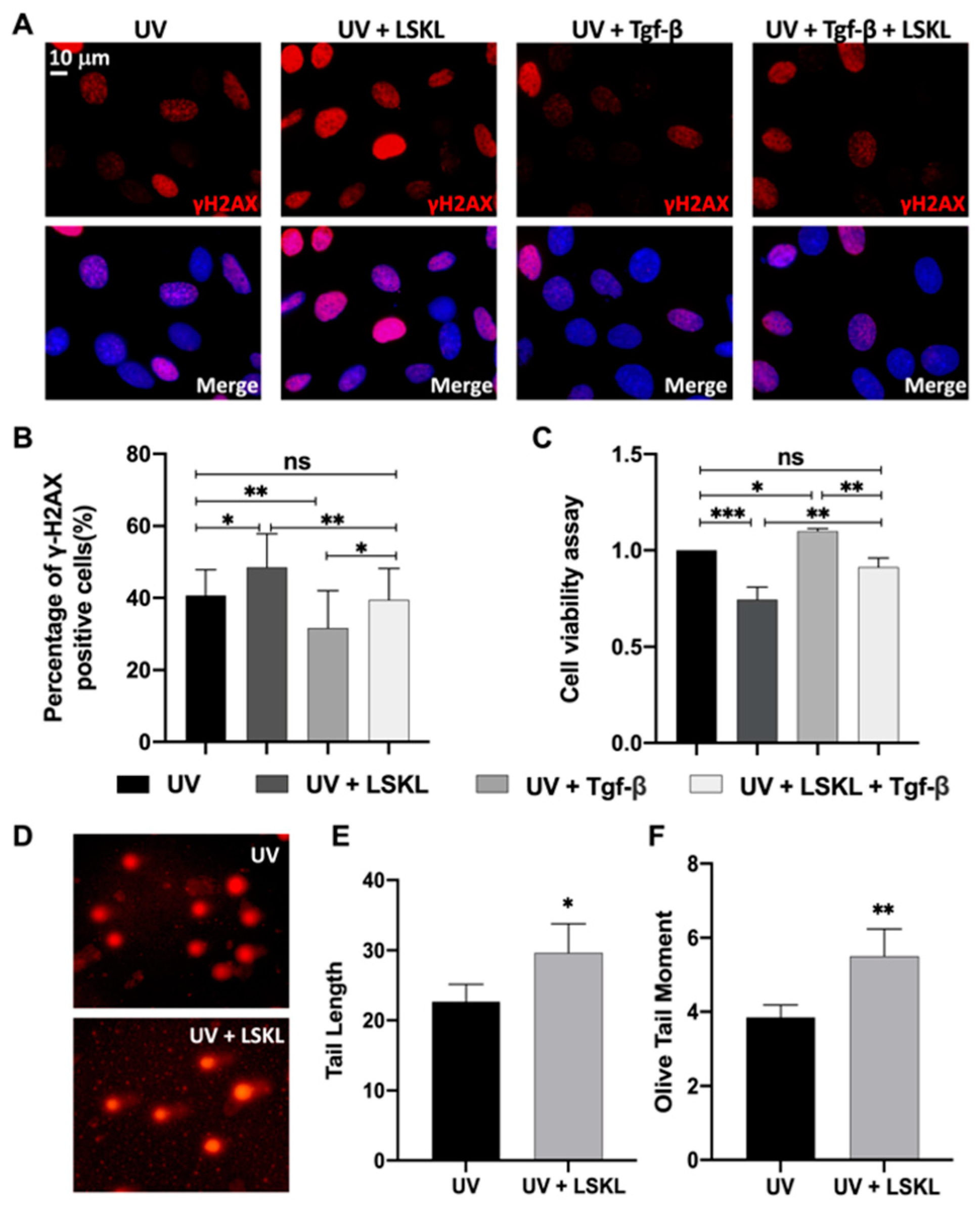
3.1. Inhibition of Tgf-β Enhanced DNA Damage and Decreased Cell Viability in 661W Cells
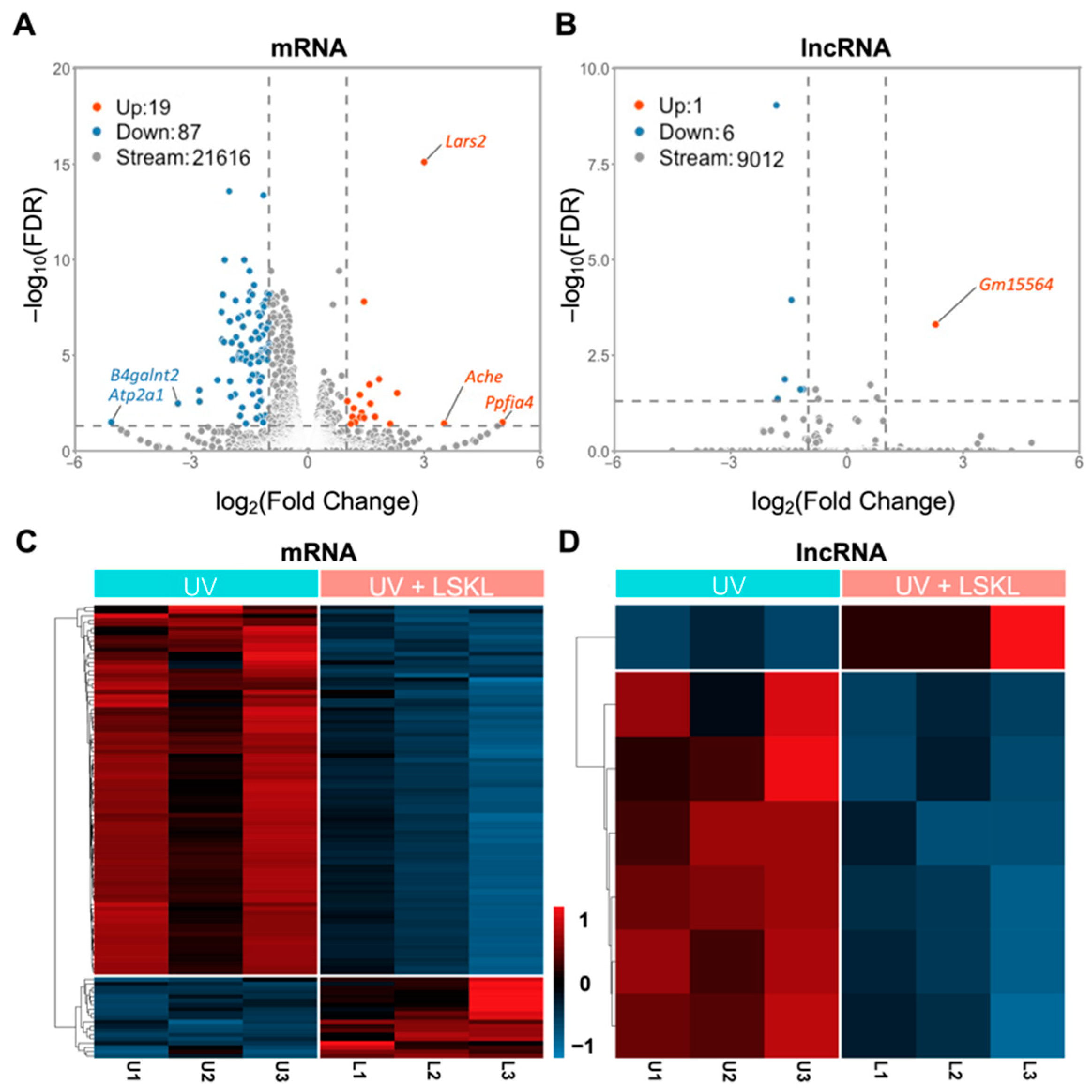
3.2. Identification of DE-mRNAs and DE-lncRNAs between UV/Ctrl and UV/LSKL 661W Cells
3.3. Enrichment Analysis and PPI Network Establishment Revealed Mechanisms and Pathways Associated with LSKL Prompting DNA Damage
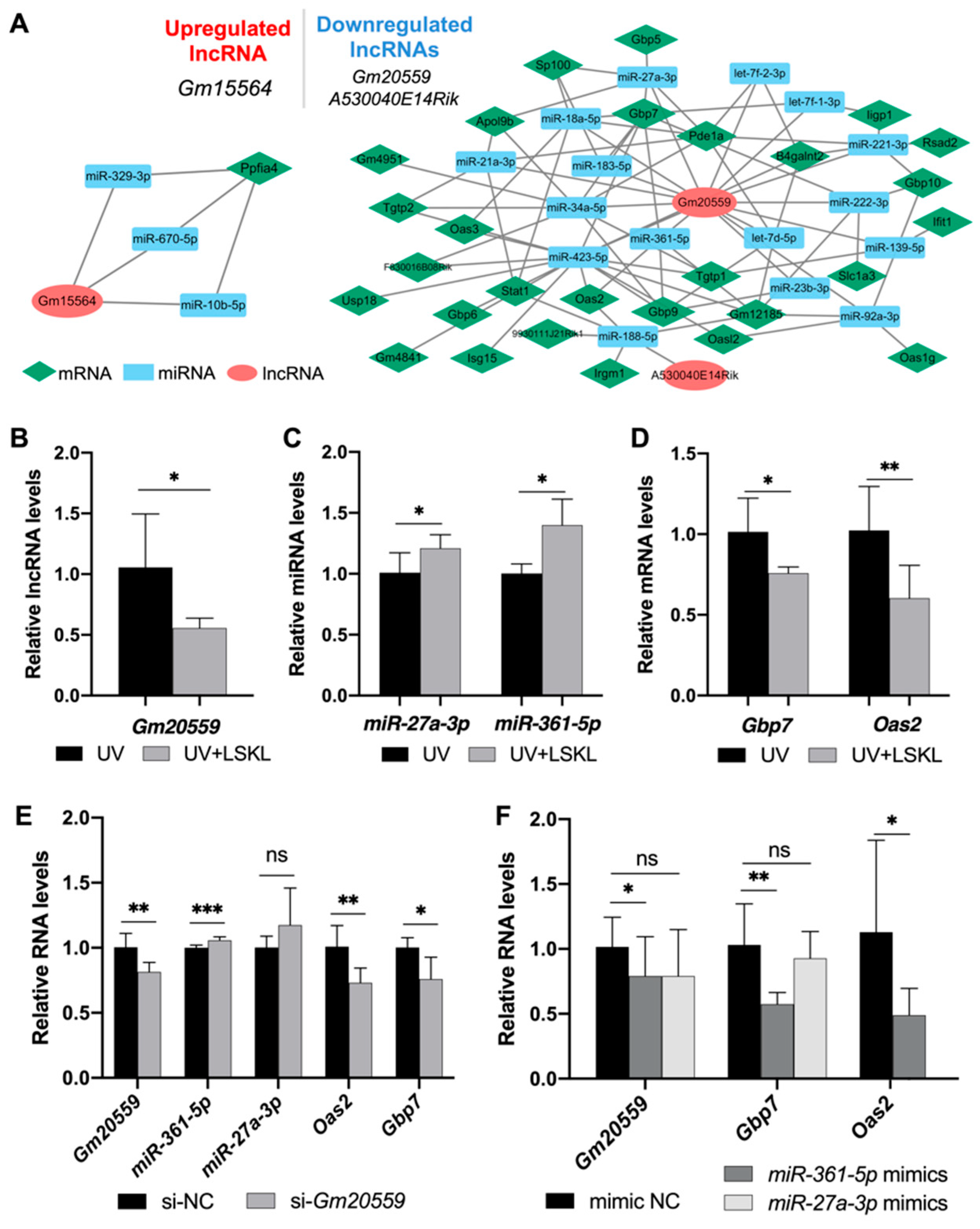
3.4. Establishment and Verification of a lncRNA-miRNA-mRNA ceRNA Network
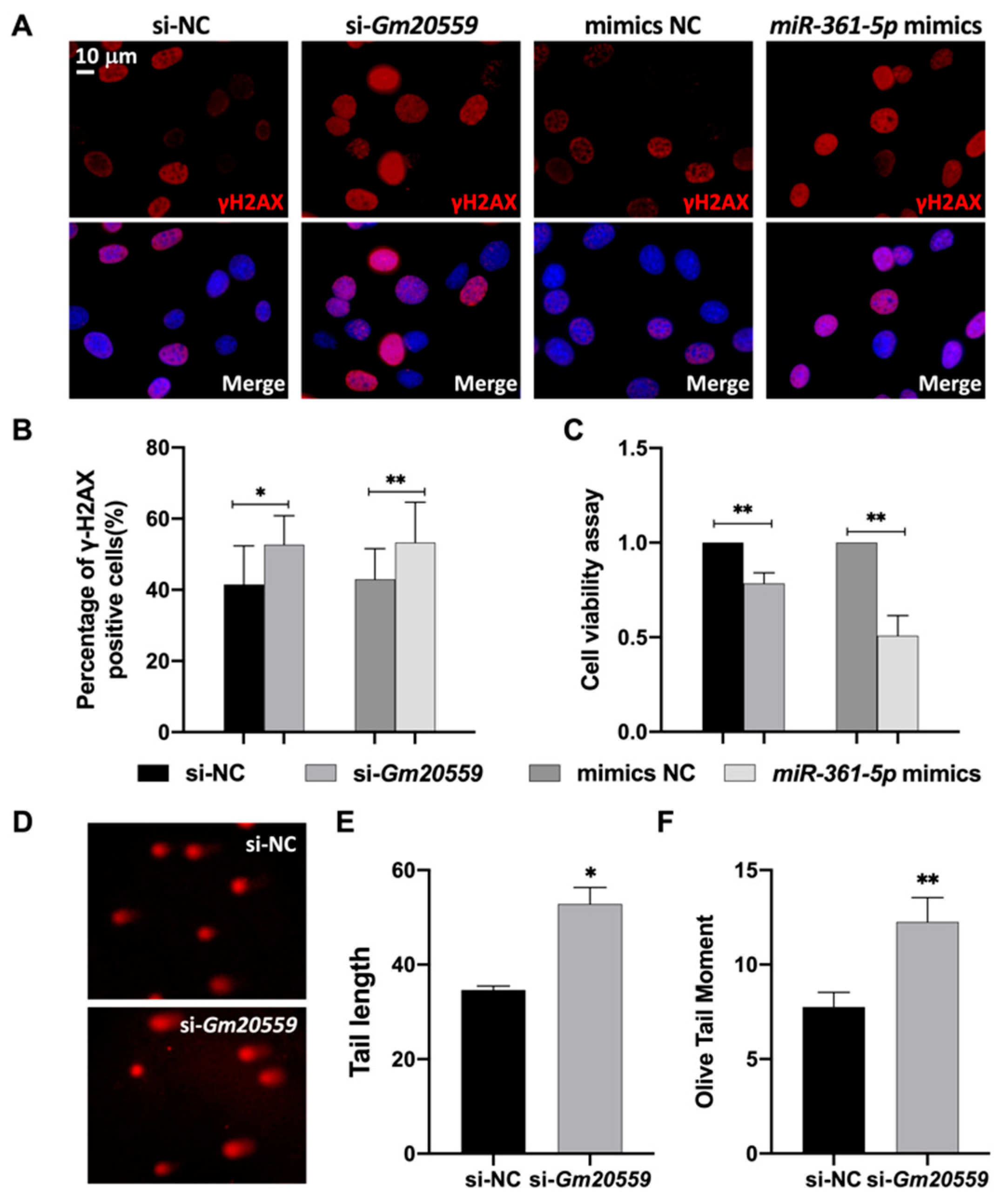
3.5. Functional Analysis of the ceRNA Network
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gallenga, C.E.; Lonardi, M.; Pacetti, S.; Violanti, S.S.; Tassinari, P.; Di Virgilio, F.; Tognon, M.; Perri, P. Molecular Mechanisms Related to Oxidative Stress in Retinitis Pigmentosa. Antioxidants 2021, 10, 848. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Cai, B.; Feng, Y.; Chen, J.; Wu, Y.; Zhuang, J.; Liu, Z.; Wu, Y. Activation of JNK signaling promotes all-trans-retinal-induced photoreceptor apoptosis in mice. J. Biol. Chem. 2020, 295, 6958–6971. [Google Scholar] [CrossRef] [PubMed]
- Toma, C.; De Cillà, S.; Palumbo, A.; Garhwal, D.P.; Grossini, E. Oxidative and Nitrosative Stress in Age-Related Macular Degeneration: A Review of Their Role in Different Stages of Disease. Antioxidants 2021, 10, 653. [Google Scholar] [CrossRef]
- Szaflik, J.P.; Janik-Papis, K.; Synowiec, E.; Ksiazek, D.; Zaras, M.; Wozniak, K.; Szaflik, J.; Blasiak, J. DNA damage and repair in age-related macular degeneration. Mutat. Res. 2009, 669, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Mencl, S.; Sahaboglu, A.; Farinelli, P.; van Veen, T.; Zrenner, E.; Ekström, P.; Paquet-Durand, F.; Arango-Gonzalez, B. Calpain and PARP activation during photoreceptor cell death in P23H and S334ter rhodopsin mutant rats. PLoS ONE 2011, 6, e22181. [Google Scholar] [CrossRef]
- Chen, P.; Liu, C.; Zhang, J.; Chen, X.; Liu, X.; He, S.; He, A.; Chen, S.; Qiu, J.; Li, Y.; et al. Tsp-1 is involved in DNA stability through Tgf-β1 activation domain in cone photoreceptor 661 W cells. Cell Tissue Res. 2022, 388, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Hegde, M.L.; Bohr, V.A.; Mitra, S. DNA damage responses in central nervous system and age-associated neurodegeneration. Mech. Ageing Dev. 2017, 161, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kinner, A.; Wu, W.; Staudt, C.; Iliakis, G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008, 36, 5678–5694. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, N.; Tian, S.; Li, F.; Hu, H.; Chen, P.; Cai, X.; Xu, L.; Zhang, J.; Chen, Z.; et al. Lithium promotes DNA stability and survival of ischemic retinal neurocytes by upregulating DNA ligase IV. Cell Death Dis. 2016, 7, e2473. [Google Scholar] [CrossRef]
- Liu, Q.; Lopez, K.; Murnane, J.; Humphrey, T.; Barcellos-Hoff, M.H. Misrepair in Context: TGFβ Regulation of DNA Repair. Front. Oncol. 2019, 9, 799. [Google Scholar] [CrossRef]
- Liu, B.; Sun, X.; Suyeoka, G.; Garcia, J.G.; Leiderman, Y.I. TGFβ signaling induces expression of Gadd45b in retinal ganglion cells. Investig. Ophthalmol. Vis. Sci. 2013, 54, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Chiang, Y.J.; Huang, F.; Li, Y.; Li, X.; Ning, Y.; Zhang, W.; Deng, H.; Chen, Y.G. DNA Damage Activates TGF-β Signaling via ATM-c-Cbl-Mediated Stabilization of the Type II Receptor TβRII. Cell Rep. 2019, 28, 735–745.e734. [Google Scholar] [CrossRef] [PubMed]
- Zi, Z. Molecular Engineering of the TGF-β Signaling Pathway. J. Mol. Biol. 2019, 431, 2644–2654. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Chen, Y.G. TGF-β Signaling from Receptors to Smads. Cold Spring Harb. Perspect. Biol. 2016, 8, a022061. [Google Scholar] [CrossRef] [PubMed]
- An, Y.S.; Kim, M.R.; Lee, S.S.; Lee, Y.S.; Chung, E.; Song, J.Y.; Lee, J.; Yi, J.Y. TGF-β signaling plays an important role in resisting γ-irradiation. Exp. Cell Res. 2013, 319, 466–473. [Google Scholar] [CrossRef]
- Liang, J.; Liao, J.; Liu, T.; Wang, Y.; Wen, J.; Cai, N.; Huang, Z.; Xu, W.; Li, G.; Ding, Z.; et al. Comprehensive analysis of TGF-β-induced mRNAs and ncRNAs in hepatocellular carcinoma. Aging 2020, 12, 19399–19420. [Google Scholar] [CrossRef]
- Lai, X.N.; Li, J.; Tang, L.B.; Chen, W.T.; Zhang, L.; Xiong, L.X. MiRNAs and LncRNAs: Dual Roles in TGF-β Signaling-Regulated Metastasis in Lung Cancer. Int. J. Mol. Sci. 2020, 21, 1193. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Zheng, H.; Jarvis, I.W.H.; Bottai, M.; Dreij, K.; Stenius, U. TGF beta promotes repair of bulky DNA damage through increased ERCC1/XPF and ERCC1/XPA interaction. Carcinogenesis 2019, 40, 580–591. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. a-Stat. Soc. 2011, 174, 245. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics-A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wang, W.; Lou, W.; Ding, B.; Yang, B.; Lu, H.; Kong, Q.; Fan, W. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging-Us 2019, 11, 2610–2627. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Tastsoglou, S.; Skoufos, G.; Karavangeli, A.; Pierros, V.; Zacharopoulou, E.; Hatzigeorgiou, A.G. DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020, 48, D101–D110. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New featuRes. for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.D.; Miller, M.W. Ethanol-induced DNA repair in neural stem cells is transforming growth factor β1-dependent. Exp. Neurol. 2019, 317, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, M.R.; Kim, H.J.; An, Y.S.; Yi, J.Y. TGF-β1 accelerates the DNA damage response in epithelial cells via Smad signaling. BioChem. Biophys. Res. Commun. 2016, 476, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Palomero, L.; Moore, J.; Guix, I.; Espín, R.; Aytés, A.; Mao, J.H.; Paulovich, A.G.; Whiteaker, J.R.; Ivey, R.G.; et al. Loss of TGFβ signaling increases alternative end-joining DNA repair that sensitizes to genotoxic therapies across cancer types. Sci. Transl. Med. 2021, 13, eabc4465. [Google Scholar] [CrossRef] [PubMed]
- Braunger, B.M.; Pielmeier, S.; Demmer, C.; Landstorfer, V.; Kawall, D.; Abramov, N.; Leibinger, M.; Kleiter, I.; Fischer, D.; Jägle, H.; et al. TGF-β signaling protects retinal neurons from programmed cell death during the development of the mammalian eye. J. Neurosci. 2013, 33, 14246–14258. [Google Scholar] [CrossRef] [PubMed]
- Henrich-Noack, P.; Prehn, J.H.; Krieglstein, J. TGF-beta 1 protects hippocampal neurons against degeneration caused by transient global ischemia. Dose-response relationship and potential neuroprotective mechanisms. Stroke 1996, 27, 1609–1614; discussion 1615. [Google Scholar] [CrossRef]
- Tesseur, I.; Nguyen, A.; Chang, B.; Li, L.; Woodling, N.S.; Wyss-Coray, T.; Luo, J. Deficiency in Neuronal TGF-β Signaling Leads to Nigrostriatal Degeneration and Activation of TGF-β Signaling Protects against MPTP Neurotoxicity in Mice. J. Neurosci. 2017, 37, 4584–4592. [Google Scholar] [CrossRef]
- Bielmeier, C.B.; Schmitt, S.I.; Kleefeldt, N.; Boneva, S.K.; Schlecht, A.; Vallon, M.; Tamm, E.R.; Hillenkamp, J.; Ergün, S.; Neueder, A.; et al. Deficiency in Retinal TGFβ Signaling Aggravates Neurodegeneration by Modulating Pro-Apoptotic and MAP Kinase Pathways. Int. J. Mol. Sci. 2022, 23, 2626. [Google Scholar] [CrossRef]
- Conedera, F.M.; Quintela Pousa, A.M.; Presby, D.M.; Mercader, N.; Enzmann, V.; Tschopp, M. Diverse Signaling by TGFβ Isoforms in Response to Focal Injury is Associated with Either Retinal Regeneration or Reactive Gliosis. Cell Mol. NeuroBiol. 2021, 41, 43–62. [Google Scholar] [CrossRef]
- Zhang, H.; Kozono, D.E.; O’Connor, K.W.; Vidal-Cardenas, S.; Rousseau, A.; Hamilton, A.; Moreau, L.; Gaudiano, E.F.; Greenberger, J.; Bagby, G.; et al. TGF-β Inhibition Rescues Hematopoietic Stem Cell Defects and Bone Marrow Failure in Fanconi Anemia. Cell Stem Cell 2016, 18, 668–681. [Google Scholar] [CrossRef]
- Ranjitkar, S.; Duan, J.E.; Srirattana, K.; Alqahtani, F.; Tulman, E.R.; Mandoiu, I.; Venkitanarayanan, K.; Tian, X. Transcriptomic Responses of Mycoplasma bovis Upon Treatments of trans-Cinnamaldehyde, Carvacrol, and Eugenol. Front. MicroBiol. 2022, 13, 888433. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.H.; Niebuhr, M.; Aulabaugh, A.; Tsai, M.D. Solution structuRes. of 2:1 and 1:1 DNA polymerase-DNA complexes probed by ultracentrifugation and small-angle X-ray scattering. Nucleic Acids Res. 2008, 36, 849–860. [Google Scholar] [CrossRef]
- Miyazono, K.I.; Ishino, S.; Makita, N.; Ito, T.; Ishino, Y.; Tanokura, M. Crystal structure of the novel lesion-specific endonuclease PfuEndoQ from Pyrococcus furiosus. Nucleic Acids Res. 2018, 46, 4807–4818. [Google Scholar] [CrossRef] [PubMed]
- Sandy, Z.; da Costa, I.C.; Schmidt, C.K. More than Meets the ISG15: Emerging Roles in the DNA Damage Response and Beyond. Biomolecules 2020, 10, 1557. [Google Scholar] [CrossRef] [PubMed]
- Malakhov, M.P.; Malakhova, O.A.; Kim, K.I.; Ritchie, K.J.; Zhang, D.E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2002, 277, 9976–9981. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.Y.; Kang, J.S.; Hwang, Y.S.; Kim, Y.J. Oligoadenylate synthase-like (OASL) proteins: Dual functions and associations with diseases. Exp. Mol. Med. 2015, 47, e144. [Google Scholar] [CrossRef]
- Jia, J.; Ouyang, Z.; Wang, M.; Ma, W.; Liu, M.; Zhang, M.; Yu, M. MicroRNA-361-5p slows down gliomas development through regulating UBR5 to elevate ATMIN protein expression. Cell Death Dis. 2021, 12, 746. [Google Scholar] [CrossRef]
- Tommasi, S.; Pinto, R.; Danza, K.; Pilato, B.; Palumbo, O.; Micale, L.; De Summa, S. miR-151-5p, targeting chromatin remodeler SMARCA5, as a marker for the BRCAness phenotype. Oncotarget 2016, 7, 80363–80372. [Google Scholar] [CrossRef]
- Li, C.G.; Mahon, C.; Sweeney, N.M.; Verschueren, E.; Kantamani, V.; Li, D.; Hennigs, J.K.; Marciano, D.P.; Diebold, I.; Abu-Halawa, O.; et al. PPARγ Interaction with UBR5/ATMIN Promotes DNA Repair to Maintain Endothelial Homeostasis. Cell Rep. 2019, 26, 1333–1343.e1337. [Google Scholar] [CrossRef]
- Wan, G.; Zhou, W.; Hu, Y.; Ma, R.; Jin, S.; Liu, G.; Jiang, Q. Transcriptional Regulation of lncRNA Genes by Histone Modification in Alzheimer’s Disease. Biomed Res. Int. 2016, 2016, 3164238. [Google Scholar] [CrossRef]
- Zhao, P.; Liu, S.; Zhong, Z.; Jiang, T.; Weng, R.; Xie, M.; Yang, S.; Xia, X. Analysis of expression profiles of long noncoding RNAs and mRNAs in brains of mice infected by rabies virus by RNA sequencing. Sci. Rep. 2018, 8, 11858. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Shenoy, A.R.; Kumar, P.; Das, R.; Tiwari, S.; MacMicking, J.D. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science 2011, 332, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Osaki, J.H.; Espinha, G.; Magalhaes, Y.T.; Forti, F.L. Modulation of RhoA GTPase Activity Sensitizes Human Cervix Carcinoma Cells to γ-Radiation by Attenuating DNA Repair Pathways. Oxidative Med. Cell Longev. 2016, 2016, 6012642. [Google Scholar] [CrossRef] [PubMed]
- Matos, P. Small GTPases in Cancer: Still Signaling the Way. Cancers 2021, 13, 1500. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Qian, W.; Bambouskova, M.; Collins, P.L.; Porter, S.I.; Byrum, A.K.; Zhang, R.; Artyomov, M.; Oltz, E.M.; Mosammaparast, N.; et al. Barrier-to-Autointegration Factor 1 Protects against a Basal cGAS-STING Response. mBio 2020, 11, e00136-20. [Google Scholar] [CrossRef]
- Cheon, H.; Holvey-Bates, E.G.; McGrail, D.J.; Stark, G.R. PD-L1 sustains chronic, cancer cell-intrinsic responses to type I interferon, enhancing resistance to DNA damage. Proc. Natl. Acad. Sci. USA 2021, 118, e2112258118. [Google Scholar] [CrossRef]
- Hasipek, M.; Guan, Y.; Grabowski, D.; Gu, X.; Saunthararajah, Y.; Silverman, R.; Stark, G.; Maciejewski, J.; Jha, B. Role of Oligoadenylate Synthetases in Myeloid Neoplasia. Blood 2020, 136, 29–30. [Google Scholar] [CrossRef]

| DEmRNAs | DElncRNAs | |
|---|---|---|
| Upregulated | (Count: 19) Lars2, Prl2c3, Rcan2, Ephx2, Tafa5, Sbsn, Tnnt2, Tagln, Id2, Sh3tc1, Sh3d21, Inpp4b, Actg2, Dnase1l2, Inka2, Ppfia4, Ache, Slurp1, Atoh8 | (Count: 1) Gm15564 |
| Downregulated | (Count: 87) Cxcl10, Olfr56, Gbp3, Ifit3, Isg15, Gbp7, Ifit1, Irgm1, Samd9l, Tgtp1, Igtp, Gbp6, Usp18, Gask1b, Rnf213, Xaf1, Trim12c, Ifit3b, Irf7, Trim34a, Oas1a, Herc6, Postn, Ifi44, Gvin1, Oasl1, Stat1, Phf11d, Gbp9, Sp100, Oasl2, Stat2, Zbp1, Gm4841, Ifit1bl2, Tnfsf10, 9930111J21Rik1, Iigp1, Trim30d, Slfn2, Apol9b, Trim12a, Trim30a, Rtp4, H2-T23, Avil, Cmpk2, H2-T24, Ifi27l2a, Irgm2, Gm4951, Phf11b, Gm4070, Rsad2, F830016B08Rik, Ifi203, Oas2, Parp14, Tgtp2, Dhx58, Acox2, Gbp5, Slfn8, Ddx60, Sp110, Misp, Oas3, Mpeg1, Oas1g, Batf2, Ifi47, Slc1a3, Ranbp3l, Perm1, Car6, Gbp4, Pde1a, Ly6f, B4galnt2, AA467197, Gm12185, Rgcc, Klf15, Zfp781, Sema3d, Atp2a1, Gbp10 | (Count: 6) Gm2619, Gm45418, AW011738, Gm9801, Gm20559, A530040E14Rik |
| Rank | Name | Score |
|---|---|---|
| 1 | Ifit1 | 1810 |
| 2 | Ifit3 | 1809 |
| 3 | Usp18 | 1806 |
| 4 | Irf7 | 1705 |
| 5 | Rsad2 | 1568 |
| 6 | Oasl2 | 1561 |
| 7 | Ifi44 | 848 |
| 8 | Oasl1 | 720 |
| 9 | Isg15 | 248 |
| 10 | Stat1 | 134 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Chen, X.; Jiang, Z.; Luo, Q.; Wan, L.; Hou, X.; Yu, K.; Zhuang, J. Transcriptome Sequencing Reveals Tgf-β-Mediated Noncoding RNA Regulatory Mechanisms Involved in DNA Damage in the 661W Photoreceptor Cell Line. Genes 2022, 13, 2140. https://doi.org/10.3390/genes13112140
Huang Y, Chen X, Jiang Z, Luo Q, Wan L, Hou X, Yu K, Zhuang J. Transcriptome Sequencing Reveals Tgf-β-Mediated Noncoding RNA Regulatory Mechanisms Involved in DNA Damage in the 661W Photoreceptor Cell Line. Genes. 2022; 13(11):2140. https://doi.org/10.3390/genes13112140
Chicago/Turabian StyleHuang, Yuke, Xi Chen, Zhigao Jiang, Qian Luo, Linxi Wan, Xiangtao Hou, Keming Yu, and Jing Zhuang. 2022. "Transcriptome Sequencing Reveals Tgf-β-Mediated Noncoding RNA Regulatory Mechanisms Involved in DNA Damage in the 661W Photoreceptor Cell Line" Genes 13, no. 11: 2140. https://doi.org/10.3390/genes13112140
APA StyleHuang, Y., Chen, X., Jiang, Z., Luo, Q., Wan, L., Hou, X., Yu, K., & Zhuang, J. (2022). Transcriptome Sequencing Reveals Tgf-β-Mediated Noncoding RNA Regulatory Mechanisms Involved in DNA Damage in the 661W Photoreceptor Cell Line. Genes, 13(11), 2140. https://doi.org/10.3390/genes13112140







