Identification and Characterization of the HD-Zip Gene Family and Dimerization Analysis of HB7 and HB12 in Brassica napus L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of HD-Zip Genes in B. napus
2.2. Phylogenetic Analysis
2.3. Gene Structure and Conserved Motif Analysis
2.4. Chromosomal Locations, Synteny, and Gene Replication Analysis
2.5. Expression Analysis from RNA-Seq Data
2.6. Plant Materials, Growth Conditions, and Treatments
2.7. Quantitative Real-Time PCR
2.8. Subcellular Localization
2.9. Yeast Two-Hybrid (Y2H) Assay
3. Results
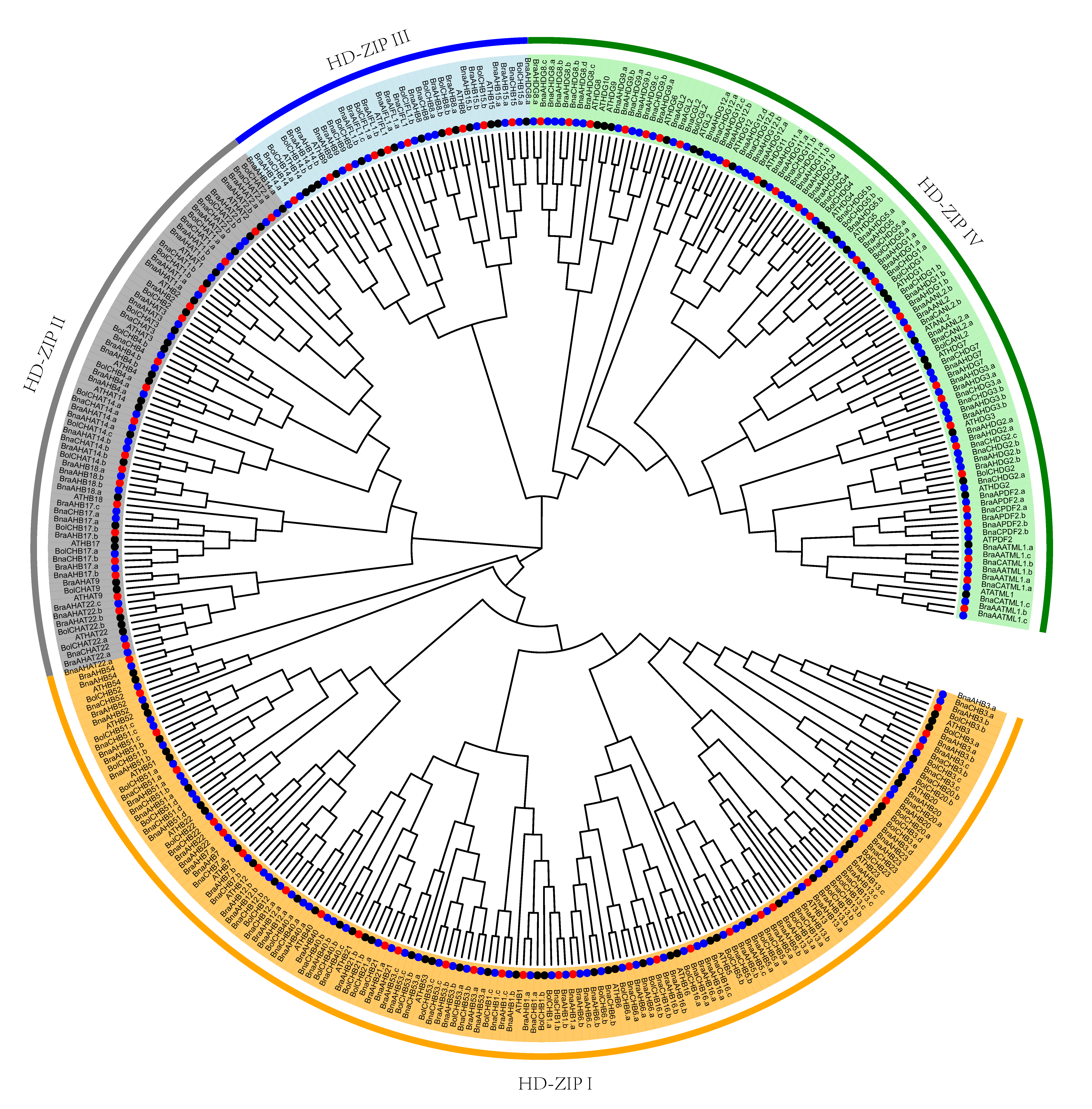
3.1. Identification and Phylogenetic Analyses of HD-Zip Proteins
3.2. Gene Structure and Conserved Motif Analysis
3.3. Chromosomal Location and Gene Duplication Analysis
3.4. Synteny Gene Analysis of HD-Zip Genes
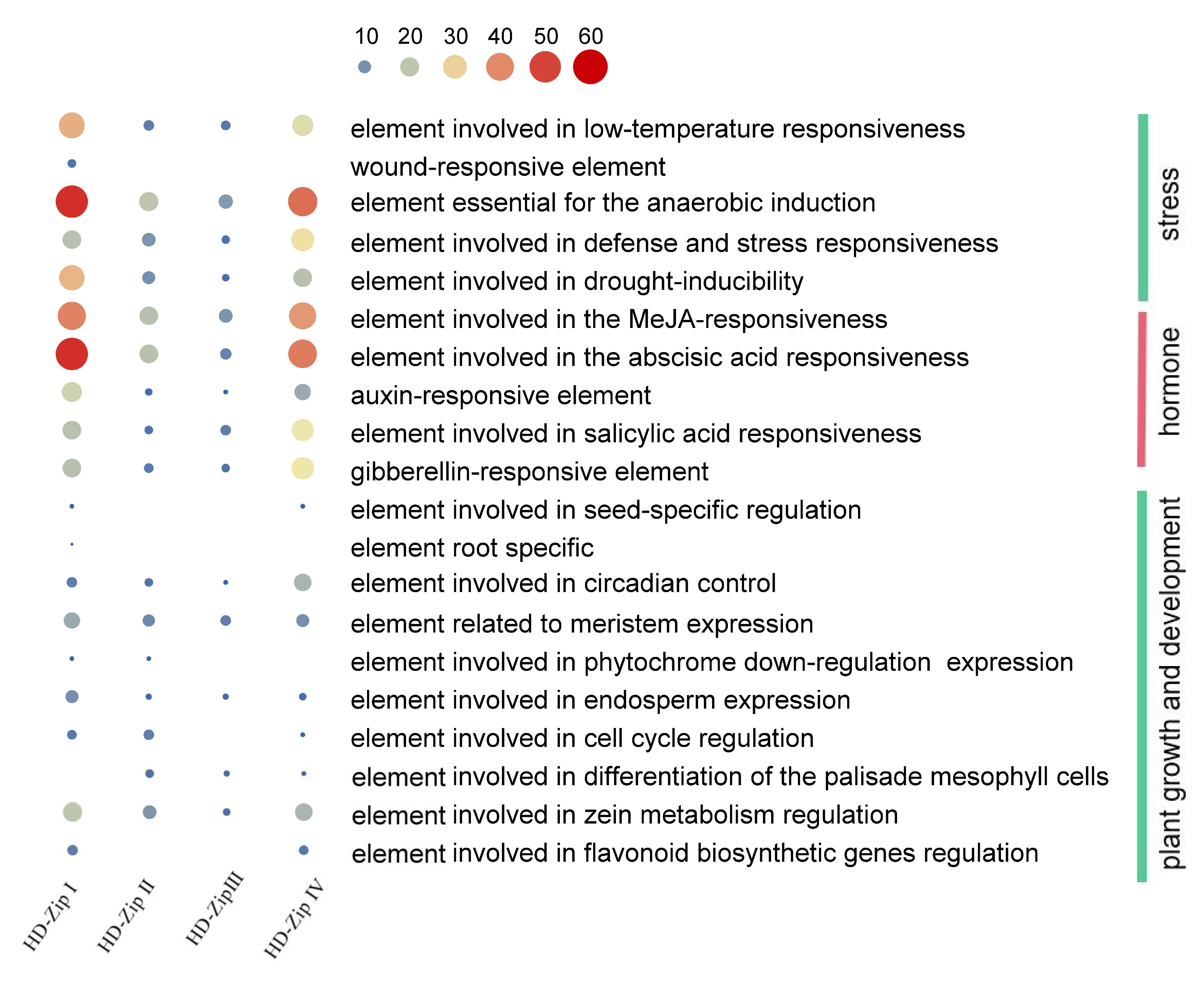
3.5. The Promoter of BnHD-Zip Genes
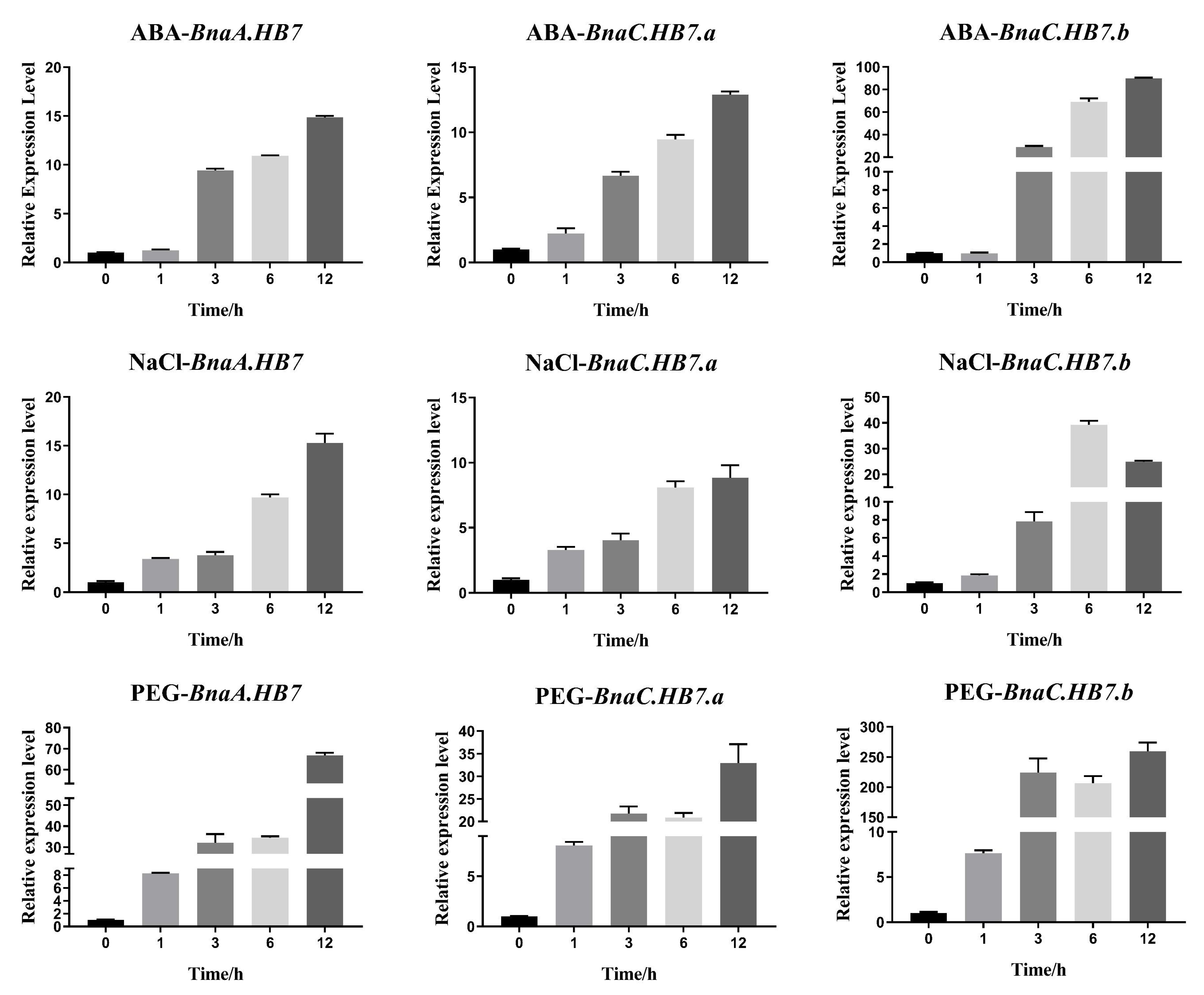
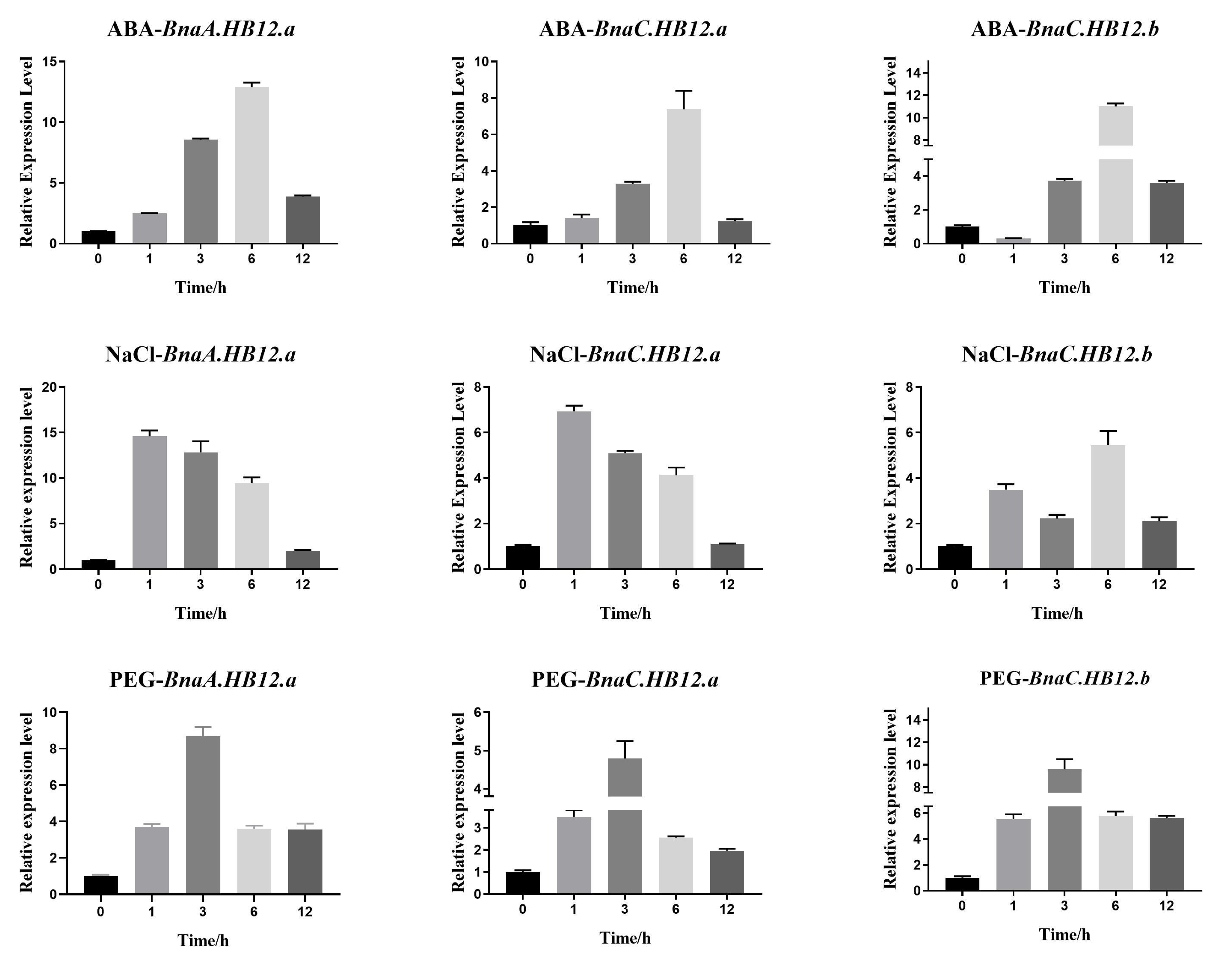
3.6. Abiotic Stress Expression Analysis
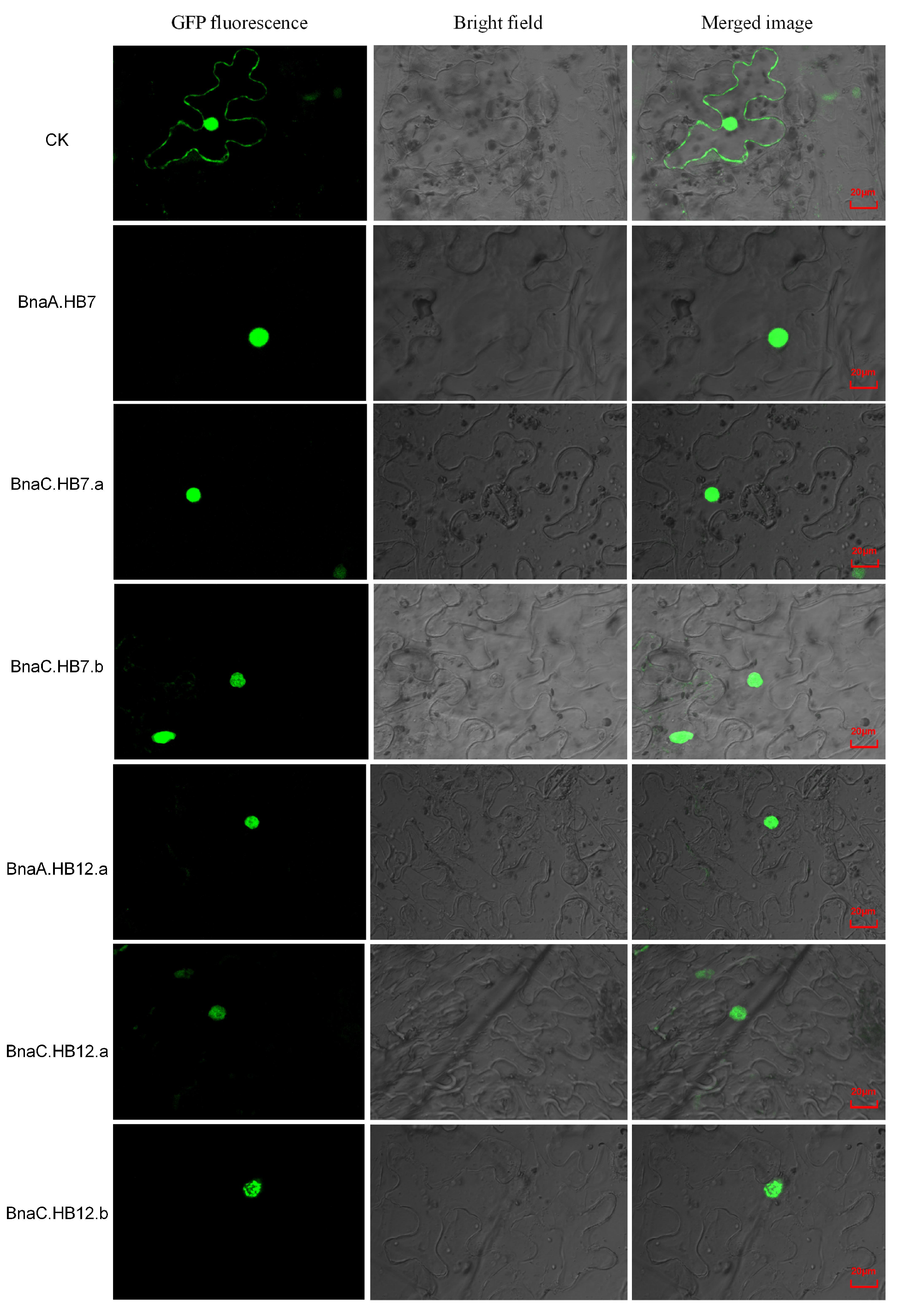
3.7. Subcellular Localization Analysis
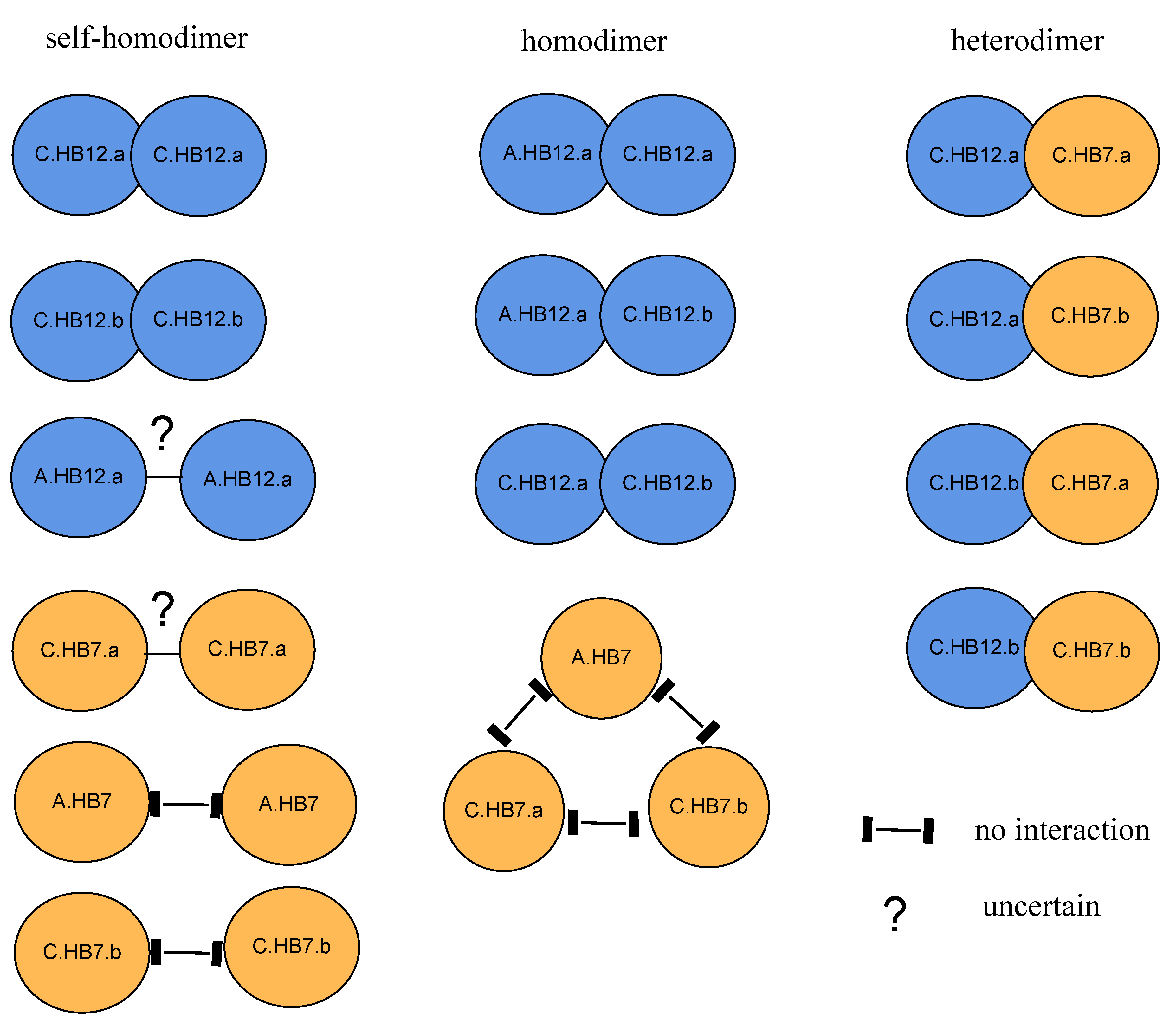
3.8. BnHB7 and BnHB12 Might Act in Homo- or Heterodimers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruberti, I.; Sessa, G.; Lucchetti, S.; Morelli, G. A Novel Class of Plant Proteins Containing a Homeodomain with a Closely Linked Leucine Zipper Motif. EMBO J. 1991, 10, 1787–1791. [Google Scholar] [CrossRef]
- Elhiti, M.; Stasolla, C. Structure and Function of Homodomain-Leucine Zipper (HD-Zip) Proteins. Plant Signal. Behav. 2009, 4, 86–88. [Google Scholar] [CrossRef]
- Sessa, G.; Steindler, C.; Morelli, G.; Ruberti, I. The Arabidopsis Athb-8, -9 and -14 Genes Are Members of a Small Gene Family Coding for Highly Related HD-ZIP Proteins. Plant Mol. Biol. 1998, 38, 609–622. [Google Scholar] [CrossRef]
- Sessa, G.; Morelli, G.; Ruberti, I. The Athb-1 and -2 HD-Zip Domains Homodimerize Forming Complexes of Different DNA Binding Specificities. EMBO J. 1993, 12, 3507–3517. [Google Scholar] [CrossRef]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The True Story of the HD-Zip Family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Henriksson, E.; Olsson, A.S.B.; Johannesson, H.; Johansson, H.; Hanson, J.; Engström, P.; Söderman, E. Homeodomain Leucine Zipper Class I Genes in Arabidopsis. Expression Patterns and Phylogenetic Relationships. Plant Physiol. 2005, 139, 509–518. [Google Scholar] [CrossRef]
- Schrick, K.; Nguyen, D.; Karlowski, W.M.; Mayer, K.F.X. START Lipid/Sterol-Binding Domains Are Amplified in Plants and Are Predominantly Associated with Homeodomain Transcription Factors. Genome Biol. 2004, 5, R41. [Google Scholar] [CrossRef]
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. HD-ZIP Gene Family: Potential Roles in Improving Plant Growth and Regulating Stress-Responsive Mechanisms in Plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef]
- Roodbarkelari, F.; Groot, E.P. Regulatory Function of Homeodomain-Leucine Zipper (HD-ZIP) Family Proteins during Embryogenesis. New Phytol. 2017, 213, 95–104. [Google Scholar] [CrossRef]
- Agalou, A.; Purwantomo, S.; Overnäs, E.; Johannesson, H.; Zhu, X.; Estiati, A.; de Kam, R.J.; Engström, P.; Slamet-Loedin, I.H.; Zhu, Z.; et al. A Genome-Wide Survey of HD-Zip Genes in Rice and Analysis of Drought-Responsive Family Members. Plant Mol. Biol. 2008, 66, 87–103. [Google Scholar] [CrossRef]
- Song, A.; Li, P.; Xin, J.; Chen, S.; Zhao, K.; Wu, D.; Fan, Q.; Gao, T.; Chen, F.; Guan, Z. Transcriptome-Wide Survey and Expression Profile Analysis of Putative Chrysanthemum HD-Zip I and II Genes. Genes 2016, 7, 19. [Google Scholar] [CrossRef]
- Ding, Z.; Fu, L.; Yan, Y.; Tie, W.; Xia, Z.; Wang, W.; Peng, M.; Hu, W.; Zhang, J. Genome-Wide Characterization and Expression Profiling of HD-Zip Gene Family Related to Abiotic Stress in Cassava. PLoS ONE 2017, 12, e0173043. [Google Scholar] [CrossRef]
- Jing, Z.; Duan, W.; Song, X.; Wu, P.; Tang, J.; Wang, Z.; Wei, Y.; Wang, C.; Hou, X. Preferential Retention, Expression Profile and Potential Functional Diversity Analysis of HD-Zip Gene Family in Brassica Rapa. Plant Growth Regul. 2017, 82, 421–430. [Google Scholar] [CrossRef]
- Yue, H.; Shu, D.; Wang, M.; Xing, G.; Zhan, H.; Du, X.; Song, W.; Nie, X. Genome-Wide Identification and Expression Analysis of the HD-Zip Gene Family in Wheat (Triticum aestivum L.). Genes 2018, 9, 70. [Google Scholar] [CrossRef]
- Li, Y.; Bai, B.; Wen, F.; Zhao, M.; Xia, Q.; Yang, D.-H.; Wang, G. Genome-Wide Identification and Expression Analysis of HD-ZIP I Gene Subfamily in Nicotiana tabacum. Genes 2019, 10, 575. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, J.; Bao, X.; Liang, M.; Lou, H.; Zhao, J.; Sun, M.; Liang, J.; Jin, L.; Li, G.; et al. Genome-Wide Identification and Expression Profile of HD-ZIP Genes in Physic Nut and Functional Analysis of the JcHDZ16 Gene in Transgenic Rice. BMC Plant Biol. 2019, 19, 298. [Google Scholar] [CrossRef]
- Sharif, R.; Xie, C.; Wang, J.; Cao, Z.; Zhang, H.; Chen, P.; Yuhong, L. Genome Wide Identification, Characterization and Expression Analysis of HD-ZIP Gene Family in Cucumis sativus L. under Biotic and Various Abiotic Stresses. Int. J. Biol. Macromol. 2020, 158, 502–520. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, J.; Guo, M.; Aslam, M.; Wang, Q.; Ma, H.; Li, S.; Zhang, X.; Cao, S. Genome-Wide Characterization and Expression Profiling of Eucalyptus Grandis HD-Zip Gene Family in Response to Salt and Temperature Stress. BMC Plant Biol. 2020, 20, 451. [Google Scholar] [CrossRef]
- Yang, Q.; Xiang, W.; Li, Z.; Nian, Y.; Fu, X.; Zhou, G.; Li, L.; Zhang, J.; Huang, G.; Han, X.; et al. Genome-Wide Characterization and Expression Analysis of HD-ZIP Gene Family in Dendrobium Officinale. Front. Genet. 2022, 13, 797014. [Google Scholar] [CrossRef]
- Mao, H.; Yu, L.; Li, Z.; Liu, H.; Han, R. Molecular Evolution and Gene Expression Differences within the HD-Zip Transcription Factor Family of Zea mays L. Genetica 2016, 144, 243–257. [Google Scholar] [CrossRef]
- Qiu, X.; Wang, G.; Abou-Elwafa, S.F.; Fu, J.; Liu, Z.; Zhang, P.; Xie, X.; Ku, L.; Ma, Y.; Guan, X.; et al. Genome-Wide Identification of HD-ZIP Transcription Factors in Maize and Their Regulatory Roles in Promoting Drought Tolerance. Physiol. Mol. Biol. Plants 2022, 28, 425–437. [Google Scholar] [CrossRef]
- Kuang, J.-F.; Chen, J.-Y.; Liu, X.-C.; Han, Y.-C.; Xiao, Y.-Y.; Shan, W.; Tang, Y.; Wu, K.-Q.; He, J.-X.; Lu, W.-J. The Transcriptional Regulatory Network Mediated by Banana (Musa acuminata) Dehydration-Responsive Element Binding (MaDREB) Transcription Factors in Fruit Ripening. New Phytol. 2017, 214, 762–781. [Google Scholar] [CrossRef]
- Li, L.; Zheng, T.; Zhuo, X.; Li, S.; Qiu, L.; Wang, J.; Cheng, T.; Zhang, Q. Genome-Wide Identification, Characterization and Expression Analysis of the HD-Zip Gene Family in the Stem Development of the Woody Plant Prunus mume. PeerJ 2019, 7, e7499. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant Genetics. Early Allopolyploid Evolution in the Post-Neolithic Brassica napus Oilseed Genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An Upgraded Brassicaceae Database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef]
- Ostergaard, L.; King, G.J. Standardized Gene Nomenclature for the Brassica Genus. Plant Methods 2008, 4, 10. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An Online Visualization and Management Tool for Customized and Annotated Phylogenetic Trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, P.; Yang, C.; Chen, H.; Luo, L.; Leng, Q.; Li, S.; Han, Z.; Li, X.; Song, C.; Zhang, X.; et al. Exploring Transcription Factors Reveals Crucial Members and Regulatory Networks Involved in Different Abiotic Stresses in Brassica napus L. BMC Plant Biol. 2018, 18, 202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kim, C.K.; Chung, J.D.; Park, S.H.; Burrell, A.M.; Kamo, K.K.; Byrne, D.H. Agrobacterium Tumefaciens-Mediated Transformation of Rosa Hybrida Using the Green Fluorescent Protein (GFP) Gene. Plant Cell Tissue Organ Cult. 2004, 78, 107–111. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, J.; Guo, S.; Yuan, X.; Zhao, S.; Tian, H.; Dai, S.; Kong, X.; Ding, Z. AtHB7/12 Regulate Root Growth in Response to Aluminum Stress. IJMS 2020, 21, 4080. [Google Scholar] [CrossRef]
- Wang, H.; Shao, W.; Yan, M.; Xu, Y.; Liu, S.; Wang, R. Genome-Wide Analysis and Expression Profiling of HD-ZIP III Genes in Three Brassica Species. Diversity 2021, 13, 684. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.P.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea Genome Reveals the Asymmetrical Evolution of Polyploid Genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Babula-Skowrońska, D. Functional Divergence of Brassica napus BnaABI1 Paralogs in the Structurally Conserved PP2CA Gene Subfamily of Brassicaceae. Genomics 2021, 113, 3185–3197. [Google Scholar] [CrossRef]
- Town, C.D.; Cheung, F.; Maiti, R.; Crabtree, J.; Haas, B.J.; Wortman, J.R.; Hine, E.E.; Althoff, R.; Arbogast, T.S.; Tallon, L.J.; et al. Comparative Genomics of Brassica oleracea and Arabidopsis thaliana Reveal Gene Loss, Fragmentation, and Dispersal after Polyploidy. Plant Cell 2006, 18, 1348–1359. [Google Scholar] [CrossRef]
- Wei, L.; Zhu, Y.; Liu, R.; Zhang, A.; Zhu, M.; Xu, W.; Lin, A.; Lu, K.; Li, J. Genome Wide Identification and Comparative Analysis of Glutathione Transferases (GST) Family Genes in Brassica napus. Sci. Rep. 2019, 9, 9196. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative Genomic and Transcriptomic Analyses of Family-1 UDP Glycosyltransferase in Three Brassica Species and Arabidopsis Indicates Stress-Responsive Regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef]
- Hu, M.; Li, M.; Wang, J. Comprehensive Analysis of the SUV Gene Family in Allopolyploid Brassica napus and Its Diploid Ancestors. Genes 2021, 12, 1848. [Google Scholar] [CrossRef]
- Sessa, G.; Carabelli, M.; Possenti, M.; Morelli, G.; Ruberti, I. Multiple Links between HD-Zip Proteins and Hormone Networks. Int. J. Mol. Sci. 2018, 19, 4047. [Google Scholar] [CrossRef]
- Gong, S.; Ding, Y.; Hu, S.; Ding, L.; Chen, Z.; Zhu, C. The Role of HD-Zip Class I Transcription Factors in Plant Response to Abiotic Stresses. Physiol. Plant 2019, 167, 516–525. [Google Scholar] [CrossRef]
- Ré, D.A.; Capella, M.; Bonaventure, G.; Chan, R.L. ArabidopsisAtHB7 and AtHB12 evolved Divergently to Fine Tune Processes Associated with Growth and Responses to Water Stress. BMC Plant Biol. 2014, 14, 150. [Google Scholar] [CrossRef]
- Zhang, S.; Haider, I.; Kohlen, W.; Jiang, L.; Bouwmeester, H.; Meijer, A.H.; Schluepmann, H.; Liu, C.-M.; Ouwerkerk, P.B.F. Function of the HD-Zip I Gene Oshox22 in ABA-Mediated Drought and Salt Tolerances in Rice. Plant Mol. Biol. 2012, 80, 571–585. [Google Scholar] [CrossRef]
- Johannesson, H.; Wang, Y.; Engström, P. DNA-Binding and Dimerization Preferences of Arabidopsis Homeodomain-Leucine Zipper Transcription Factors In Vitro. Plant Mol. Biol. 2001, 45, 63–73. [Google Scholar] [CrossRef]
- Meijer, A.H.; Scarpella, E.; van Dijk, E.L.; Qin, L.; Taal, A.J.; Rueb, S.; Harrington, S.E.; McCouch, S.R.; Schilperoort, R.A.; Hoge, J.H. Transcriptional Repression by Oshox1, a Novel Homeodomain Leucine Zipper Protein from Rice. Plant J. 1997, 11, 263–276. [Google Scholar] [CrossRef]
- Harris, J.C.; Sornaraj, P.; Taylor, M.; Bazanova, N.; Baumann, U.; Lovell, B.; Langridge, P.; Lopato, S.; Hrmova, M. Molecular Interactions of the γ-Clade Homeodomain-Leucine Zipper Class I Transcription Factors during the Wheat Response to Water Deficit. Plant Mol. Biol. 2016, 90, 435–452. [Google Scholar] [CrossRef]




| Group | Darmor-bzh | Quinta | Tapidor | Shengli | Zheyou7 | ZS11 | Gangan | No2127 | Westar |
|---|---|---|---|---|---|---|---|---|---|
| I | 69 | 60 | 72 | 64 | 66 | 70 | 75 | 72 | 68 |
| II | 26 | 38 | 37 | 34 | 37 | 38 | 38 | 33 | 37 |
| III | 13 | 20 | 18 | 19 | 19 | 19 | 20 | 17 | 20 |
| IV | 57 | 50 | 52 | 52 | 34 | 53 | 42 | 51 | 46 |
| Total | 165 | 168 | 179 | 169 | 156 | 180 | 175 | 173 | 171 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, X.; Wu, D.; Zhang, X.; Liu, Q.; Lu, Q.; Song, M. Identification and Characterization of the HD-Zip Gene Family and Dimerization Analysis of HB7 and HB12 in Brassica napus L. Genes 2022, 13, 2139. https://doi.org/10.3390/genes13112139
Peng X, Wu D, Zhang X, Liu Q, Lu Q, Song M. Identification and Characterization of the HD-Zip Gene Family and Dimerization Analysis of HB7 and HB12 in Brassica napus L. Genes. 2022; 13(11):2139. https://doi.org/10.3390/genes13112139
Chicago/Turabian StylePeng, Xiangyong, Di Wu, Xin Zhang, Qingwei Liu, Qiuli Lu, and Min Song. 2022. "Identification and Characterization of the HD-Zip Gene Family and Dimerization Analysis of HB7 and HB12 in Brassica napus L." Genes 13, no. 11: 2139. https://doi.org/10.3390/genes13112139
APA StylePeng, X., Wu, D., Zhang, X., Liu, Q., Lu, Q., & Song, M. (2022). Identification and Characterization of the HD-Zip Gene Family and Dimerization Analysis of HB7 and HB12 in Brassica napus L. Genes, 13(11), 2139. https://doi.org/10.3390/genes13112139





