Transcription Analysis of Liver and Muscle Tissues from Landrace Finishing Pigs with Different Feed Conversion Ratios
Abstract
1. Introduction
2. Materials & Methods
2.1. Animals, Phenotype Selection, and Sample Collection
2.2. RNA Isolation
2.3. RNA Library Preparation and Sequencing
2.4. Read Processing and Mapping
2.5. Differential Gene Expression Analysis and qPCR Validation
2.6. DEGs Functional Annotation Clustering
3. Results
3.1. RNA-Seq Data
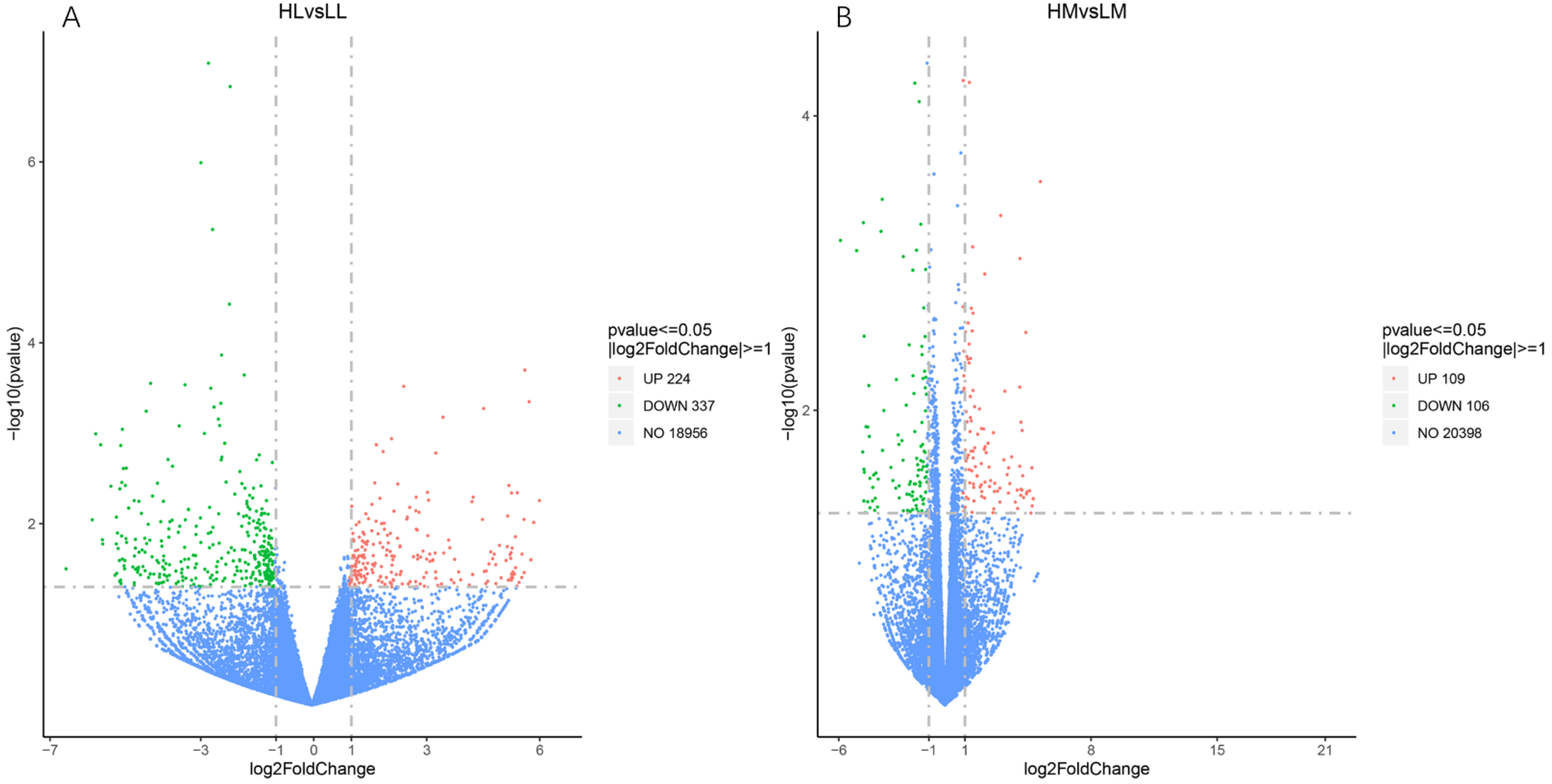
3.2. Gene Expression Analysis
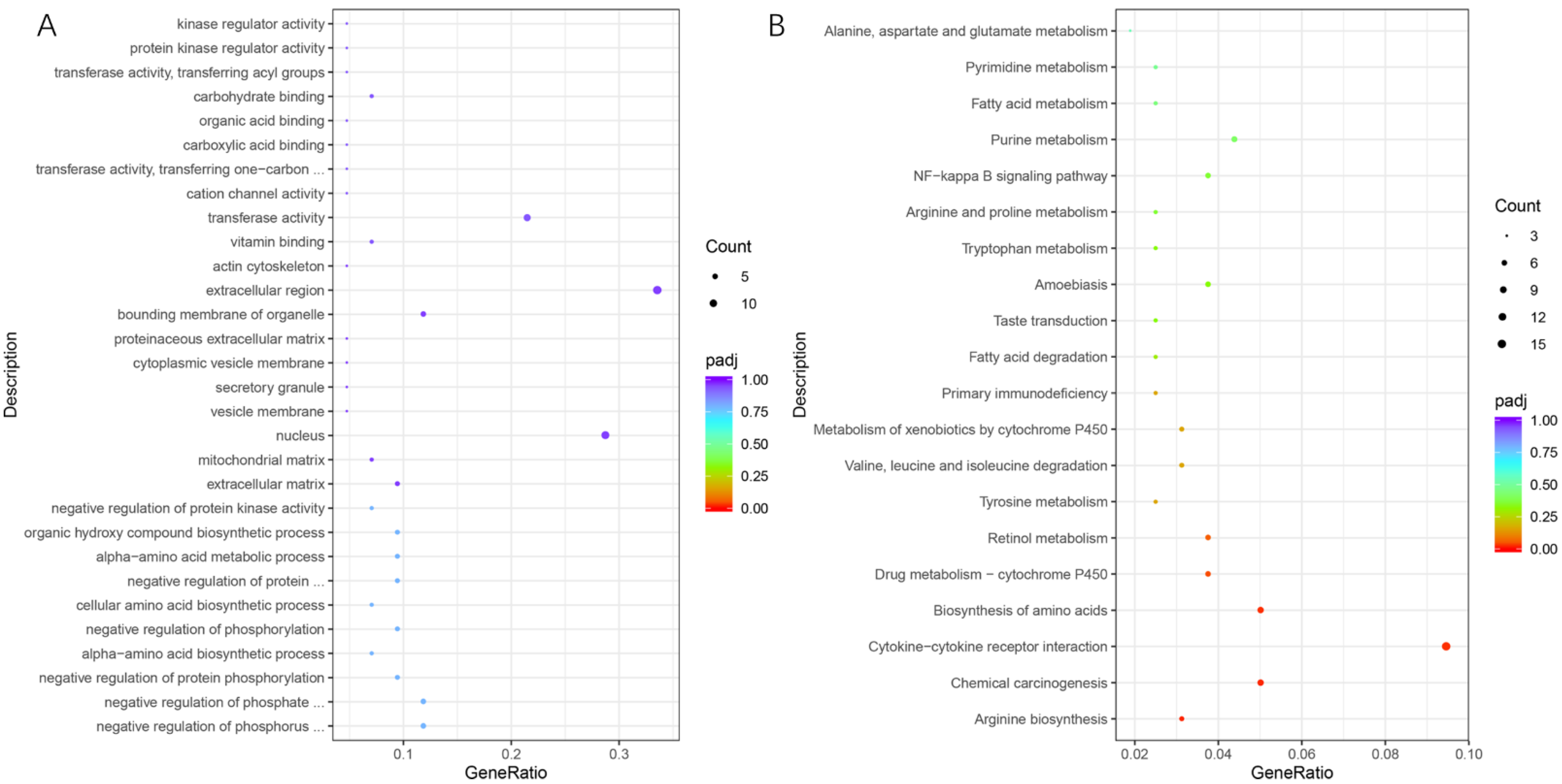
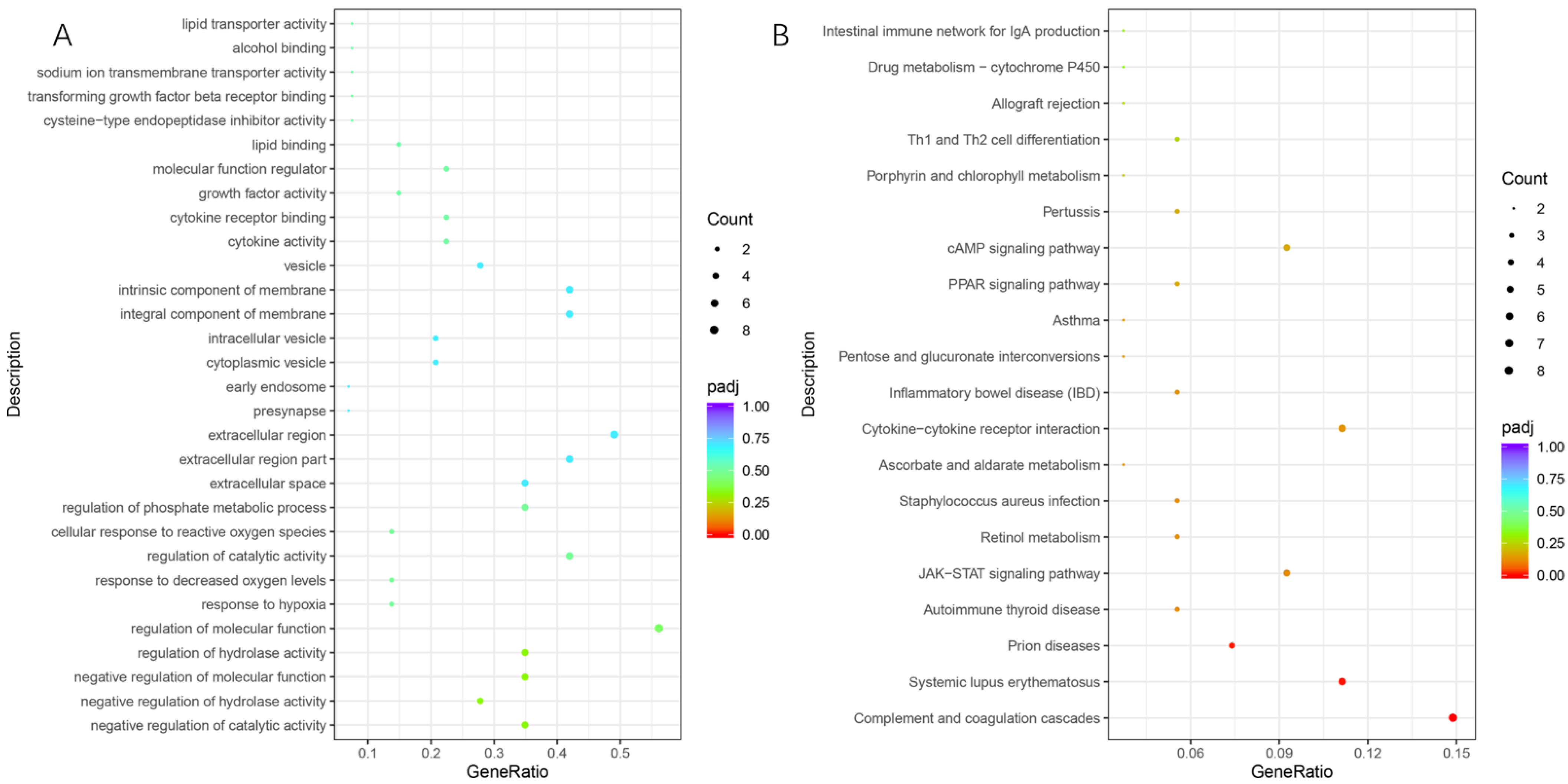
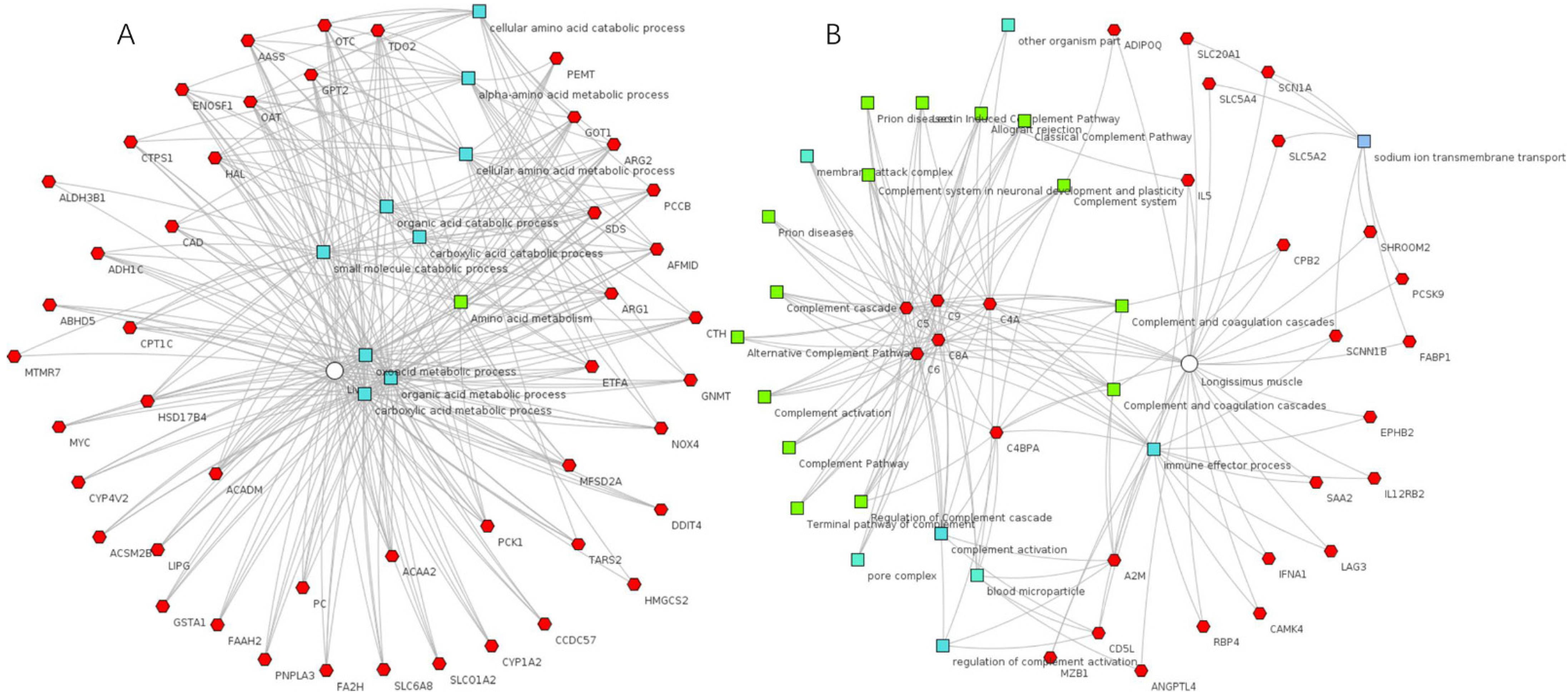
3.3. Functional Annotation Clustering of DEGs
3.4. Validation of DEGs Using qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aggrey, S.E.; Karnuah, A.B.; Sebastian, B.; Anthony, N.B. Genetic properties of feed efficiency parameters in meat-type chickens. Genet Sel. Evol. 2010, 42, 25. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Casey, D.S.; Dekkers, J.C. Selection response and genetic parameters for residual feed intake in Yorkshire swine. J. Anim. Sci. 2008, 86, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.A.; Kadowaki, H.; Shibata, T.; Oikawa, T.; Suzuki, K. Genetic parameters for measures of residual feed intake and growth traits in seven generations of Duroc pigs. Livest Sci. 2009, 121, 45–49. [Google Scholar] [CrossRef]
- Do, D.N.; Strathe, A.B.; Jensen, J.; Mark, T.; Kadarmideen, H.N. Genetic parameters for different measures of feed efficiency and related traits in boars of three pig breeds. J. Anim. Sci. 2013, 91, 4069–4079. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Ostersen, T.; Strathe, A.B.; Mark, T.; Jensen, J.; Kadarmideen, H.N. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 2014, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Strathe, A.B.; Ostersen, T.; Pant, S.D.; Kadarmideen, H.N. Genome-wide association and pathway analysis of feed efficiency in pigs reveal candidate genes and pathways for residual feed intake. Front. Genet. 2014, 5, 307. [Google Scholar] [CrossRef]
- Sahana, G.; Kadlecova, V.; Hornshoj, H.; Nielsen, B.; Christensen, O.F. A genome-wide association scan in pig identifies novel regions associated with feed efficiency trait. J. Anim. Sci. 2013, 91, 1041–1050. [Google Scholar] [CrossRef]
- Ding, R.R.; Yang, M.; Wang, X.W.; Quan, J.P.; Zhuang, Z.W.; Zhou, S.P.; Li, S.Y.; Xu, Z.; Zheng, E.Q.; Cai, G.Y.; et al. Genetic Architecture of Feeding Behavior and Feed Efficiency in a Duroc Pig Population. Front. Genet. 2018, 9, 220. [Google Scholar] [CrossRef]
- Gondret, F.; Vincent, A.; Houee-Bigot, M.; Siegel, A.; Lagarrigue, S.; Causeur, D.; Gilbert, H.; Louveau, I. A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs. BMC Genom. 2017, 18, 244. [Google Scholar] [CrossRef]
- Vincent, A.; Louveau, I.; Gondret, F.; Trefeu, C.; Gilbert, H.; Lefaucheur, L. Divergent selection for residual feed intake affects the transcriptomic and proteomic profiles of pig skeletal muscle. J. Anim. Sci. 2015, 93, 2745–2758. [Google Scholar] [CrossRef]
- Horodyska, J.; Hamill, R.M.; Reyer, H.; Trakooljul, N.; Lawlor, P.G.; McCormack, U.M.; Wimmers, K. RNA-Seq of Liver From Pigs Divergent in Feed Efficiency Highlights Shifts in Macronutrient Metabolism, Hepatic Growth and Immune Response. Front. Genet. 2019, 10, 117. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, S.; Wu, J.; Ding, R.; Quan, J.; Zheng, E.; Yang, J.; Wu, Z. A Transcriptome Analysis Identifies Biological Pathways and Candidate Genes for Feed Efficiency in DLY Pigs. Genes 2019, 10, 725. [Google Scholar] [CrossRef] [PubMed]
- Messad, F.; Louveau, I.; Renaudeau, D.; Gilbert, H.; Gondret, F. Analysis of merged whole blood transcriptomic datasets to identify circulating molecular biomarkers of feed efficiency in growing pigs. BMC Genom. 2021, 22, 501. [Google Scholar] [CrossRef] [PubMed]
- Lkhagvadorj, S.; Qu, L.; Cai, W.; Couture, O.P.; Barb, C.R.; Hausman, G.J.; Nettleton, D.; Anderson, L.L.; Dekkers, J.C.; Tuggle, C.K. Gene expression profiling of the short-term adaptive response to acute caloric restriction in liver and adipose tissues of pigs differing in feed efficiency. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, R494–R507. [Google Scholar] [CrossRef]
- Jing, L.; Hou, Y.; Wu, H.; Miao, Y.; Li, X.; Cao, J.; Brameld, J.M.; Parr, T.; Zhao, S. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs. Sci. Rep. 2015, 5, 11953. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Hou, Y.; Liu, F.; Liu, A.; Jing, L.; Zhao, C.Z.; Luan, Y.; Miao, Y.X.; Zhao, S.H.; Li, X.Y. Transcriptome Analysis Reveals that Vitamin A Metabolism in the Liver Affects Feed Efficiency in Pigs. G3-Genes Genom. Genet. 2016, 6, 3615–3624. [Google Scholar] [CrossRef]
- Louveau, I.; Vincent, A.; Tacher, S.; Gilbert, H.; Gondret, F. Increased expressions of genes and proteins involved in mitochondrial oxidation and antioxidant pathway in adipose tissue of pigs selected for a low residual feed intake. J. Anim. Sci. 2016, 94, 5042–5054. [Google Scholar] [CrossRef]
- Song, D.; Peng, Q.; Chen, Y.; Zhou, X.; Zhang, F.; Li, A.; Huang, D.; Wu, Q.; Ye, Y.; He, H.; et al. Altered Gut Microbiota Profiles in Sows and Neonatal Piglets Associated with Porcine Epidemic Diarrhea Virus Infection. Sci. Rep. 2017, 7, 17439. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Ballester, M.; Sanchez, J.P.; Gonzalez-Rodriguez, O.; Revilla, M.; Reyer, H.; Wimmers, K.; Torrallardona, D.; Quintanilla, R. Integrative approach using liver and duodenum RNA-Seq data identifies candidate genes and pathways associated with feed efficiency pigs. Sci. Rep. 2018, 8, 558. [Google Scholar] [CrossRef]
- Rui, L.Y. Energy Metabolism in the Liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar] [CrossRef]
- Shimizu, N.; Maruyama, T.; Yoshikawa, N.; Matsumiya, R.; Ma, Y.; Ito, N.; Tasaka, Y.; Kuribara-Souta, A.; Miyata, K.; Oike, Y.; et al. A muscle-liver-fat signalling axis is essential for central control of adaptive adipose remodelling. Nat. Commun. 2015, 6, 6693. [Google Scholar] [CrossRef] [PubMed]
- Reyer, H.; Oster, M.; Magowan, E.; Dannenberger, D.; Ponsuksili, S.; Wimmers, K. Strategies towards Improved Feed Efficiency in Pigs Comprise Molecular Shifts in Hepatic Lipid and Carbohydrate Metabolism. Int. J. Mol. Sci. 2017, 18, 1674. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Xu, Y.Y.; Hou, Y.; Qi, X.L.; Zhou, L.; Liu, H.Y.; Luan, Y.; Jing, L.; Miao, Y.X.; Zhao, S.H.; et al. Proteomic analysis indicates that mitochondrial energy metabolism in skeletal muscle tissue is negatively correlated with feed efficiency in pigs. Sci. Rep. 2017, 7, 45291. [Google Scholar] [CrossRef] [PubMed]
- Vigors, S.; O’Doherty, J.V.; Bryan, K.; Sweeney, T. A comparative analysis of the transcriptome profiles of liver and muscle tissue in pigs divergent for feed efficiency. BMC Genom. 2019, 20, 461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X.; He, Y.Z.; Wang, C.D.; Ao, H.; Tan, Z.; Xing, K. Variations in Microbial Diversity and Metabolite Profiles of Female Landrace Finishing Pigs With Distinct Feed Efficiency. Front. Vet. Sci. 2021, 8, 702931. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Wang, L.G.; Nie, J.F.; Sicotte, H.; Li, Y.; Eckel-Passow, J.E.; Dasari, S.; Vedell, P.T.; Barman, P.; Wang, L.W.; Weinshiboum, R.; et al. Measure transcript integrity using RNA-seq data. BMC Bioinform. 2016, 17, 58. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Xing, K.; Zhu, F.; Zhai, L.; Liu, H.; Wang, Y.; Wang, Z.; Chen, S.; Hou, Z.; Wang, C. Integration of transcriptome and whole genomic resequencing data to identify key genes affecting swine fat deposition. PLoS ONE 2015, 10, e0122396. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Wang, Y.; Yang, T.; Xing, K.; Ao, H.; Chen, S.K.; Zhang, F.X.; Zhao, X.T.; Liu, J.F.; Wang, C.D. Differentially expressed genes in the caecal and colonic mucosa of Landrace finishing pigs with high and low food conversion ratios. Sci. Rep. 2017, 7, 14886. [Google Scholar] [CrossRef]
- Xing, K.; Zhu, F.; Zhai, L.W.; Chen, S.K.; Tan, Z.; Sun, Y.Y.; Hou, Z.C.; Wang, C.D. Identification of genes for controlling swine adipose deposition by integrating transcriptome, whole-genome resequencing, and quantitative trait loci data. Sci. Rep. 2016, 6, 23219. [Google Scholar] [CrossRef] [PubMed]
- Xing, K.; Zhao, X.; Ao, H.; Chen, S.; Yang, T.; Tan, Z.; Wang, Y.; Zhang, F.; Liu, Y.; Ni, H.; et al. Transcriptome analysis of miRNA and mRNA in the livers of pigs with highly diverged backfat thickness. Sci. Rep. 2019, 9, 16740. [Google Scholar] [CrossRef]
- Grubbs, J.K.; Dekkers, J.C.; Huff-Lonergan, E.; Tuggle, C.K.; Lonergan, S.M. Identification of potential serum biomarkers to predict feed efficiency in young pigs. J. Anim. Sci. 2016, 94, 1482–1492. [Google Scholar] [CrossRef]
- Gilbert, H.; Billon, Y.; Brossard, L.; Faure, J.; Gatellier, P.; Gondret, F.; Labussiere, E.; Lebret, B.; Lefaucheur, L.; Le Floch, N.; et al. Review: Divergent selection for residual feed intake in the growing pig. Animal 2017, 11, 1427–1439. [Google Scholar] [CrossRef]
- Le Naou, T.; Le Floc’h, N.; Louveau, I.; Gilbert, H.; Gondret, F. Metabolic changes and tissue responses to selection on residual feed intake in growing pigs. J. Anim. Sci. 2012, 90, 4771–4780. [Google Scholar] [CrossRef]
- Yang, L.; He, T.; Xiong, F.; Chen, X.; Fan, X.; Jin, S.; Geng, Z. Identification of key genes and pathways associated with feed efficiency of native chickens based on transcriptome data via bioinformatics analysis. BMC Genom. 2020, 21, 292. [Google Scholar] [CrossRef]
- Alexandre, P.A.; Kogelman, L.J.; Santana, M.H.; Passarelli, D.; Pulz, L.H.; Fantinato-Neto, P.; Silva, P.L.; Leme, P.R.; Strefezzi, R.F.; Coutinho, L.L.; et al. Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle. BMC Genom. 2015, 16, 1073. [Google Scholar] [CrossRef] [PubMed]
- Miltiadou, D.; Hager-Theodorides, A.L.; Symeou, S.; Constantinou, C.; Psifidi, A.; Banos, G.; Tzamaloukas, O. Variants in the 3’ untranslated region of the ovine acetyl-coenzyme A acyltransferase 2 gene are associated with dairy traits and exhibit differential allelic expression. J. Dairy Sci. 2017, 100, 6285–6297. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fang, X.; Yang, R.; Yu, H.; Jiang, P.; Sun, B.; Zhao, Z. MiR-152 Regulates Apoptosis and Triglyceride Production in MECs via Targeting ACAA2 and HSD17B12 Genes. Sci. Rep. 2018, 8, 417. [Google Scholar] [CrossRef] [PubMed]
- Bacher, A.; Eberhardt, S.; Fischer, M.; Kis, K.; Richter, G. Biosynthesis of vitamin b2 (riboflavin). Annu. Rev. Nutr. 2000, 20, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, M.; Ishizaki, T.; Kimura, T. GTP- and GDP-Dependent Rab27a Effectors in Pancreatic Beta-Cells. Biol. Pharm. Bull. 2015, 38, 663–668. [Google Scholar] [CrossRef]
- Suhy, A.M.; Webb, A.; Papp, A.C.; Geier, E.G.; Sadee, W. Expression and splicing of ABC and SLC transporters in the human blood-brain barrier measured with RNAseq. Eur. J. Pharm. Sci. 2017, 103, 47–51. [Google Scholar] [CrossRef]
- Al Shawaf, E.; Abu-Farha, M.; Devarajan, S.; Alsairafi, Z.; Al-Khairi, I.; Cherian, P.; Ali, H.; Mathur, A.; Al-Mulla, F.; Al Attar, A.; et al. ANGPTL4: A Predictive Marker for Diabetic Nephropathy. J. Diabetes Res. 2019, 2019, 4943191. [Google Scholar] [CrossRef]
- Fernandez-Hernando, C.; Suarez, Y. ANGPTL4: A multifunctional protein involved in metabolism and vascular homeostasis. Curr. Opin. Hematol. 2020, 27, 206–213. [Google Scholar] [CrossRef]
- Ermert, D.; Blom, A.M. C4b-binding protein: The good, the bad and the deadly. Novel functions of an old friend. Immunol. Lett. 2016, 169, 82–92. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; He, Y.; Tan, Z. Transcription Analysis of Liver and Muscle Tissues from Landrace Finishing Pigs with Different Feed Conversion Ratios. Genes 2022, 13, 2067. https://doi.org/10.3390/genes13112067
Wang Z, He Y, Tan Z. Transcription Analysis of Liver and Muscle Tissues from Landrace Finishing Pigs with Different Feed Conversion Ratios. Genes. 2022; 13(11):2067. https://doi.org/10.3390/genes13112067
Chicago/Turabian StyleWang, Zhixin, Yingzhi He, and Zhen Tan. 2022. "Transcription Analysis of Liver and Muscle Tissues from Landrace Finishing Pigs with Different Feed Conversion Ratios" Genes 13, no. 11: 2067. https://doi.org/10.3390/genes13112067
APA StyleWang, Z., He, Y., & Tan, Z. (2022). Transcription Analysis of Liver and Muscle Tissues from Landrace Finishing Pigs with Different Feed Conversion Ratios. Genes, 13(11), 2067. https://doi.org/10.3390/genes13112067





