Abstract
MADS domain transcription factors play roles throughout the whole lifecycle of plants from seeding to flowering and fruit-bearing. However, systematic research into MADS-box genes of the economically important vegetable crop pepper (Capsicum spp.) is still lacking. We identified 174, 207, and 72 MADS-box genes from the genomes of C. annuum, C. baccatum, and C. chinense, respectively. These 453 MADS-box genes were divided into type I (Mα, Mβ, Mγ) and type II (MIKC* and MIKCC) based on their phylogenetic relationships. Collinearity analysis identified 144 paralogous genes and 195 orthologous genes in the three Capsicum species, and 70, 114, and 10 MADS-box genes specific to C. annuum, C. baccatum, and C. chinense, respectively. Comparative genomic analysis highlighted functional differentiation among homologous MADS-box genes during pepper evolution. Tissue expression analysis revealed three main expression patterns: highly expressed in roots, stems, leaves, and flowers (CaMADS93/CbMADS35/CcMADS58); only expressed in roots; and specifically expressed in flowers (CaMADS26/CbMADS31/CcMADS11). Protein interaction network analysis showed that type II CaMADS mainly interacted with proteins related to flowering pathway and flower organ development. This study provides the basis for an in-depth study of the evolutionary features and biological functions of pepper MADS-box genes.
1. Introduction
MADS-box genes comprise a large family of genes distributed throughout the plant kingdom and therefore occupy an important position in plant growth and development. The MADS acronym is composed of the initials of four proteins: a yeast transcription factor (MCM1), the Arabidopsis thaliana floral homozygote AGAMOUS (AG), an Antirrhinum majus floral homozygote (DEFICIENS), and human serum response factor (SRF) [1,2]. All four proteins have a highly conserved region consisting of 56–58 amino acids called the MADS domain [3,4]. Approximately one billion years ago, a duplication event occurred in the common ancestor of MADS-box genes, resulting in two distinct branches, type I and type II [5]. Type I proteins contain mainly SRF structural domains, and type I MADS-box genes can be further divided into Mα, Mβ, and Mγ; only a few types I genes have a biological function [6]. Type II genes are divided into MIKCC and MIKC* subtypes based on their structural features [7].
Replication and evolution of the type I MADS-box genes appear to be faster than those of type II genes, but these observations are based on few studies, mainly on the function of type II MADS-box gene in flower development [8]. According to the classical model of flower development “ABCDE”: class A genes regulate the formation of sepals; class A and B genes together regulate petals; class B and C genes control the differentiation of stamens; class C and D genes are mainly involved in the formation of ovules; class E genes are involved in the regulation of flower organs during the formation process [9,10,11]. In A.thaliana, APETALA1 (AP1) represents a class A gene [12]; APTALA3 (AP3) and PISTILATA (PI) genes belong to class B [13]; AG is a representative gene with class C function [14]; SEPALLATA (SEP) genes belong to class E [15], including SEP1, SEP2, SEP3, and SEP4; and class D genes SEEDSTICK (STK) and SHATTERPROOF1 (SHP1) [16]. In addition, the functions of some genes regulating flowering time, such as FLOWING LOCUS C (FLC), SHORT VIRAL PHASE (SVP), and SUPPORT OF OVEREXPRESSION OF CONSTANS 1 (SCO1), have been confirmed in A. thaliana, rice (Oryza sativa), and wheat (Triticum aestivum) [17,18,19].
MADS-box genes are involved in many plants’ growth and development processes. MADS-box genes play important regulatory roles in fruit growth and development, such as the FOREVER YOUNG FLOWER (SlFYFL) gene, the SEP-type SlCMB1 gene, and auxin-related SlIAA9 in tomato (Solanum lycopersicon) [20,21,22]; PaMADS7 of cherry (Cerasus pseudocerasus) [23]; MA-MADS5 of banana (Musa nana) [24]; and VEGETATIVE TO PRODUCTIVE TRANSITION 2 (TRV2) in wheat (Triticum aestivum) [25]. MADS-box genes are also involved in the plant stress response, such as Zymoseptoria tritici ZtRlm1; AtAGL16; and SiMADS51 in Setaria italica [26,27,28]. Therefore, the MADS-box family is one of the driving factors in plant diversity and plays an important role in growth and development [29,30].
Pepper (Capsicum spp.) originated in Central America and the Andes mountains, growing in tropical and temperate environments [31]. At present, 27 species of Capsicum have been identified [32], with five species cultivated long-term: Capsicum annuum, C. baccatum, C. chinese, C. frutescens, and C. pubescens [33]. Evolutionary analysis shows that C. annuum differentiated from C. chinense around 1.14 million years ago, and C. baccatum differentiated from C. annuum and C. chinense 1.7 million years ago [34]. At present, there is little research on the MADS-box gene family in pepper. A CaMADS-box gene is involved positively in low-temperature, salt, and osmotic stress signaling pathways [35]. CanMADS1 and CanMADS6 genes are expressed in flower buds and fruits and are highly expressed at 2 days after flowering, suggesting involvement in regulating pepper fruit development [36]. However, the members of the MADS-box gene family in pepper have not been systematically identified or analyzed. In this study, we carried out genome-wide identification of the MADS-box gene family from C. annuum, C. baccatum, and C. chinense genome data [34,37,38]. The objectives of this study are to identify and chacterize the MADS-box genes from three Capsicum species: C. annuum, C. baccatum, and C. chinense using genome-wide survey. As a result, 174, 207, and 72 MADS-box genes were identified from above three Capsicum species, respectively. Collinearity analysis identified 144 paralogous genes and 195 orthologous genes in the three peppers, and MADS-box genes underwent genome replication events in three pepper species during evolution. Protein interaction network analysis revealed that CaAP1 and CaAG were at the core of the network. These results revealed that type II MADS-box genes play vital roles in flower organ development of pepper. This study provides a theoretical basis for further study of revealing the functions of MADS-box genes in peppers and for the molecular breeding of peppers.
2. Material and Methods
2.1. Plant Materials
The seeds of C. annuum were collected in Sep 2019 from the Fangyang (32°37′ N and 87°46′ E) of the mid-reach of Huaihe River in Anhui, China. C. annuum seeds planted in pots containing soil:vermiculite:perlite (2:1:1) and placed in a growth chamber under long-day conditions (16 h−1 light/8 h−1 dark, 23/20 °C day/light, 150 µmol m–2 s–1). For tissue expression analysis, roots, stems, leaves were collected at the third true-leaf expanding stage, flower, sepal, petals, stamens, and pistils were harvested at flowering stage. Pepper fruits that grew to 3 cm long were sampled. All samples were immediately snap-frozen with liquid nitrogen and then stored in a −80 °C refrigerator until RNA extraction.
2.2. Identification and Naming of MADS-box Family Genes in Three Peppers
MADS-box protein sequences of A. thaliana were downloaded from the TAIR database (http://www.arabidopsis.org, accessed on 6 November 2021), and genomic data of Capsicum annuum, Capsicum baccatum and Capsicum chinense were downloaded from the Pepper Genome Platform (http://peppergenome.snu.ac.kr/, accessed on 6 November 2021) [34]. Algorithm-based BLASTP was performed using the MADS-box protein sequence of A. thaliana as the query in the protein databases of C. annuum, C. baccatum and C. chinense, with an E-value < 1 × 10−5 and other parameters as default values. The obtained candidate protein sequences were compared with Pfam (http://pfam.xfam.org/, accessed on 6 November 2021) database using HMMER (http://www.hmmer.org/, accessed on 6 November 2021). The MADS-box domain based on SRF domain (PF01486) and K domain (PF00319) was used for further comparison and screening, and the parameters were the default values. Thus, the MADS-box gene family members of three species were identified and named according to the order of their position on the chromosome. In order to view the distribution of MADS-box on the chromosomes of three pepper more directly, the online website MG2C (http://mg2c.iask.iN/mg2c_v2.0/, accessed on 6 November 2021) was used to draw the chromosome location map. The theoretical molecular weights and isoelectric points of MADS-box proteins were computed by the ExPASy (https://web.expasy.org/protparam/, accessed on 6 November 2021) proteomics server. The subcellular localization of CaMADS-box, CbMADS-box and CcMADS-box proteins were predicted by the ProtComp 9.0 (http://liNux1.softberry.com/berry.phtml, accessed on 6 November 2021) server.
2.3. Phylogenetic Tree Construction, Gene Structure and Protein Motif Analysis
The ClustalW program was used to perform multiple sequence alignments between the MADS-box gene family protein sequences of C. annuum, C. baccatum, C. chinense, and A. thaliana, with the default parameters [39]. MEGA11 was used to construct Maximum Likelihood phylogenetic trees and analyze the evolutionary relationship of MADS-box gene families among different species [40]. The phylogenetic trees were visualized using the EvolView server (https://www.evolgenius.info/evolview/#/treeview, accessed on 6 November 2021) [41]. Analysis of MADS-box gene exon-intron distribution based on C. annuum, C. baccatum, and C. chinense genome gff3 files using GSDS (http://gsds.cbi.pku.edu.cn/, accessed on 6 November 2021) visualization server. The conserved motifs of the CaMADS-box, CbMADS-box, and CcMADS-box family were identified using the MEME website (http://meme-suite.org/tools/meme, accessed on 6 November 2021). The motif length range was set to 10–60 amino acid residues, the maximum number of motif discoveries was set to 10 and other parameters were set to default values.
2.4. Selective Pressure Analysis
Paralogous and orthologous of MADS-box genes in C. annuum, C. baccatum, and C. chinense were identified using the Ortho Venn2 online website (https://orthovenn2.bioinfotoolkits.net/home, accessed on 6 November 2021) [42]. DNaSP 6.0 software was used to calculate the non-synonymous substitution rate (Ka) and the synonymous substitution rate (Ks) [43], and the selection pressure of replicated gene pairs during evolution was evaluated by calculating the ratio Ka/Ks. Ka/Ks > 1, <1 or =1 represent positive, negative or neutral evolution, respectively [44]. Tbtools [45] was used to visualize the collinearity relationship among gene members of paralogous and orthologous MADS-box genes in three Capsicum species.
2.5. Pepper MADS-box Gene Expression Analysis and qPCR Validation
RNA-seq data of C. annuum, C. baccatum, and C. chinense transcriptomic were obtained from the BioProject database (https://www.ncbi.nlm.nih.gov/bioproject, accessed on 6 November 2021), and the transcriptome accession number of the three Capsicum species was PRJNA223222, PRJNA308879 and PRJNA331024, respectively. [34]. The fastp [46] and RSEM tools [47] were used to filter and compare sequencing data, and the comparison was achieved using the Bowtie2 tool [48]. Parameters are set to default values. The results were standardized using the fragments per kilobase of transcript per million mapped reads (FPKM) of a gene. After the FPKM value was converted by log2(FPKM + 1), a heat map was created using the TBtools software [45], and the expression of CaMADS-box, CbMADS-box and CcMADS-box genes in different tissue was analyzed.
RNA from each tissue was extracted using TRIzol kit (Life Technologies, Carlsbad, CA, USA); reverse transcription was performed using HiScript III RT SuperMix for qPCR (Vazyme, Nanjing, China) kit; qPCR analysis was performed using ChamQ Universal SYBR qPCR (Vazyme, Nanjing) reagent. The PCR instrument was an ABI ViiA7 real-time fluorescence quantitative PCR machine (Life Technologies, USA). The Primer 3.0 tools (https://bioinfo.ut.ee/primer3−0.4.0/, accessed on 6 November 2021) were used to design CaMADS-box gene-specific amplification primers, using CaUBI3 as the reference gene [49] (Table S1). The qPCR primers are listed in Table S1. All qPCR assays were performed using three independent biological replicates, each with three technical replicates. The PCR conditions consisted of 2 min of initial denaturation at 94 °C followed by 40 cycles of 30 s at 94 °C, 45 s at 62 °C, 30 s at 72 °C, and a final 5 min extension at 72 °C. The PCR conditions and the calculation method of relative gene expression were the same as before [50].
2.6. Protein Interaction Network Prediction and Validation
The AraNetV2 tool (http://www.inetbio.org/aranet/, accessed on 6 November 2021) and the STRING (http://string-db.org/cgi, accessed on 6 November 2021) databases were used to identify the orthologous pairs between type II CaMADS-box and AtMADS-box genes. Then, the predicted protein–protein interaction network was displayed through Cytoscape software [51]. The yeast two-hybrid (Y2H) assays were performed in accordance with a method described previously [52,53]. The full-length CDSs of CaAG and CaAP1 were separately cloned into a pGBKT7-GAL4 vector (Clontech, Mountain View, CA, USA) as bait plasmids. The full-length CDSs of CaSEP3 and CaSVP were separately cloned into pGADT7 as the prey constructs (primers in Table S1). The resulting prey and bait constructs were cotransformed into the yeast strain AH109 as per the description of BD library construction and screening kit (Clontech, USA). The transformants were grown on SD/-Leu/-Trp medium and then transferred to SD/-Leu/-Trp/-His-Ade medium with 3 mM 3-AT to detect the interaction.
3. Results
3.1. Genome-Wide Identification and Characterization of the MADS-box Transcription Factor Gene Family from Capsicum L.
We identified 174, 207, and 72 MADS-box gene family members from three Capsicum species: C. annuum, C. baccatum, and C. chinense, respectively (Table S2), by referring to the amino acid sequences of A. thaliana MADS-box proteins, local BLAST comparison, and screening using the Pfam (http://pfam.xfam.org/, accessed on 6 November 2021) website. The number of genes in C. annuum and C. baccatum was more than that in C. chinense. All 453 MADS-box proteins possessed conserved SRF and K domains; the genes encoding these proteins were named CaMADS1-CaMADS174, CbMADS1-CbMADS207, and CcMADS1-CcMADS72, respectively. Analysis of physicochemical properties of MADS-box proteins in the three pepper types found that the amino acid (aa) lengths of CaMADS-box, CbMADS-box, and CcMADS-box proteins were 59–661, 49–660, and 78–1100 aa, respectively (Table S2). Their molecular weights were 6813–74021.51, 5691.74–74516.8 and 9052.62–125758.3 KDa, respectively. Isoelectric points ranged from 4.51 to 10.94, 4.89 to 10.23 and 4.46 to 10, respectively (Table S2). Prediction of subcellular localization showed that most MADS-box proteins were located in the nucleus, with a few located outside the cellular (Table S2).
3.2. Categorization, Structural Classification, and Structure of MADS-box Genes in Pepper
A phylogenetic tree was reconstructed using 105 AtMADS-box and 453 pepper MADS-box genes to further study the phylogenetic relationships of MADS-box genes. Following the classification and structure of the A. thaliana MADS-box family, we reconstructed two separate phylogenetic trees of type I (Figure S1) and type II (Figure 1) genes, respectively. The results showed that type I MADS-box genes were divided into three subfamilies (Mα, Mβ, and Mγ). There were 99, 6, and 16 members of Mα, Mβ, and Mγ, respectively, in C. annuum; 134, 8, and 7 members, respectively, in C. baccatum; and 34, 3, and 6 members, respectively, in C. chinense (Figure S1). The Mα subfamily in C. baccatum was considerably expanded, while the Mβ subfamily in C. chinense was substantially contracted. Type II MADS-box genes were further divided into 10 subfamilies: SEP, AGL16, AP1, STK/AG, AGL12, SCO1, SVP, PI/AP3, FLC/MAF, and MIKC*. The “ABCDE” genes of flower development, such as SEP, AP1, AG, PI, AP3, and STK, were amplified among the type II genes of the three pepper species (Figure 1, Table S2).
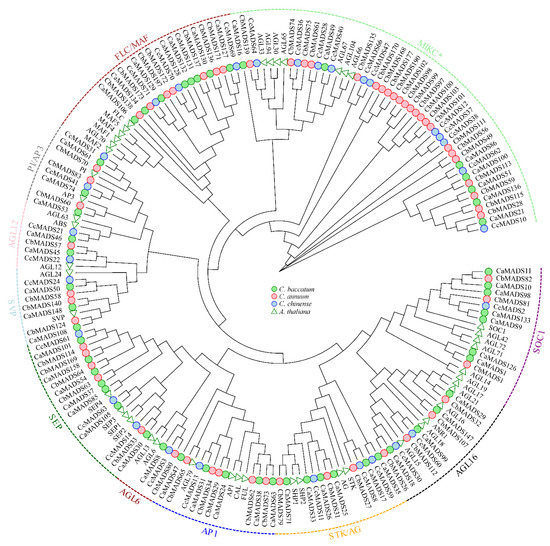
Figure 1.
Phylogenetic tree of type II MADS-box genes in Arabidopsis thaliana, Capsicum annuum, C. baccatum, and C. chinense.
Conserved motifs of MADS-box family proteins were analyzed using the online website MEME. A total of 10 motifs with a length of 15–41 amino acids were predicted (Table S3), and their distribution trend was conserved within every subfamily (Figures S2A, S3A and S4A). Motifs 1 and 3 were common to most MADS-box transcription factors, but motif 6 was unique to type II protein members. Motif 5 was also a unique domain for MIKC-type protein members.
Most MADS-box genes belonging to the same subfamily exhibited the same pattern of gene structure, but there were great differences among different members (Figures S2B, S3B and S4B). Most type I genes was composed of one exon, but CbMADS28, CbMADS81, CbMADS124, and CcMADS47 had two exons. Meanwhile, type II genes contained multiple introns and exons. Compared with type II genes, type I genes had suffered intron loss. Differences in exon and intron structure between type I and type II genes may be one of the reasons for the increase in MADS-box gene family members during evolution.
3.3. Phylogenetic Relationships of MADS-box Genes in Pepper
To investigate homologous MADS-box genes in pepper and possible gene duplication in each pepper species, we next identified the evolutionary relationships of MADS-box genes in the three capsicum species. There were 39 groups of orthologous genes, accounting for 22.9% (40/174) of MADS-box genes in C. annuum, 19.3% (40/207) of MADS-box genes in C. baccatum, and 55.5% (40/72) of MADS-box genes in C. chinense (Figure 2). This indicated that some MADS-box genes were preserved during evolution of C. annuum and C. baccatum, while MADS-box genes were highly conserved during the evolution of C. chinense. In addition to 40 groups of orthologous genes shared by the three pepper species (Table S4), 193 MADS-box homologs were found between any two pepper species (Table S5). There were 85 pairs of orthologous genes between C. baccatum and C. annuum, 61 pairs of orthologous genes between C. annuum and C. chinense, and 47 pairs of orthologous genes between C. baccatum and C. chinense (Table S5). Furthermore, C. annuum, C. baccatum, and C. chinense possessed 70, 117, and 10 unique MADS-box genes, respectively (Figure 2). There were 140 pairs of paralogous MADS-box genes, of which 47 pairs were tandem repeats (Table S6). Among these duplicated genes, some displayed have one-to-many relationships, such as CaMADS40, which was the tandem repeat gene of both CaMADS39 and CaMADS41. This was reflected in C. baccatum, with tandem repeat genes of CbMADS20 including CbMADS18, CbMADS17, CbMADS9, CbMADS8, and CbMADS191. However, there were no one-to-many situations in C. chinense. This indicates that the MADS-box gene family of pepper has an obvious gene replication phenomenon, which explains why the number of MADS-box genes in C. annuum and C. baccatum is more than that in C. chinense.
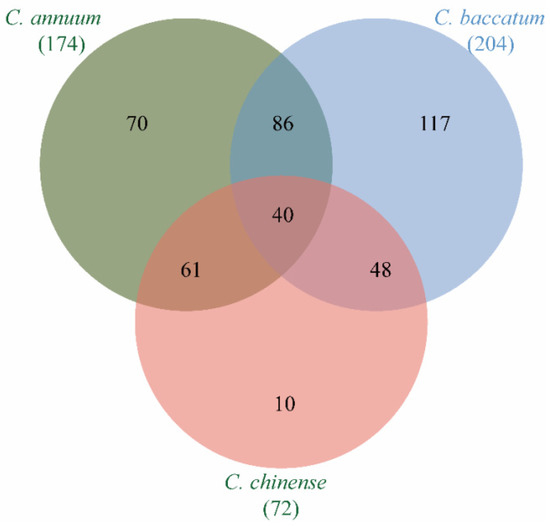
Figure 2.
Number of MADS-box orthologs in C. annuum, C. baccatum, and C. chinense.
To further explore the evolutionary mechanisms of MADS-box genes in pepper, we constructed collinear circos of the three Capsicum species based on orthologous genes and tandem repeat genes (Figure 3). The results revealed that MADS-box genes are distributed on every chromosome, mainly located at the terminus of each chromosome arm. Orthologous genes in the three Capsicum species were very close on the chromosome, anchored in a highly conserved collinearity block. Based on the genomic information available so far, we found 39 genes not localized to chromosomes in C. annuum, 69 genes not localized to chromosomes in C. baccatum, and 1 gene not localized to a chromosome in C. chinense. This phenomenon may also result from the error generated during chromosome assembly or the poor quality of assembly [34].
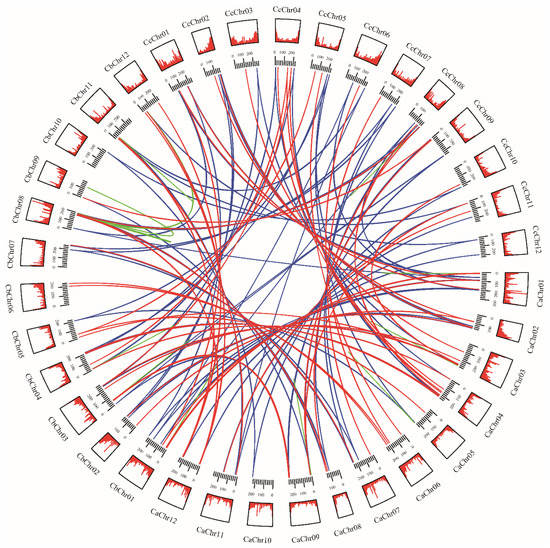
Figure 3.
Homologous MADS-box gene pairs in Capsicum annuum (Ca), C. baccatum (Cb), and C. chinense (Cc). Tracks from outside to inside are chromosomes numbers, gene density of the chromosome, and homologous gene pairs among the three Capsicum species. Blue lines connect homologous gene pairs that exist in three Capsicum species; red lines connect homologous gene pairs in two species; green lines connect paralogous genes.
In conclusion, the results revealed that the MADS-box transcription factor family of pepper is somewhat conserved. The 10 pairs of genes found in C. annuum formed linear relationships between pairs (Figure 3), indicating that duplication occurred between MADS-box genes. We calculated the selection pressure of paralogous as well as orthologous genes of the MADS-box gene family in Capsicum spp. pepper. The results showed that among the paralogous homologs (Figure 4A), Ka/Ks < 1 for all paralogous genes in C. annuum, while in C. baccatum, there were 32 pairs of paralogs with Ka/Ks > 1 (Table S7). This indicates that C. annuum was subject to strong purifying selection during its evolution, whereas C. baccatum was susceptible to environmental changes. However, no Ka/Ks values for paralogous genes were detected in C. chinense. Among the orthologs in the three Capsicum species, Capsicum annuum (Ca), C. baccatum (Cb), and C. chinense (Cc) (Figure 4B), the mean Ka/Ks values of the orthologs of Ca-Cb, Ca-Cc, and Cb-Cc were 0.6055, 0.6003, and 0.5952, respectively, with Ca-Cb having the largest mean value, implying that the MADS-box homolog of Ca-Cb was subject to greater purifying selection.
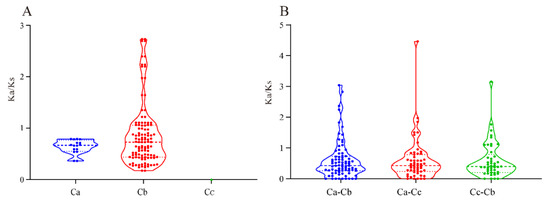
Figure 4.
Selection pressure statistics of paralogous (A) and orthologous (B) MADS-box genes in pepper. Ca, C. annuum; Cb, C. baccatum; Cc, C. chinense. The ‘dots’ reflect the maximum and minimum Ka/Ks scores.
3.4. Comparative Evolutionary Relationships of Type II MADS-box Genes in Three Capsicum Speciess
To study the contraction and expansion of type II MADS-box family members during evolution, the phylogenetic relationships among MIKC MADS-box genes in the three pepper species were explored using collinearity analysis. The results revealed 33 pairs of colinear genes between C. annuum and C. baccatum, and 18 pairs of colinear genes between C. baccatum and C. chinense (Figure 5, Table S8). Most type II genes were located at both ends of chromosomes, such as CaMADS8 on chromosome 1 and CbMADS80 on chromosome 8. In addition, the homologous genes on chromosomes 1, 2, 11, and 12 of C. annuum were distributed on chromosomes 4 and 5 of C. baccatum; the type II homologous genes on chromosomes 1 and 2 of C. annuum were located on chromosomes 6 and 3 of C. chinense. In C. baccatum, the type II homologous genes of chromosomes 1, 5, and 8 were distributed on chromosomes 1, 2, and 6 of C. chinense. There was no type II MADS-box gene on chromosome 9 in the three Capsicum species, which may be related to interchromosome 9 translocations [34]. In summary, most of the type II MADS-box genes in the three peppers showed conserved collinearity among chromosomal regions, but there was also deviation in duplicated gene pairs.
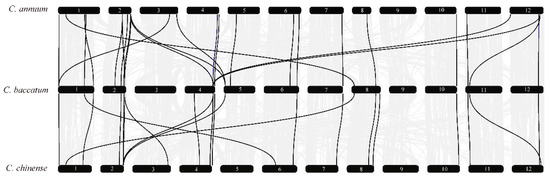
Figure 5.
Collinearity of type II MADS-box genes in C. annuum, C. baccatum, and C. chinense.
3.5. Expression Characteristics of MADS-box Genes in Different Pepper Tissues
We next analyzed the expression profiles of the MADS-box genes in root, stem, leaf, and flower tissues using RNA-seq data for the three peppers. The results showed that the expression of MADS-box genes in 73 groups of orthologous genes was considerably different among the three pepper species in the four tissues (Figure 6). Comprehensive analysis revealed that their expression patterns could mainly divide the genes into three categories: (1) genes expressed in all four tissues, such as CaMADS93/CbMADS35/CcMADS58, CaMADS82/CbMADS198/CcMADS4, and CaMADS116/CbMADS134/CcMADS66, indicating that they are widely involved in the growth and development of pepper; (2) genes with high expression during flower development, such as CaMADS26/CbMADS31/CcMADS11, CaMADS30/CbMADS33/CcMADS14, and CaMADS74/CbMADS83/CcMADS41, belonging to type II, suggesting that they play important roles in flower development; and (3) genes with high expression in roots, such as CaMADS9/CbMADS81/CcMADS2 and CaMADS68/CbMADS74/CcMADS36, indicating that these genes may be involved in root development and some physiological and biochemical processes in underground plant part. In addition to the three distinct expression patterns, most of the orthologous genes showed the same expression trend in the three pepper species, but some homologous genes displayed different expressions in the same tissue. For example, CcMADS69 was not expressed in any tissues, while its homologous gene CaMADS127 was highly expressed not only in flowers, but also in stems, indicating that orthologous genes in pepper may have gained or lost functions in the process of evolution.
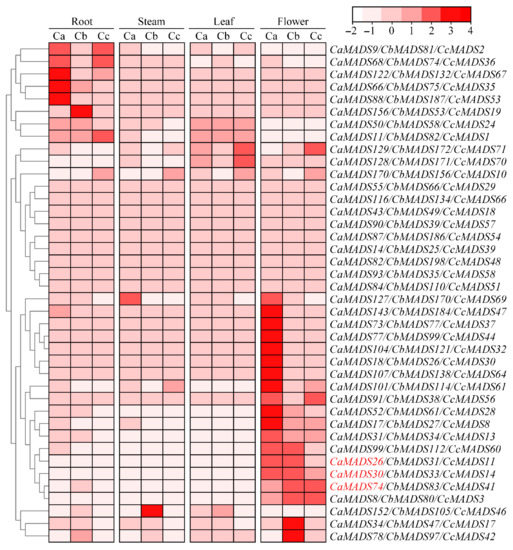
Figure 6.
Expression profiles of MADS-box genes in different tissues from Capsicum annuum (Ca), C. baccatum (Cb), and C. chinense (Cc). Color bar indicates the variation range of log10(FPKM + 1) values of MADS-box genes in different tissues. Expression of genes marked in red was verified by qPCR.
To further observe the expression of the type II MADS-box genes in different tissues of pepper, an expression heat map of 39 CaMADS-box genes in root, stem, leaf, and flower tissues was drawn (Figure S5). Six MADS-box genes, CaMADS74, CaMADS30, CaMADS61, CaMADS26, CaMADS105, and CaMADS63, were selected for analyzing expression levels in root, stem, leaf, flower, and fruit tissues using qPCR. The results revealed that the six MADS-box genes were differentially expressed in different tissues of pepper (Figure S5A), but they were highly expressed in flowers, which was consistent with the results of RNA-seq data (Figure 6). Both CaMADS26 and CaMADS30 were highly expressed in flowers, moderately expressed in fruits, and almost not expressed in other tissues. Both CaMADS61 and CaMADS74 were highly expressed in flowers, with little or no expression in other tissues, but CaMADS74 was weakly expressed in roots. Expression of CaMADS63 was the highest in flowers, followed by fruits and leaves, and low or trace expression was found in other tissues. However, the expression of CaMADS105 was higher in fruits than in flowers, and low or no expression was found in other tissues.
We next further analyzed the expression profiles of these six type II genes using qPCR in sepal, petal, stamen, and pistil tissues (Figure S5B). In the sepal, CaMADS61 expression was the highest, followed by CaMADS105 and CaMADS74. CaMADS30 was moderately expressed, while CaMADS63 and CaMADS26 were weakly expressed. CaMADS61 was highly expressed in petals, while the expression levels of CaMADS105 and CaMADS30 were relatively low. For other genes, there was little or no expression in petals. In stamen tissue, CaMADS63, CaMADS26, and CaMADS74 showed slightly expressed; CaMADS105 and CaMADS30 were moderately expressed; and CaMADS61 was highly expressed. CaMADS74 was the highest expression in the pistil, followed by CaMADS105. CaMADS30, CaMADS26, and CaMADS63 were slightly expressed, but CaMADS74 was not expressed in the pistil.
3.6. Interaction Network of Type II CaMADS Proteins
To better understand the biological functions of type II MADS-box genes in pepper, we next predicted the interaction network of CaMADS proteins. The results revealed that only 17 type II CaMADS-box members interacted with each other (Figure 7). The interacting proteins were mainly flowering pathway proteins and flower organ development proteins, of which AP1, AG, SEP3, AGL20, and AGL21 were at the core of the network. AtAP1 regulates the transition of inflorescence meristem and the morphological development of flower organs [54]. AtAG controls the stamens and carpels and inhibits the expression of AtAP1 [55]. AtSEP3 belongs to the class D gene, which involved in the process of flower development and activates the function of AtAG [56]. Moreover, Y2H assay confirmed that CaAG (CaMADS26) interacted with CaSVP (CaMADS50), and CaSEP3 (CaMADS105) could interact with CaAP1 (CaMADS24) and CaAG, respectively (Figure 8).
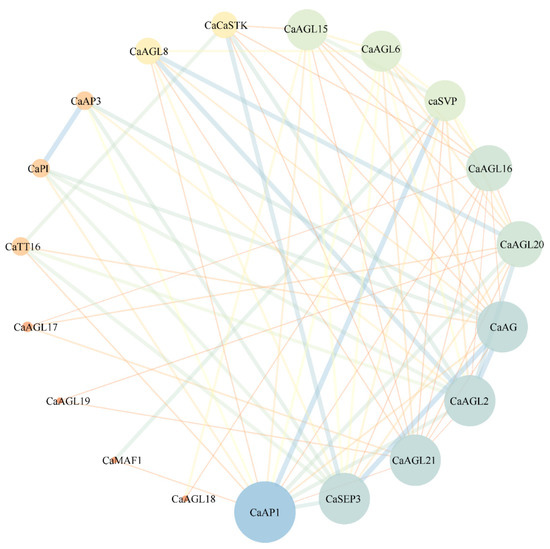
Figure 7.
Type II CaMADS-box Protein Interaction Network Diagram.
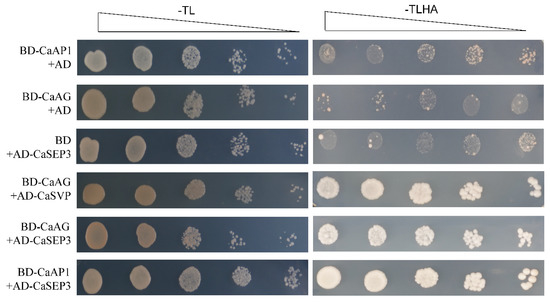
Figure 8.
Yeast two-hybrid assays of interactions among CaAP1, CaAG, CaSVP, and CaSEP3 proteins.
Connection between nodes varies with the combined_score value (representing the reliability of the predicted interaction between the two proteins, ranging between 0 and 1), thickening with an increase in score value. The size of nodes increases with the number of proteins interacting with node proteins.
Triangles represent a 10-fold dilution, with T, L, H, and A representing Tryptophan, Leucine, Histidine, and Adenine, respectively.
4. Discussion
Gene duplication often accompanies plant evolution and is an important reason for the expansion of gene families [57]. The MADS-box family is one of the largest transcription factor families and plays an important role in growth and development, and signal transduction [4,58]. With the development of sequencing technology, MADS-box gene family members have been identified in a variety of plants in varying numbers, such as 107 MADS-box gene members in A. thaliana [7], 83 in Camellia sinensis [59], 44 in Nelumbo nucifera [60], 44 in Erigeron breviscapus [61], 131 in Solanum lycopersicum [62], 54 in Morella rubra [63], 42 Phyllostachys heterocycle [64], 80 in Triticum aestivum [65], 54 in Ziziphus jujuba [66], 144 in Raphanus sativus [67], 82 in Lactuca sativa [68], 160 in Brassica rapa [69], 78 in Callicarpa americana [70], and 108 in Chrysanthemum nankingense [71]. These studies indicate that MADS-box genes have undergone obvious amplification and contraction, and the number and distribution in different subfamilies are also different. We study identified 174, 207, and 72 MADS-box genes from C. annuum, C. baccatum, and C. chinense, respectively (Figure 1; Table S1), in line with this trend. Moreover, the number of MADS-box family genes in C. baccatum was more than that in C. annuum and C. chinense, which may be due to the expansion of the C. baccatum genome caused by the amplification of retrotransposons [34]. The number of MADS-box genes of type I and MIKC subfamilies in C. annuum and C. baccatum was more than that of the model plant A. thaliana and the related species tomato. These results indicate that the MADS-box genes in C. annuum and C. baccatum have significantly expanded, but strangely, the number of genes identified in C. chinense was lower than that in C. annuum and C. baccatum, suggesting that a large number of genes have been lost during evolution. Among the MADS-box family members, those belonging to the same subfamily possessed similar motif composition and gene structure, but there was a unique motif composition and gene structure between type I and type II genes. The MADS-box genes of type I, including Mα, Mβ, and Mγ, generally contained no or few exons (Figures S2–S4) and may have lost multiple introns during the diversification of the MADS-box family. In addition, the distribution of introns in pepper MADS-box genes was also different. MIKC–type genes had more introns than those of type I, which are also found in A. thaliana, tomato, and rice [72], indicating that evolution between species is conserved. However, some genes of the same subfamily showed different intron and exon arrangements, indicating the complexity of gene structure evolution, which needs further study. The same conserved motifs in the same subfamily (Figures S2B, S3B and S4B) suggest that these motifs play an important role in gene functional specificity. Analyses of gene structure and conserved motifs provide clues for the expansion and contraction of the MADS-box gene family in pepper.
In eukaryotes, gene replication plays an important role in amplifying the number of transcription factor families and genomic complexity [57,62]. Previous studies confirmed that C. annuum diverged from C. chinense 1.14 million years ago and C. baccatum diverged from C. chinense and C. annuum 1.7 million years ago [34]. Our study revealed only 47 groups of orthologous genes among three pepper species, while other orthologous genes were lost to a certain extent (Figure 2). We also identified 144 groups of paralogous genes and 195 groups of orthologous genes (Figure 3, Tables S4–S6). However, some homologous genes were lost in the three species of pepper. These results further demonstrate both obvious contraction and expansion trends that the different subfamily members of the MADS-box gene family during the process of pepper evolution.
Genome sequencing revealed a dynamic rearrangement of chromosomes 3, 5, and 9 in C. baccatum, namely, translocation [34]. MADS-box genes at these loci were also changed, such as CbMADS35 on chromosome 3 of C. baccatum, and its homologous gene CcMADS58 on chromosome 9. In addition, some homologous genes were also located on different chromosomes. Moreover, most of the orthologous genes with Ka/Ks greater than 1 displayed positive selection and may show positive changes in function under the influence of the environment (Figure 4).
Most MADS-box genes were differentially expressed in different tissues of pepper (Figure 6 and Figure S5), indicating their functional diversity in different tissues. Some MADS-box genes showed tissue-specific expression, such as CaMADS9/CbMADS81/CcMADS2 and CaMADS66/CbMADS75/CcMADS35, which were mainly specifically expressed in roots, and are important candidate genes for further functional analysis. Some MADS-box genes were highly expressed in fruits (Figure S5), such as CaMADS30, CaMASDS61, CaMADS63, and CaMADS105, suggesting important roles in controlling fruit development. Several studies have proved that MADS-box genes play an important role in the morphogenesis and growth of roots and fruits [73,74]. CaMADS105 were expressed in sepal, petal, stamen, and pistil (Figure S5B), suggesting that they play vital roles in all stages of flower development. MIKC MADS-box genes play a central role in plant development [75].
In this study, we predicted the possible key genes in pepper flower organ development through phylogenetic relationships, such as class A genes (CcMADS13/CaMADS31/CbMADS34), class B genes (CcMADS41/CaMADS74/CbMADS83), class C genes (CcMADS11/CaMADS26/CbMADS31), class D genes (CcMADS8/CaMADS17/CbMADS27), and class E genes (CcMADS14/CaMADS30/CbMADS33, CcMADS61/CaMADS101/CbMADS114). Protein interaction network analysis revealed (Figure 7) that type II MADS-box proteins were mainly proteins related to flowering regulation and flower organ development. Y2H assay confirmed that CaAG interacted with CaSEP3 and CaSVP, and CaSEP3 interacted with CaAP1 and CaAG, suggesting that CaAG, CaSEP3, CaSVP, and CaAP1 play important roles in flowering, and the formations of sepal, petal, carpel, and stamen. AtAG is the only C functional gene in Arabidopsis, which interacts genetically with other allotypic genes to identify floral organs [76]. In summary, we comprehensively identified the MADS-box gene members of the three Capsicum species and analyzed their structural characteristics and evolutionary rules. The MIKC MADS-box genes identified in this study should be candidate genes for pepper breeding and improvement.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13112047/s1, Table S1: Primer information used in this study; Table S2: Information of MADS-box genes identified in three Capsicum species; Table S3: Conserved amino acid sequences in MADS-box genes of three Capsicum species identified by MEME; Table S4: The identified orthologous genes in three Capsicum species; Table S5: The identified orthologous genes between two Capsicum species; Table S6: The identified paralogous genes among in Capsicum species; Table S7: The Ka/Ks ratios for the paralogous and orthologous gene pairs of MADS-box genes in C. annuum, C. baccatum, and C. chinense; Table S8: Homologous II type MADS-box gene pairs in C. annuum, C. baccatum, and C. chinense; Figure S1: Phylogenetic tree of type I MADS-box genes in Arabidopsis thaliana, Capsicum annuum, C. baccatum, and C. chinense; Figure S2: Gene structure and motifs analysis of MADS-box genes in C. annuum; Figure S3: Gene structure and motifs analysis of MADS-box genes in C. baccatum; Figure S4: Gene structure and motifs analysis of MADS-box genes in Capsicum chinense; Figure S5: Tissue expression profiles of six MADS-box genes using qPCR A: Expression patterns of six MADS-box genes in root, stem, leaf, flower, and fruit tissues; B: Expression patterns of six MADS-box genes in sepal, petal, stamen, and pistil tissues.
Author Contributions
Z.G. performed the genomic analysis. Z.G. and X.W. performed all the experiments in the lab. S.A.M.B. and T.F. helped Z.G. with the genomic analysis and qPCR validation. N.H. and X.L. were in charge of the pepper field management. X.H. and R.L. designed and discussed the experimental work. Z.G. has written the original manuscript, X.H, R.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Talent Introduction Start-up Fund Project of Anhui Science and Technology University (NXYJ202001), the University Discipline (Major) Top Talent Cultivation Funding Project (gxbjZD2021072), the Postgraduate Science Research Project of Anhui Science and Technology University (YK202115), and the Natural Science Foundation of Anhui (2108085QC125).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
MADS-box gene members of A. thaliana were downloaded from the TAIR database (http://www.arabidopsis.org, accessed on 6 November 2021). Genomic data of C. annuum, C. baccatum and C. chinense were downloaded from the Pepper Genome Platform (http://peppergenome.snu.ac.kr/, accessed on 6 November 2021). C. annuum, C. baccatum and C. chinense transcriptome sequencing data were downloaded from the BioProject database (https://www.ncbi.nlm.nih.gov/bioproject, accessed on 3 April 2021).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shore, P.; Sharrocks, A.D. The MADS-box Family of Transcription Factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lawton-Rauh, A.L.; Alvarez-Buylla, E.R.; Purugganan, M.D. Molecular Evolution of Flower Development. Trends Ecol. Evol. 2000, 15, 144–149. [Google Scholar] [CrossRef]
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Theissen, G. The Major Clades of MADS-box Genes and Their Role in The Development and Evolution of Flowering Plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; de Pouplana, L.R.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and Phylogenetic Analyses of The Complete MADS-box Transcription Factor Family in Arabidopsis: New Openings to The MADS World. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Grimplet, J.; Martínez-Zapater, J.M.; Carmona, M.J. Structural and Functional Annotation of the MADS-box Transcription Factor Family in Grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef]
- Weigel, D.; Meyerowitz, E.M. The ABCs of Floral Homeotic Genes. Cell 1994, 78, 203–209. [Google Scholar] [CrossRef]
- Theissen, G.; Kim, J.T.; Saedler, H. Classification and Phylogeny of the MADS-box Multigene Family Suggest Defined Roles of MADS-box Gene Subfamilies in The Morphological Evolution of Eukaryotes. J. Mol. Evol. 1996, 43, 484–516. [Google Scholar] [CrossRef]
- Ferrario, S.; Immink, R.G.; Angenent, G.C. Conservation and Diversity in Flower Iand. Curr. Opin. Plant Biol. 2004, 7, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Irish, V.F.; Sussex, I.M. Function of The Apetala-1 Gene During Arabidopsis Floral Development. Plant Cell 1990, 2, 741–753. [Google Scholar] [PubMed]
- Jack, T.; Brockman, L.L.; Meyerowitz, E.M. The Homeotic Gene APETALA3 of Arabidopsis thaliana Encodes a MADS box and is Expressed in Petals and Stamens. Cell 1992, 68, 683–697. [Google Scholar] [CrossRef]
- Mizukami, Y.; Ma, H. Ectopic Expression of The Floral Homeotic Gene AGAMOUS in Transgenic Arabidopsis Plants Alters Floral Organ Identity. Cell 1992, 71, 119–131. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C Floral Organ Identity Functions Require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box Protein Complexes Control Carpel and Ovule Development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jeong, D.H.; An, G. A Possible Working Mechanism for Rice SVP-Group MADS-box Proteins as Negative Regulators of Brassinosteroid Responses. Plant Signal. Behav. 2008, 3, 471–474. [Google Scholar] [CrossRef] [PubMed]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-Induced Silencing by Long Antisense Transcripts of an Arabidopsis Polycomb Target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Shimada, S.; Ogawa, T.; Kitagawa, S.; Suzuki, T.; Ikari, C.; Shitsukawa, N.; Abe, T.; Kawahigashi, H.; Kikuchi, R.; Handa, H.; et al. A Genetic Network of Flowering-Time Genes in Wheat Leaves, in Which an APETALA1/FRUITFULL-Like Gene, VRN1, is Upstream of FLOWERING LOCUS T. Plant J. 2009, 58, 668–681. [Google Scholar] [CrossRef]
- Xie, Q.; Hu, Z.; Zhu, Z.; Dong, T.; Zhao, Z.; Cui, B.; Chen, G. Overexpression of a Novel MADS-box Gene SlFYFL delays Senescence, Fruit Ripening and Abscission in Tomato. Sci. Rep. 2014, 4, 4367. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Z.; Yao, Q.; Guo, X.; Nguyen, V.; Li, F.; Chen, G. A Tomato MADS-box Protein, SlCMB1, Regulates Ethylene Biosynthesis and Carotenoid Accumulation During Fruit Ripening. Sci. Rep. 2018, 8, 3413. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Dusi, V.; Pennisi, F.; Pandolfini, T. How Hormones and MADS-box Transcription Factors are Involved in Controlling Fruit Set and Parthenocarpy in Tomato. Genes 2020, 11, 1441. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Liu, C.; Song, L.; Li, M. Pamads7, A MADS-box Transcription Factor, Regulates Sweet Cherry Fruit Ripening and Softening. Plant Sci. 2020, 301, 110634. [Google Scholar] [CrossRef] [PubMed]
- Roy Choudhury, S.; Roy, S.; Nag, A.; Singh, S.K.; Sengupta, D.N. Characterization of an AGAMOUS-like MADS box Protein, a Probable Constituent of Flowering and Fruit Ripening Regulatory System in Banana. PLoS ONE 2012, 7, e44361. [Google Scholar] [CrossRef] [PubMed]
- Adamski, N.M.; Simmonds, J.; Brinton, J.F.; Backhaus, A.E.; Chen, Y.; Smedley, M.; Hayta, S.; Florio, T.; Crane, P.; Scott, P.; et al. Ectopic Expression of Triticum polonicum VRT-A2 Underlies Elongated Glumes and Grains in Hexaploid Wheat in a Dosage-Dependent Manner. Plant Cell 2021, 33, 2296–2319. [Google Scholar] [CrossRef]
- Mohammadi, N.; Mehrabi, R.; Gohari, A.M.; Roostaei, M.; Goltapeh, E.M.; Safaie, N.; Kema, G.H.J. MADS-Box Transcription Factor ZtRlm1 Is Responsible for Virulence and Development of the Fungal Wheat Pathogen Zymoseptoria tritici. Front. Microbiol. 2020, 11, 1976. [Google Scholar] [CrossRef]
- Zhao, P.X.; Miao, Z.Q.; Zhang, J.; Chen, S.Y.; Liu, Q.Q.; Xiang, C.B. Arabidopsis MADS-box Factor AGL16 Negatively Regulates Drought Resistance Via Stomatal Density and Stomatal Movement. J. Exp. Bot. 2020, 71, 6092–6106. [Google Scholar] [CrossRef]
- Lai, D.; Yan, J.; He, A.; Xue, G.; Yang, H.; Feng, L.; Wei, X.; Li, L.; Xiang, D.; Ruan, J.; et al. Genome-wide Identification, Phylogenetic and Expression Pattern Analysis of MADS-box Family Genes in Foxtail Millet (Setaria italica). Sci. Rep. 2022, 12, 4979. [Google Scholar] [CrossRef]
- Theissen, G.; Saedler, H. Plant Biology. Floral Quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Hirano, H.Y. Function and Diversification of MADS-box Genes in Rice. Sci. World J. 2006, 6, 1923–1932. [Google Scholar] [CrossRef]
- Aguilar-Meléndez, A.; Morrell, P.L.; Roose, M.L.; Kim, S.C. Genetic Diversity and Structure in Semiwild and Domesticated Chiles (Capsicum annuum; Solanaceae) from Mexico. Am. J. Bot. 2009, 96, 1190–1202. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, E.; Zhang, D.; Mays, A.D.; Saftner, R.A.; Stommel, J.R. Genetic Diversity in Capsicum baccatum is Significantly Influenced by its Ecogeographical Distribution. BMC Genet. 2012, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Troconis-Torres, I.G.; Rojas-López, M.; Hernández-Rodríguez, C.; Villa-Tanaca, L.; Maldonado-Mendoza, I.E.; Dorantes-Álvarez, L.; Tellez-Medina, D.; Jaramillo-Flores, M.E. Biochemical and Molecular Analysis of Some Commercial Samples of Chilli Peppers From Mexico. J. Biomed. Biotechnol. 2012, 2012, 873090. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, J.; Yeom, S.I.; Kim, Y.M.; Seo, E.; Kim, K.T.; Kim, M.S.; Lee, J.M.; Cheong, K.; Shin, H.S.; et al. New Reference Genome Sequences of Hot Pepper Reveal The Massive Evolution of Plant Disease-Resistance Genes by Retroduplication. Genome Biol. 2017, 18, 210. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Ma, J.; Luo, D.; Hou, X.; Ma, F.; Zhang, Y.; Meng, Y.; Zhang, H.; Guo, W. CaMADS, a MADS-box transcription factor from pepper, plays an important role in the response to cold, salt, and osmotic stress. Plant Sci. 2019, 280, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.K.; Moon, Y.H.; Chung, J.E.; Lee, S.Y.; Park, H.G.; An, G. Characterization of MADS box Genes from Hot Pepper. Mol Cells. 2001, 11, 352–359. [Google Scholar]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome Sequencing of Cultivated and Wild Peppers Provides Insights into Capsicum Domestication and Specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Zhu, Z.; Liu, Y.; Chen, J.; Zhou, Y.; Liu, F.; Lei, J.; Gaut, B.S.; Cao, B.; et al. The 3D Architecture of The Pepper Genome and its Relationship to Function and Evolution. Nat. Commun. 2022, 13, 3479. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2–3. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview, An Online Tool for Visualizing, Annotating and Managing Phylogenetic Trees. Nucleic. Acids. Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. Orthovenn2: A Web Server for Whole-genome Comparison and Annotation of Orthologous Clusters Across Multiple Species. Nucleic. Acids. Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. Dnasp 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Yadav, C.B.; Bonthala, V.S.; Muthamilarasan, M.; Pandey, G.; Khan, Y.; Prasad, M. Genome-wide Development of Transposable Elements-based Markers in Foxtail Millet and Construction of an Integrated Database. DNA Res. 2015, 22, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification From RNA-seq Data with or Without a Reference Genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Wan, H.; Yuan, W.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Zhao, J.; Liu, S.; et al. Identification of Reference Genes for Reverse Transcription Quantitative Real-time PCR Normalization in Pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 2011, 416, 24–30. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, F.; Huang, W.; Sun, Q.; Huang, X. Identification of Reliable Reference Genes for qRT-PCR in the Ephemeral Plant Arabidopsis pumila Based on Full-length Transcriptome Data. Sci. Rep. 2019, 9, 8408. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef]
- Si, Z.; Liu, H.; Zhu, J.; Chen, J.; Wang, Q.; Fang, L.; Gao, F.; Tian, Y.; Chen, Y.; Chang, L.; et al. Mutation of SELF-PRUNING Homologs in Cotton Promotes Short-Branching Plant Architecture. J. Exp. Bot. 2018, 5269, 2543–2553. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, X.Z.; Ma, B.; Zhang, T.T.; Sang, N.; Lu, Z.; Zhu, J.B. Components and Functional Diversification of Florigen Activation Complexes in Cotton. Plant Cell Physiol. 2021, 62, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- Mandel, M.A.; Yanofsky, M.F. A Gene Triggering Flower Formation in Arabidopsis. Nature 1995, 377, 522–524. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.; Muiño, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-domain Transcription Factor Complexes in Arabidopsis Flower Development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, C.; Romera-Branchat, M.; Pelaz, S. A New Role of The Arabidopsis SEPALLATA3 Gene Revealed by its Constitutive Expression. Plant J. 2005, 43, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Kim, S.; Koh, J.; Yoo, M.J.; Kong, H.; Hu, Y.; Ma, H.; Soltis, P.S.; Soltis, D.E. Expression of Floral MADS-box Genes in Basal Angiosperms: Implications for the Evolution of Floral Regulators. Plant J. 2005, 43, 724–744. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Jin, Y.J.; Wan, H.H.; Cheng, L.; Feng, Z.G. Genome-wide Identification and Expression Analysis of the MADS-box Transcription Factor Family in Camellia sinensis. J. Appl. Genet. 2021, 62, 249–264. [Google Scholar] [CrossRef]
- Lin, Z.; Cao, D.; Damaris, R.N.; Yang, P. Genome-wide Identification of MADS-box Gene Family in Sacred Lotus (Nelumbo nucifera) Identifies a SEPALLATA Homolog Gene Involved in Floral Development. BMC Plant Biol. 2020, 20, 497. [Google Scholar] [CrossRef]
- Tang, W.; Tu, Y.; Cheng, X.; Zhang, L.; Meng, H.; Zhao, X.; Zhang, W.; He, B. Genome-wide Identification and Expression Profile of the MADS-box Gene Family in Erigeron breviscapus. PLoS ONE 2019, 14, e0226599. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-wide Analysis of the MADS-box Transcription Factor Family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.B.; Jia, H.M.; Wang, Y.; Wang, G.Y.; Zhou, C.C.; Jia, H.J.; Gao, Z.S. Genome-wide Identification and Analysis of the MADS-box Gene Family and its Potential Role in Fruit Development and Ripening in Red Bayberry (Morella rubra). Gene 2019, 717, 144045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, D.; Lin, X.; Ding, M.; Tong, Z. Genome-wide Identification of MADS-box Family Genes in Moso Bamboo (Phyllostachys edulis) and a Functional Analysis of Pemads5 in Flowering. BMC Plant Biol. 2018, 18, 176. [Google Scholar] [CrossRef]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P.; et al. Genome-wide Identification and Analysis of the MADS-box Gene Family in Bread Wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0181443. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Feng, C.; Liu, M.; Wang, J.; Hu, Y. Genome-Wide Identification, Characterization of the MADS-box Gene Family in Chinese Jujube and their Involvement in Flower Development. Sci. Rep. 2017, 7, 1025. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Xu, L.; Nie, S.; Chen, Y.; Liang, D.; Sun, X.; Karanja, B.K.; Luo, X.; Liu, L. Genome-wide Characterization of the MADS-box Gene Family in Radish (Raphanus sativus L.) and Assessment of its Roles in Flowering and Floral Organogenesis. Front. Plant Sci. 2016, 7, 1390. [Google Scholar] [CrossRef]
- Ning, K.; Han, Y.; Chen, Z.; Luo, C.; Wang, S.; Zhang, W.; Li, L.; Zhang, X.; Fan, S.; Wang, Q. Genome-Wide Analysis of MADS-box Family Genes During Flower Development in Lettuce. Plant Cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef]
- Duan, W.; Song, X.; Liu, T.; Huang, Z.; Ren, J.; Hou, X.; Li, Y. Genome-Wide Analysis of the MADS-box Gene Family in Brassica rapa (Chinese cabbage). Mol. Genet. Genomics 2015, 290, 239–255. [Google Scholar] [CrossRef]
- Alhindi, T.; Al-Abdallat, A.M. Genome-Wide Identification and Analysis of the MADS-box Gene Family in American Beautyberry (Callicarpa americana). Plants 2021, 10, 1805. [Google Scholar] [CrossRef]
- Won, S.Y.; Jung, J.A.; Kim, J.S. Genome-wide Analysis of the MADS-box Gene Family in Chrysanthemum. Comput. Biol. Chem. 2021, 90, 107424. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ming, M.; Li, J.; Shi, D.; Qiao, X.; Li, L.; Zhang, S.; Wu, J. Genome-wide Identification of the MADS-box Transcription Factor Family in Pear (Pyrus bretschneideri) Reveals Evolution and Functional Divergence. PeerJ 2017, 5, e3776. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Krizek, B.A.; Meyerowitz, E.M. Dimerization Specificity of Arabidopsis MADS Domain Homeotic Proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 1996, 93, 4793–4798. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Yanofsky, M.F. Function and Evolution of the Plant MADS-box Gene Family. Nat. Rev. Genet 2001, 2, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-box Genes and Crop Domestication: The Jack of all Traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef]
- Honma, T.; Goto, K. Complexes of MADS-box Proteins are Sufficient to Convert Leaves into Floral Organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).