Abstract
The rapid rate of virus transmission and pathogen mutation and evolution highlight the necessity for innovative approaches to the diagnosis and prevention of infectious diseases. Traditional technologies for pathogen detection, mostly PCR-based, involve costly/advanced equipment and skilled personnel and are therefore not feasible in resource-limited areas. Over the years, many promising methods based on clustered regularly interspaced short palindromic repeats and the associated protein systems (CRISPR/Cas), i.e., orthologues of Cas9, Cas12, Cas13 and Cas14, have been reported for nucleic acid detection. CRISPR/Cas effectors can provide one-tube reaction systems, amplification-free strategies, simultaneous multiplex pathogen detection, visual colorimetric detection, and quantitative identification as alternatives to quantitative PCR (qPCR). This review summarizes the current development of CRISPR/Cas-mediated molecular diagnostics, as well as their design software and readout methods, highlighting technical improvements for integrating CRISPR/Cas technologies into on-site applications. It further highlights recent applications of CRISPR/Cas-based nucleic acid detection in livestock industry, including emerging infectious diseases, authenticity and composition of meat/milk products, as well as sex determination of early embryos.
1. Introduction
As in humans, large-scale recurring epidemics dramatically affect livestock populations, especially food-producing animals such as pigs and cattle. Infectious diseases caused by pathogens such as African swine fever virus (ASFV, family Asfarviridae) [1], porcine reproductive and respiratory syndrome virus (PRRSV, family Arteriviridae) [2], porcine epidemic diarrhea virus (PEDV, family Coronaviridae) [3], encephalomyocarditis virus (EMCV, family Picornaviridae) [4] and classical swine fever virus (CSFV, family Flaviviridae) [5] have induced reproductive failures, high mortality and trade restrictions, bringing serious commercial damages to the swine industry [6]. Bovine viral diarrhea virus (BVDV, family Flaviviridae) and lumpy skin disease virus (LSDV, a member of the genus Capripoxvirus and family Poxviridae), etc., are economically important infectious agents of cattle worldwide. In addition, pathogenic bacteria and parasites affect animal production and animal welfare, causing significant economic losses to the livestock industry [7,8,9,10]. Livestock production in large communities promote spreading and maintaining huge pathogen populations and facilitate mutation and evolution of pathogens [11,12,13], which further complicates the prevention and control of infectious diseases. Several molecular diagnostic approaches have been developed and applied for identifying and quantifying a wide range of pathogens. Reverse transcription polymerase chain reaction (RT-PCR), reverse transcription quantitative polymerase chain reaction (RT-qPCR) and droplet digital PCR (ddPCR) are effective methods of pathogen detection [14,15,16]; however, these are not suitable for use in the field due to their dependence on sophisticated/expensive equipment, long testing time and highly skilled personnel. Isothermal diagnostic approaches, such as reverse transcription loop-mediated isothermal amplification (RT-LAMP) and recombinase polymerase amplification (RPA) [17,18,19], have the following advantages: amplification at single temperature eliminates the need of bulky, advanced and expensive equipment, making them cost effective and potential for detection at the point of care (POC); compared to RT-PCR (2–4 h), the turnaround time (~1 h) is less; these methods enable naked eye visualization as well as real-time monitoring [19,20]. Nevertheless, detailed analyses of these isothermal amplification methods indicate their low sensitivity, low specificity, and low throughput [21,22,23]. The multiplexed primer pairs in LAMP may yield false-positive assay results [24,25]. LAMP is less sensitive to complex samples such as blood and tissues due to possible inhibitors of Bst polymerase [26]. To effectively prevent and control infectious diseases, it is desirable to establish a feasible, sensitive, specific, and reliable on-site diagnostic strategy for nucleic acid detection.
First described in bacterial genomes 30 years ago [27], the clustered regularly interspaced short palindromic repeats and the associated protein (CRISPR/Cas) system was reported in 2005 to operate as a natural defense mechanism of bacteria and archaea against viral and plasmid DNA infections [28]. CRISPR/Cas systems can be categorized into Class 1/Class 2. The Class 1 systems, including type I, type III and type IV, are characterized by multi-subunit-protein complexes. The Class 2 systems (type II and type V and type VI) consist of a single effector protein (known as “Cas”), which is large, polydomain and polyfunctional, as well as a guide CRISPR RNA (crRNA). CRISPR/Cas Type II is distinguished by the existence of Cas9 and a chimeric single guide RNA (sgRNA). The ribonucleoprotein (sgRNA + Cas9) identifies a protospacer adjacent motif (PAM) positioned at the G-rich 3′ end of a double-stranded DNA (dsDNA) target, which activates Cas9 nuclease for the induction of a blunt-end double-strand break (DSB) on the target DNA [29]. CRISPR/Cas type V systems are primarily characterized by a set of effector proteins Cas12 (Cas12a/Cpf1, Cas12b/C2c1, etc.) and crRNA, which increases the multiplexing capacity of the type V. The crRNA + Cas12 complex recognizes a T-rich 5′-end-localized PAM (not G-rich PAM) and generates staggered 5′-end dsDNA breaks (cis-cleavage activity) [30]. The target binding of crRNA + Cas12 also induces non-specific trans-cleavage of single-stranded DNA (ssDNA) [31]. CRISPR/Cas type VI systems (A, B, C and D subtypes) have Cas13 as the effector protein. Upon recognition of a single-stranded RNA (ssRNA) target, crRNA + Cas13 complex induces a blunt-end break (cis-cleavage activity) and indiscriminate degradation of any neighboring ssRNA (trans-cleavage activity) [32]. Since the first CRISPR/Cas-based diagnostic tool was reported in 2016, a large number of CRISPR Class 2-based diagnostic systems have been emerging, especially during the COVID-19 pandemic in 2020–2022 (Table 1). Likewise, since 2020, the “collateral” cleavage properties of CRISPR/Cas12 and CRISPR/Cas13 have been further widely used for in vitro detection of different pathogens, including viruses, bacteria and parasites in livestock (Table 2).

Table 1.
Summary of reported CRISPR/Cas-based diagnostic platforms and their detection modes.

Table 2.
Overview of applications of CRISPR-based nucleic acid detection in livestock.
This review summarizes research efforts to improve the sensitivity, speed, affordability and field deployability of CRISPR/Cas-mediated diagnostics occurring worldwide. It also provides a summary of using CRISPR/Cas diagnostics to detect/diagnose/genotype various (non)pathogenic nucleic acids in livestock.
2. Development of CRISPR/Cas-Based Nucleic Acid Detection Systems
2.1. Cas Type II Based Diagnostic Platforms
Most of the early CRISPR-based diagnostic inventions, from 2016 to 2019, relied on the Cas9/type II systems (Table 1, Figure 1), which by themselves were not able to elicit a strong/specific signal when target nucleic acids exist in the sample. These technologies employ different design strategies based on sgRNA/Cas9 and require pre-amplification (paired dCas9 (PC) reporter system [35]) or post-amplification (CRISPR-typing PCR (ctPCR) [37,40], CRISPR- or Cas9/sgRNAs-associated reverse PCR (CARP) [39], finding low abundance sequences by hybridization-next generation sequencing (FLASH-NGS) [41]) for target nucleic acids by PCR. These approaches compromise the vast promise of CRISPR diagnostics. Since the complex, specific and detailed steps involved in the methods make single-tube detection almost impossible, resulting in a possible chance of contamination during the diagnosis. Although PCR is the most famous amplification technique, the requirement for thermal cycling limits the non-laboratory applications of these CRISPR/Cas9-based methods.
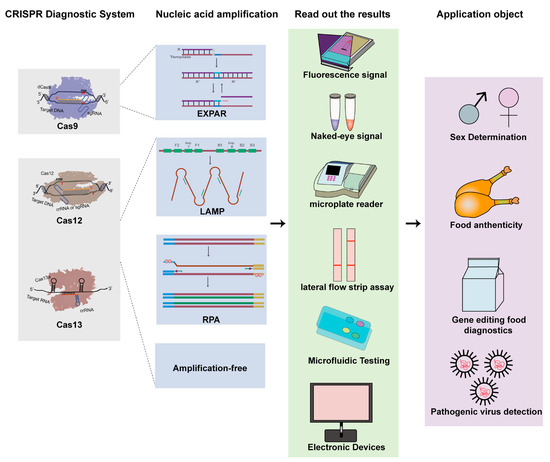
Figure 1.
Schematic principle of clustered regularly interspaced short palindromic repeats (CRISPR) Class 2 systems in nucleic acid detection and the applications of CRISPR and the associated protein systems (CRISPR/Cas)-based diagnostics in livestock industry.
Additional CRISPR/Cas9-mediated diagnostics such as nucleic acid sequence based amplification-CRISPR cleavage (NASBACC) [33], CRISPR/Cas9-triggered nicking endonuclease-mediated strand displacement amplification method (CRISDA) [38], and CRISPR/Cas9 triggered isothermal exponential amplification reaction (CAS-EXPAR) [36] involved isothermal amplification, avoiding the need for thermal cycler in conventional PCR and thus taking a major step towards POC diagnostics. NASBACC can distinguish viral strains with single-base resolution, relying on isothermal RNA amplification combined with toehold switch sensors [33]. Unlike traditional amplification reactions, such as in CRISDA, CAS-EXPAR does not need any exogenous primers and thus has been shown to be more specific for mutant targets [36]. The pioneering success story of CRISPR/Cas9-mediated diagnostics has inspired other CRISPR/Cas systems, such as lateral flow-based and fluorometer-based diagnostics [106,107].
2.2. Cas Type V and VI Based Diagnostic Platforms
Preliminarily in 2016, C2c2 (now known as Cas13a) was observed to catalyze ssRNA cleavage in presence of single crRNA and complementary protospacer [32]. Furthermore, in 2017, the Gootenberg group confirmed that Cas13a (previously known as C2c2) exhibits RNA targeting collateral activity which can be conducted at isothermal conditions and based on Cas13a developed a novel system named specific high-sensitivity enzymatic reporter unlocking (SHERLOCK), used for point-care diagnostics [65]. It can detect target RNA or DNA (in vitro transcription required), with attomolar sensitivity and specificity of single-base mispairs [65]. One year later, the second version (SHERLOCKv2) was reported with advances: quad-channel monoreactive multiplexing with orthogonal CRISPR/Cas; quantitative measuring for ~2 attomoles inputs; 3.5-fold improvement of sensitivity by combining Cas13 with Csm6; and lateral flow readout without any additional device [66].
In 2018, Li et al. developed the one-hour low-cost multipurpose highly efficient system (HOLMES) using the incidental cleavage activity of Cas12a on non-target ssDNA and a fluorescent reporter linked to ssDNA [43]. The authors demonstrated that when combined with PCR, HOLMES achieved SHERLOCK sensitivity (attomolar level), better than PCR or qPCR alone [43]. HOLMES was validated for the detection of DNA and RNA (requiring reverse transcription into cDNA) viruses [43]. Its version 2 (HOLMESv2) replaces Cas12a with Cas12b, enabling not only molecular diagnostics but also epigenetic applications, such as quantification of DNA methylation level [31,47]. The combination of Cas12a ssDNase activation with isothermal amplification was validated in 2018 and named as DNA endonuclease-targeted CRISPR trans reporter (DETECTR), also with attomolar sensitivity [31]. In the same year, a diverse CRISPR family containing Cas14 was identified to be similar to type V [64]. Unlike Cas12 with low fidelity in discriminating against ssDNA substrates, Cas14 requires complementarity in the seed region to recognize ssDNA substrates, which property raises its possibility to detect SNPs without the constraint of a PAM sequence [64].
One-tube reaction system: In spite of the impressiveness and rapid development of CRISPR/Cas-based detectors, shift of reactants to another tube and the reliance on nucleic acid isolation may result in a high chance of contamination during the test process and false positives. One-tube detection platform by RT-RPA and CRISPR/Cas12 (DETECTR) was deployed for COVID-19 detection [55]. The platform achieved single-tube testing by physically separating the two reaction components during amplification and mixing them by centrifugation afterwards [55]. A single-tube assay platform with RPA and CRISPR/Cas13a (SHERLOCK) also works [55]. A primary comparison of DETECTR and SHERLOCK indicated that their testing effectiveness was essentially similar, despite the more complex components of SHERLOCK [55]. Nucleic acid extraction, however, was still required prior to these single-tube assays.
Heating unextracted diagnostic samples to bliterate nucleases (HUDSON) is a methodology using heat and chemical reduction to lyse virus particle and deactivate high-level RNases present in body fluids [67]. Combining HUDSON with SHERLOCK, field-deployable diagnostic platforms were developed for the detection of viruses from body fluids without the need for a nucleic acid extraction step [67,69]. The in-tube fluorescence readout method also reduced the risk of contamination as the reaction tubes remained closed [69]. SHERLOCK testing in one pot (STOP) platform also combines LAMP and CRISPR/Cas12b-based detection in a single tube, but requires an extraction step using magnetic beads prior to the testing, which raises the cost and handling time, and is still unable to improve the sensitivity (200 copies/reaction) to the level of RT-qPCR (20 copies/reaction) [50]. Instead of using canonical PAM, a suboptimal PAM-mediated method (sPAMC) reported in 2022 appears to be the first real one-tube detection methodology without the need for RNA extraction [62]. The decreased binding affinity of CRISPR/Cas12a to the suboptimal PAM substrate diminished its cis-cleavage activity, facilitating the shift of equilibrium to isothermal amplification and thereby leading to stronger fluorescence. The test time is within 20 min and the sensitivity is similar that of RT-qPCR [62].
Amplification-free strategies: Some novel amplification-free detection strategies, based on CRISPR/Cas12a or CRISPR/Cas13a, were reported as potentially more suitable POC sensors for viral nucleic acids. In 2019, an electrochemical biosensor (E-CRISPR) was reported based on the trans-cleavage activity of Cas12a and electrode consisting non-specific ssDNA, to convert target recognizing into electrochemical signal. Not only for nucleic acid sensing, the system can also be utilized for protein sensing [45]. Another E-CRISPR was developed in 2020 with immobilized low surface coverage and morphologically uncompact hpDNA which supplies approachable substrates for effective Cas12a cleavage, resulting in higher sensitivity than that of conventional ssDNA [53]. Additionally in 2021, an amplification-free fluorescent biosensor was created via a metal-enhanced fluorescence (MEF) with DNA-functionalized Au nanoparticles (AuNP). MEF color changes from purple to red-purple when target DNA-activated CRISPR/Cas12a degrades ssDNA between AuNP and fluorophore [57]. Enhanced analysis of nucleic acids with CrRNA expansions (CRISPR-ENHANCE), another CRISPR/Cas12a-based detection system, employs genetically engineered crRNA with specific 7-mer’-expansions and optimized working conditions. Without the need for target pre-amplification, it achieves femtomolar level sensitivity [59].
In addition, Fozouni et al. developed a CRISPR/Cas13a-multiple crRNAs assay for rapid, POC detection, which improved sensitivity not by target amplification, but by activating more Cas13a per target RNA, enabling direct conversion of the fluorescent signal to viral load [71]. Ultralocalized Cas13a detection confines the RNA-activated CRISPR/Cas13a system to cell-sized reactors by droplet microfluidics to simultaneously increase target and reporter local concentrations. By comparison with the bulk Cas13a assay, it realizes a >10,000-fold sensitivity improvement and achieves absolute digital single-molecule RNA quantification [72]. In 2022, a dual enzyme amplification scheme that combines target-induced Cas activation with a following release of the enzymatic reporter—horseradish peroxidase (HRP)—into solution, has achieved quick, convenient (25 °C) and sensitive (~10 fM) detection of nucleic acids, without the need for PCR [74]. CRISPR/Cas13a-graphene field-effect transistors (gFETs) method, may be one of the most sensitive amplification-free diagnostic systems so far and has been validated for the detecting of SARS-CoV-2 and respiratory syncytial virus as low as 1 aM [73].
Multiplexed pathogen detection system: The combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN) platform was reported in 2020 for scalable multiplexed pathogen detection. Nanoliter droplets of CRISPR/Cas13/crRNA reaction reagents are self-organized in a micropole array, paired with amplified sample droplets, to detect each sample repeatedly for each crRNA [68]. CARMEN system combined with Cas13 has been reported to effectively screen over 4500 crRNA candidates against desired targets through a single array platform. CARMEN can be easily scaled for practical use due to its inherent multiplexing, throughput capabilities, as well as 300 times reduction in reagent cost per test due to miniaturization [68]. The authors demonstrated that CARMEN-Cas13 enables the simultaneous detection of all 169 human-related viruses, as well as subtypes of influenza A strains and HIV drug resistance variations [68]. The CARMEN panel has also been applied to simultaneously detect up to 21 respiratory viruses [108], 52 clinically relevant bacterial species as well as a number of key antibiotic resistance genes [109], demonstrating its diagnostic-grade performance in both academic and clinical settings [108].
Rapid Visual CRISPR Assays: In 2022, Xie et al. systematically screened and identified nine ssDNA-FQ reporters suitable for CRISPR/Cas12a-based visual colorimetric detection, with particularly strong performance of ROX-labeled reporter [61]. A convolutional neural network algorithm was also developed and implemented in the MagicEye mobile app to enable standardization and automation of image analytical colorimetric evaluation [61]. By Combining the DETECTR strategy [31] and the ROX-labeled reporter, RApid VIsual CRISPR (RAVI-CRISPR) has been established as a device-less (only a portable rechargeable incubator is required) single-tube colorimetric POC platform [61]. The RAVI-CRISPR has been successfully applied for detection of SARS-CoV-2 [61], ASFV [61], and JEV [86], for sex determination in pigs [101], as well as for recognition of meat species and meat products [104]. The RAVI-CRISPR in these applications has a LOD of 2–9 total copies and takes only 35–60 min using naked-eye colorimetric detection. The RAVI-CRISPR/MagicEye system appears to be a breakthrough technology for rapid pen- or bed-side detection.
2.3. Software for Designing CRISPR/Cas-Based Nucleic Acid Assays
An increasing number of novel RNA-guided CRISPR endonucleases, such as Cas9 from various types of bacteria, Cpf1 nuclease, C2c1, C2c2, and C2c3 systems, have been discovered with a different PAM. The guide RNA sequence directly affects the target-induced cleavage efficiency and non-intentional off-target binding and cleavage. Therefore, the design of efficient and specific guide RNA is crucial for the successful application of CRISPR/Cas-mediated diagnostics. A list of CRISPR design tools have been created and some popular guide RNA design tools, such as CRISPOR [110], CHOPCHOP [111], Off-Spotter [112], Cas-OFFinder [113], CRISPR-Era [114], and E-CRISP [115], are available with GUIs for ease of use. Some of these tools, such as Off-Spotter [112] and Cas-OFFinder [113], were developed specifically for detecting potential off-target editing. Other tools, such as CHOPCHOP [111] and CRISPR-OFFinder [116], are not only for Cas9 but also provide options for alternative Cas nucleases and PAM recognition. CRISPR-OFFinder is a versatile tool to rapidly design sgRNAs for different CRISPR/Cas systems with minimal off-target effects, particularly for Cpf1 and C2c1 [116]. User-defined PAM and sgRNA length are supported to enhance targeting specificity [116]. While CRISPR-RT [117] and CRISPR-DT [118] were developed to help scientists design gRNAs for the CRISPR/Cas13a/C2c2 and CRISPR/Cas12a/Cpf1 systems, respectively. To quickly discover or score hundreds of CRISPR targets, command-line tools like FlashFry [119] are also available. They are fast and flexible with the output presented in easy-to-operate text files.
2.4. Readout Methods
Many CRISPR/Cas-based diagnostics have been developed with different readout methods, and major attempts have been made to achieve POC testing. The fluorescence and lateral flow assay are so far the most used readout methods in CRISPR/Cas-based detection platforms. Other signal detection methods have also been reported, such as using electrochemical biosensors (E-CRISPR [45,95], PGMs-CRISPR [54], MOECS [120]), chemiluminescence enhancement biosensors (CRICED [121], CLE-CRISPR [122]), toehold switch-linked colorimetry (NASBACC [33]), and potentiometry (CRISPR-Chip [42]). Some CRISPR/Cas-based diagnostic technologies allow for fluorescent signals to be read by the naked eye under blue light (CRISPR-Cas12a-NER [48], opvCRISPR [56], CASdetec [49]) or to be measured with a mobile phone (multiple enhanced CRISPR-Cas13 assay [71], SHINE [69]). Naked-eye colorimetric detection (RAVI-CRISPR [61], CRISPR-Cas13a/HRP assay [74]) reported in 2022 is probably simplest-to-date reading method for molecular POC testing, without fluorescence detector or mobile phone required for visualization.
3. Current Applications of CRISPR/Cas-Based Nucleic Acid Detection Technologies in Livestock
3.1. CRISPR/Cas-Based Detection of Pathogenic Viruses in Livestock
ASFV is a nucleocytoplasmic large DNA virus that is highly infectious and pathogenic [123]. To date, there is no available vaccine or antiviral drug against ASFV, but CRISPR-based methods of ASFV detection to control ASF transmission. Lin et al. established CRISPR/Cas9 eraser-based PAM-implanted PCR combined with lateral flow endpoint detection method [75] (Table 2, Figure 1). Pretreatment of PCR mixture with Cas9/sgRNA to selective clean up contamination amplicons abolishes false-positive amplification. However, if the source of contamination is unknown, sequencing must be employed prior to designing new primers and sgRNAs [75]. The CRISPR/Cas9 erase method requires costly instrumentation and specialized handling system, therefore remains unsuitable for fast clinical testing [75]. Reported AssSFV diagnostic assays combing the incidental cleavage activity of Cas13a (CRISPR/Cas13a-LFD [78]) or Cas12a (RAVI-CRISPR [61,76]) with isothermal amplification solved this problem. A water bath/portable rechargeable hand warmer and a pipette are the main devices required to perform these assays, indicating their potential in-field applicability for ASFV detection. These methods have high sensitivity (LoD: 7–10 copies/µL) and high specificity [61,76,78]. The single-tube reaction in these methods reduces the likelihood of contamination. Compared to CRISPR/Cas13a-LFD [78], RAVI-CRISPR [61,76] is even more advanced and cheaper, as it does not require lateral flow strip or fluorescence detector. RAVI-CRISPR achieves accurate colorimetric naked-eye detection using a ROX-labeled reporter [61,76]. The CRISPR/Cas12a/multiplex-crRNA system was designed for direct detection of ASFV DNA without nucleic-acid preamplification [79]. Its detection limit (~1 pM) is 6–64 times stronger than that of the conventional single-crRNA CRISPR/Cas12a system [79]. It also reduces the possibility of detecting losses due to naturally occurring mutations in viral genes [79]. In the future, the CRISPR/Cas12a/multiplex-crRNA method could be further combined with naked-eye colorimetric detection using a ROX-labeled reporter [61,76], making the diagnostic platform more deployable in the field. In reality, ASFV, CSFV, and APPV are co-endemic in many areas, causing highly similar clinical symptoms. So far, a multiplex RT-PCR assay [124] is available to test these viruses simultaneously, but it requires specialized instruments and skilled personnel. The multiplex isothermal amplification in combination with CRISPR/Cas12a assay, which has successfully distinguished PEDV, TGEV, PDCoV, and SADS-CoV [83] would have the potential to simultaneously and differentially detect these three viruses in the field.
PRRSV is a positive-sense RNA virus, and can cause abortion of pregnant sows, and respiratory symptoms and death in pigs [125]. Visual nucleic acid detection methods based on CRISPR/Cas13a [25] or CRISPR/Cas12a [80], respectively, have been established, since PRRSV detection approaches based on antigen-antibody response [126], PCR or RT-qPCR [127] are not applicable for poorly equipped laboratories or on-site diagnostics with high sensitivity. The sensitivity of the CRISPR/Cas13a-based method for PRRSV detection is 172 copies/reaction, similar to that of RT-qPCR [128,129], while the CRISPR/Cas12a assay is much more sensitive and can reach the sensitivity of one copy/reaction within 25 min [80]. Both CRISPR/Cas-based assays have been successfully deployed in clinical samples from diverse farms [25,80].
PEDV, an enveloped, positive RNA virus, induces acute intestinal infections manifested by severe dehydration, diarrhea, nausea and high rate of mortality in piglets [130,131]. PEDV can be classified into genotypes GI and GII according to mutations in spike (S) gene [132]. The CV777 vaccine strain has been created based on PEDV GI genotype and widely used to control GI PEDV infection in pigs [133]. Whereas, the GII genotype is commonly reported in cases of immune failure and has become a main prevalent PEDV strain [134,135]. For distinguishing PEDV-attenuated vaccine strains and wild-type virus strains, an RT-ERA-CRISPR/Cas12a assay was developed with high sensitivity (LOD of two copies) based on a 51nt deletion mutation in the open reading frame 3 (ORF3) gene [81]. To decide if a pig should be immunized with the CV777 vaccine, an RT-RAA-CRISPR/Cas12a platform targeting the S gene was created for the detection of GII PEDV [82]. Additionally, Liu et al. (2022) developed a single-tube multiplex RT-LAMP-Cas12a diagnostics to simultaneously detect TGEV, PDCoV, SADS-CoV, and PEDV, although it cannot recognize different PEDV strains [83]. With naked-eye colorimetric detection, it has a LOD of one copy and takes only 25 min [83]. These CRISPR/Cas12a-based assays, with different detection purposes, have promising potential for the prevent and control of PEDV worldwide.
Besides, the strategy of combining isothermal amplification with the collateral cleavage activity of CRISPR/Cas12a (DETECTR [31]) or CRISPR/Cas13a (SHERLOCK [65]) has also been applied for the detection of PCV3 (ssDNA circovirus, ERA-CRISPR/Cas12a) [84], PPV (non-enveloped DNA virus, ERA-CRISPR/Cas12a) [85], EMCV (non-enveloped ssRNA virus, RAA-CRISPR/Cas13a) [87], JEV (ssRNA virus, RT-LAMP-CRISPR/Cas12a) [86], LSDV (linear dsDNA virus, RPA-Cas12a) [88], BVDV (positive-sense ssRNA virus, CRISPR-Cas13a-based) [89], and CaPV (LAMP-CRISPR/Cpf1) [90]. These diagnostic methods have robustness, convenience, sensitivity, specificity, affordability and potential adaptation for in-field detection or surveillance of the viruses in clinical and vector samples.
3.2. CRISPR-Based Detection of Pathogenic Bacteria and Parasites in Livestock
Compared to Cas13a-based SHERLOCK [65], Cas12a-based DETECTR [31] may be a more desirable strategy for the detection of bacterial nucleic acids without the need for in vitro transcription, since Cas12a is a DNA endonuclease. Brucella spp. can cause widespread brucellosis in cattle, sheep, pigs, as well as dogs [136]. Four humankind pathogens of Brucella spp. (Brucella abortus, Brucella melitensis, Brucella canis, and Brucella suis) can transmit between humans and animals, leading to occupational risks for livestock workers [137,138]. The development of an RPA-CRISPR/Cas12a diagnostics with both fluorescent and electrochemical signal readout methods enabled rapid and accurate detection of these four Brucella strains in blood and milk samples [95]. The dual-signal readout approach improves the accuracy of the assay, and the authors demonstrate that this method has better diagnostic performance than real-time PCR [95]. Additionally, the RPA-CRISPR/Cas12a strategy has been used for the detection of other foodborne pathogenic bacteria (Listeria monocytogenes, Yersinia enterocolitica, Staphylococcus aureus, Campylobacter jejuni, Escherichia coli, etc.) in food production animals at attomolar level [96,97,98,99,100].
Traditional parasite diagnostic approaches, such as light microscopy and immunoassay, are not reliable and require a large quantity of samples [139]. Cryptosporidium parvum is a zoonotic important intestinal protozoan parasite and can induce cryptosporidiosis in humans and domestic/wild animals all over the world [10]. A diagnostic method based on RPA-Cas12a/crRNA (ReCTC) has been validated for the detection of C. parvum subtype family IId from clinical human and bovine fecal samples [94]. In future studies, the ReCTC assay could be further optimized as a one-tube reaction for faster and simpler on-site diagnosis. Toxoplasma gondii is a globally distributed protozoan parasite, causing life-threatening consequences in immunocompromised patients [140] as well as abortions and stillbirths in livestock [141,142]. In combination with RPA or RAA, CRISPR-Cas12a/Cas13a based methods have been reported for quick in-field detection of T. gondii [91,92,93]. The authors demonstrated that the RAA-Cas12a/Cas13a systems have advantages over conventional PCR-based method in terms of conveniency and sensitivity [92,93]. In terms of reducing cross-contamination and cost, the RPA-CRISPR/Cas12a assay (single-tube strategy and 100 nM of Cas12a) is more advanced than the RAA-CRISPR/Cas12a assay (two separate processes and 800 nM of Cas12a) [91]. Applications of these novel methods potentially contribute to the control of toxoplasmosis in humans and animals.
3.3. CRISPR Assays for Sex Determination and Meat/Milk Products
Sex determination of early embryos is a requisite for the achievement of sex control and ideal male/female ratios [143]. It has a huge impact on breeding efficiency and worldwide animal production, such as milk yield and weight gain [143,144,145]. Previous reports have also indicated that pork quality depends on sex-related physiology [146,147] and sex-expressed genes [148]. A RAVI-CRISPR/LAMP-CRISPR-Cas12a system targeting zinc finger protein X-linked (ZFX) and sex-determining region Y (SRY) genes was established for sex identification of early pig embryos and pork with simplicity, cheapness, sensitivity, and specificity [101]. Compared to the traditional methods such as PCR and fluorescence-activated cell sorting (FACS), it can be easily performed in the field and does not require technical expertise. Its fluorescence signal can be checked with the naked eye, a portable UV/blue transilluminator/luminescent flashlight [101]. The RAVI-CRISPR strategy also has potential in determining the sex of other livestock species.
Meat and meat products are an indispensable part of human diet and meat/food safety is a high concern worldwide. Meat adulteration causes serious economic and health consequences globally and harms the religious beliefs of Muslim consumers [149]. Conventional methods for the identification of animal/meat products, such as enzyme-linked immunosorbent assays (ELISA) [150] and chromatographic methods [151], demand knowledgeable personnel and are expensive, inaccurate, and time-consuming. Molecular techniques, particularly CRISPR/Cas12-based methods targeting species-specific DNA have been reported for meat identification. The CRISPR/Cas based PCR DNA barcoding method (CAPCOD), integrating CRISPR/Cas12 system and PCR, can identify 0.1% (w/w) pork contamination in raw meat mixtures [102]. It has been used for identifying pork content in complex processed (non-)halal foods [102]. This method is fast and specific, but requires complex instrumentation for PCR, limiting its application in-field detection. The DETECTR strategy combining isothermal amplification (instead of PCR) and the CRISPR/Cas12a detection technique [31] can overcome this problem. An RPA-CRISPR/Cas12a method has been developed for pork detection and validated on beef and pork mixtures under raw, cooked and high-pressure conditions [103]. As low as 10−3 ng of porcine genomic DNA can be identified by the portable box in 30 min [103]. The authors confirmed that the results were consistent with the real-time PCR method. The RPA-CRISPR/Cas12a method can therefore be used for in situ pork detection with high speed, accuracy and sensitivity [103]. In addition, LAMP-CRISPR/Cas12a with a Texas red-labeled ssDNA reporter for visual colorimetric detection (RAVI-CRISPR) has been reported to detect meat species of pig, chicken and duck with high sensitivity (1.0 pg/μL) and speed (~40 min) [104]. The assay has been validated in pilot POC detection of a food processing factory, supporting its potential applications in customs, quarantine units as well as meat import or export sectors [104]. Additionally, a CRISPR/Cas12a-driven surface-enhanced Raman scattering (SERS) biosensor has been successfully developed for goat milk authenticity detection with an ultra-low detection limit of 224 aM [105]. The CRISPR/Cas12-based strategies can also potentially be applied to the detection of gene-edited foods. However, CRISPR/Cas-based methods are generally qualitative and the interpretation of results can be subjective as each person may interpret color changes differently. qPCR is a well-known method for quantifying milk/meat product components, but it requires specialized instruments and skilled personnel. Strategies like the warm-start rapid digital CRISPR approach (WS-RADICA) [63] and microfluidics-enabled digital isothermal Cas13a assay (MEDICA) [152] may hold great promise as the next-generation nucleic acid quantification approaches alternatives to qPCR [63,152], since they had lower detection limits and greater inhibitor tolerance than a bulk isothermal amplification-combined CRISPR/Cas-based assay and had similar performance to RT-dPCR and RT-qPCR [63,72,152].
4. Conclusions and Future Prospects
Rapid detection of infectious diseases is highly required in diagnosis and infection prevention, not only in humans, but also in livestock. Meat/milk authenticity and composition should also be evaluated quickly, reliably, and cost-effectively from public health perspectives and from religious perspectives. The sex determination and control of early embryos also has immense impact on global livestock production. Nucleic acid detection methods combining isothermal amplification and CRISPR/Cas systems have emerged in recent years, with robustness, convenience, sensitivity, specificity, affordability, and potential adaptation for on-site detection. The strategies mainly employ the target-activated trans-cleavage activities of Cas12 and Cas13, which can efficiently cleave ssDNA or ssRNA sequences. However, these approaches still have space for further improvement to reduce the chance of contamination and cost, and to increase speed and sensitivity. For example, the combination of using suboptimal PAM and RAVI-CRISPR could be tested to achieve nucleic acid extraction-free one-tube visual colorimetric detection. Naked-eye colorimetry of RAVI-CRISPR as a readout method could be widely applied to reduce costs, instead of using lateral flow strips or fluorescence detectors. In addition, a number of CRISPR-Cas12a/Cas13a-based amplification-free platforms have been successfully applied for human virus diagnostics, but their potential in nucleic acid detection in livestock has not yet been explored. The multiplexed pathogen detection system CARMEN also has great potential to simultaneously detect all important livestock-associated viruses and to comprehensively identify their variant subtypes.
Funding
This Open Access publication was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—491094227 “Open Access Publication Funding” and the University of Veterinary Medicine Hannover, Foundation.
Acknowledgments
The author would like to thank Xie Shengsong and Xu Bingrong for creating the figure. Many thanks also go to Magdalene Paetzold for her support.
Conflicts of Interest
The author declares no conflict of interest.
References
- Zhou, X.; Li, N.; Luo, Y.; Liu, Y.; Miao, F.; Chen, T.; Zhang, S.; Cao, P.; Li, X.; Tian, K.; et al. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018, 65, 1482–1484. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Ye, C.; Chang, X.-B.; Jiang, C.-G.; Wang, S.-J.; Cai, X.-H.; Tong, G.-Z.; Tian, Z.-J.; Shi, M.; An, T.-Q. Importation and Recombination Are Responsible for the Latest Emergence of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus in China. J. Virol. 2015, 89, 10712–10716. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Byrum, B.; Zhang, Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014, 20, 917–919. [Google Scholar] [CrossRef] [PubMed]
- Koenen, F.; De Clercq, K.; Lefebvre, J.; Strobbe, R. Reproductive failure in sows following experimental infection with a Belgian EMCV isolate. Vet. Microbiol. 1994, 39, 111–116. [Google Scholar] [CrossRef]
- Zhou, B. Classical Swine Fever in China-An Update Minireview. Front. Vet. Sci. 2019, 6, 187. [Google Scholar] [CrossRef]
- VanderWaal, K.; Deen, J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA 2018, 115, 11495–11500. [Google Scholar] [CrossRef]
- Lanyon, S.R.; Hill, F.I.; Reichel, M.P.; Brownlie, J. Bovine viral diarrhoea: Pathogenesis and diagnosis. Vet. J. 2014, 199, 201–209. [Google Scholar] [CrossRef]
- Davies, F.G. Lumpy skin disease, an African capripox virus disease of cattle. Br. Vet. J. 1991, 147, 489–503. [Google Scholar] [CrossRef]
- Uzzau, S.; Brown, D.J.; Wallis, T.; Rubino, S.; Leori, G.; Bernard, S.; Casadesús, J.; Platt, D.J.; Olsen, J.E. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 2000, 125, 229–255. [Google Scholar] [CrossRef]
- Xiao, L.; Fayer, R.; Ryan, U.; Upton, S.J. Cryptosporidium taxonomy: Recent advances and implications for public health. Clin. Microbiol. Rev. 2004, 17, 72–97. [Google Scholar] [CrossRef]
- La Rosa, G.; Fratini, M.; Della Libera, S.; Iaconelli, M.; Muscillo, M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super. Sanita 2012, 48, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Nichol, S.T.; Arikawa, J.; Kawaoka, Y. Emerging viral diseases. Proc. Natl. Acad. Sci. USA 2000, 97, 12411–12412. [Google Scholar] [CrossRef] [PubMed]
- Fournié, G.; Kearsley-Fleet, L.; Otte, J.; Pfeiffer, D.U. Spatiotemporal trends in the discovery of new swine infectious agents. Vet. Res. 2015, 46, 114. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, J.C.; Oton-Gonzalez, L.; Mazziotta, C.; Lanzillotti, C.; Iaquinta, M.R.; Tognon, M.; Martini, F. Simultaneous Detection and Viral DNA Load Quantification of Different Human Papillomavirus Types in Clinical Specimens by the High Analytical Droplet Digital PCR Method. Front. Microbiol. 2020, 11, 591452. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Tests and Collection Kits Authorized by the FDA: Infographic. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/covid-19-tests-and-collection-kits-authorized-fda-infographic (accessed on 18 October 2022).
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M. Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int. J. Mol. Med. 2021, 47, 100. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, A.; Mostafa, A.; Berger, J.; Aydin, M.Y.; Sun, F.; de Ramirez, S.A.S.; Valera, E.; Cunningham, B.T.; King, W.P.; Bashir, R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 22727–22735. [Google Scholar] [CrossRef]
- Subsoontorn, P.; Lohitnavy, M.; Kongkaew, C. The diagnostic accuracy of isothermal nucleic acid point-of-care tests for human coronaviruses: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 22349. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Martini, F.; Maritati, M.; Caselli, E.; Gallenga, C.E.; Guarino, M.; de Giorgio, R.; Mazziotta, C.; Tramarin, M.L.; Badiale, G.; et al. Advanced Molecular and Immunological Diagnostic Methods to Detect SARS-CoV-2 Infection. Microorganisms 2022, 10, 1193. [Google Scholar] [CrossRef]
- Fitri, L.E.; Widaningrum, T.; Endharti, A.T.; Prabowo, M.H.; Winaris, N.; Nugraha, R.Y.B. Malaria diagnostic update: From conventional to advanced method. J. Clin. Lab. Anal. 2022, 36, e24314. [Google Scholar] [CrossRef]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef]
- Huang, W.E.; Lim, B.; Hsu, C.; Xiong, D.; Wu, W.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M.; et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Zanoli, L.M.; Spoto, G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 2013, 3, 18–43. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef]
- Chang, Y.; Deng, Y.; Li, T.; Wang, J.; Wang, T.; Tan, F.; Li, X.; Tian, K. Visual detection of porcine reproductive and respiratory syndrome virus using CRISPR-Cas13a. Transbound. Emerg. Dis. 2020, 67, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Thekisoe, O.; Kuboki, N.; Nambota, A.; Fujisaki, K.; Sugimoto, C.; Igarashi, I.; Yasuda, J.; Inoue, N. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop. 2007, 102, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.M.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Yamano, T.; Nishimasu, H.; Zetsche, B.; Hirano, H.; Slaymaker, I.M.; Li, Y.; Fedorova, I.; Nakane, T.; Makarova, K.S.; Koonin, E.V.; et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 2016, 165, 949–962. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Guk, K.; Keem, J.O.; Hwang, S.G.; Kim, H.; Kang, T.; Lim, E.-K.; Jung, J. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens. Bioelectron. 2017, 95, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, L.; Wei, W.; Wang, Y.; Wang, B.; Lin, P.; Liu, W.; Yixuan, Y.; Li, X.; Liu, D.; et al. Paired Design of dCas9 as a Systematic Platform for the Detection of Featured Nucleic Acid Sequences in Pathogenic Strains. ACS Synth. Biol. 2017, 6, 211–216. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90, 2193–2200. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, B.; Xu, X.; Long, F.; Wang, J. CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method. Sci. Rep. 2018, 8, 14126. [Google Scholar] [CrossRef]
- Zhou, W.; Hu, L.; Ying, L.; Zhao, Z.; Chu, P.K.; Yu, X.-F. A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018, 9, 5012. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Q.; Xu, X.; Xia, Q.; Long, F.; Li, W.; Shui, Y.; Xia, X.; Wang, J. Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique. Anal. Bioanal. Chem. 2018, 410, 2889–2900. [Google Scholar] [CrossRef]
- Zhang, B.; Xia, Q.; Wang, Q.; Xia, X.; Wang, J. Detecting and typing target DNA with a novel CRISPR-typing PCR (ctPCR) technique. Anal. Biochem. 2018, 561–562, 37–46. [Google Scholar] [CrossRef]
- Quan, J.; Langelier, C.; Kuchta, A.; Batson, J.; Teyssier, N.; Lyden, A.; Caldera, S.; McGeever, A.; Dimitrov, B.; King, R.; et al. FLASH: A next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019, 47, e83. [Google Scholar] [CrossRef]
- Hajian, R.; Balderston, S.; Tran, T.; DeBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M.; et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Guo, L.; Cui, T.; Wang, X.-G.; Xu, K.; Gao, Q.; Zhou, Q.; Li, W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019, 20, 132. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Somoza, R.; Wang, L.; Welter, J.F.; Li, Y.; Caplan, A.I.; Liu, C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem. Int. Ed. 2019, 58, 17399–17405. [Google Scholar] [CrossRef]
- English, M.A.; Soenksen, L.R.; Gayet, R.V.; de Puig, H.; Angenent-Mari, N.M.; Mao, A.S.; Nguyen, P.Q.; Collins, J.J. Programmable CRISPR-responsive smart materials. Science 2019, 365, 780–785. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.-P.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, M.; Liu, Y.; Ma, P.; Dang, L.; Meng, Q.; Wan, W.; Ma, X.; Liu, J.; Yang, G.; et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci. Bull. 2020, 65, 1436–1439. [Google Scholar] [CrossRef]
- Guo, L.; Sun, X.; Wang, X.; Liang, C.; Jiang, H.; Gao, Q.; Dai, M.; Qu, B.; Fang, S.; Mao, Y.; et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell. Discov. 2020, 6, 34. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.; Woolley, A.E.; Segel, M.; Barretto, R.P.J.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef]
- Lu, S.; Li, F.; Chen, Q.; Wu, J.; Duan, J.; Lei, X.; Zhang, Y.; Zhao, D.; Bu, Z.; Yin, H. Rapid detection of African swine fever virus using Cas12a-based portable paper diagnostics. Cell Discov. 2020, 6, 18. [Google Scholar] [CrossRef]
- Hu, M.; Yuan, C.; Tian, T.; Wang, X.; Sun, J.; Xiong, E.; Zhou, X. Single-Step, Salt-Aging-Free, and Thiol-Free Freezing Construction of AuNP-Based Bioprobes for Advancing CRISPR-Based Diagnostics. J. Am. Chem. Soc. 2020, 142, 7506–7513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yan, Y.; Que, H.; Yang, T.; Cheng, X.; Ding, S.; Zhang, X.; Cheng, W. CRISPR/Cas12a-Mediated Interfacial Cleaving of Hairpin DNA Reporter for Electrochemical Nucleic Acid Sensing. ACS Sens. 2020, 5, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Shi, Z.; Qian, J.; Bi, K.; Fang, M.; Xu, Z. A CRISPR-Cas12a-derived biosensor enabling portable personal glucose meter readout for quantitative detection of SARS-CoV-2. Biotechnol. Bioeng. 2021, 118, 1587–1596. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, L.; Liu, C.; Ye, S.; Chen, W.; Li, D.; Huang, W. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J. Transl. Med. 2021, 19, 74. [Google Scholar] [CrossRef]
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-CoV-2 detection. Biosens. Bioelectron. 2021, 172, 112766. [Google Scholar] [CrossRef]
- Choi, J.H.; Lim, J.; Shin, M.; Paek, S.; Choi, J. CRISPR-Cas12a-Based Nucleic Acid Amplification-Free DNA Biosensor via Au Nanoparticle-Assisted Metal-Enhanced Fluorescence and Colorimetric Analysis. Nano Lett. 2021, 21, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Zhang, Q.; Xia, X.; Li, C.; Ho, W.K.H.; Yan, J.; Huang, Y.; Wu, H.; Wang, P.; Yi, C.; et al. A CRISPR-Cas12a integrated SERS nanoplatform with chimeric DNA/RNA hairpin guide for ultrasensitive nucleic acid detection. Theranostics 2022, 12, 5914–5930. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Gurijala, J.; Rananaware, S.R.; Pizzano, B.L.; Stone, B.T.; Jain, P.K. CRISPR-ENHANCE: An enhanced nucleic acid detection platform using Cas12a. Methods 2022, 203, 116–124. [Google Scholar] [CrossRef]
- Chen, Z.; Li, J.; Li, T.; Fan, T.; Meng, C.; Li, C.; Kang, J.; Chai, L.; Hao, Y.; Tang, Y.; et al. A CRISPR/Cas12a-empowered surface plasmon resonance platform for rapid and specific diagnosis of the Omicron variant of SARS-CoV-2. Natl. Sci. Rev. 2022, 9, nwac104. [Google Scholar] [CrossRef]
- Xie, S.; Tao, D.; Fu, Y.; Xu, B.; Tang, Y.; Steinaa, L.; Hemmink, J.D.; Pan, W.; Huang, X.; Nie, X.; et al. Rapid Visual CRISPR Assay: A Naked-Eye Colorimetric Detection Method for Nucleic Acids Based on CRISPR/Cas12a and a Convolutional Neural Network. ACS Synth. Biol. 2022, 11, 383–396. [Google Scholar] [CrossRef]
- Lu, S.; Tong, X.; Han, Y.; Zhang, K.; Zhang, Y.; Chen, Q.; Duan, J.; Lei, X.; Huang, M.; Qiu, Y.; et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat. Biomed. Eng. 2022, 6, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chan, C.; Springs, S.L.; Lee, Y.H.; Lu, T.K.; Yu, H. A warm-start digital CRISPR/Cas-based method for the quantitative detection of nucleic acids. Anal. Chim. Acta 2022, 1196, 339494. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.-S.F.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef]
- Sheng, Y.; Zhang, T.; Zhang, S.; Johnston, M.; Zheng, X.; Shan, Y.; Liu, T.; Huang, Z.; Qian, F.; Xie, Z.; et al. A CRISPR/Cas13a-powered catalytic electrochemical biosensor for successive and highly sensitive RNA diagnostics. Biosens. Bioelectron. 2021, 178, 113027. [Google Scholar] [CrossRef]
- Fozouni, P.; Son, S.; Derby, M.D.d.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef]
- Tian, T.; Shu, B.; Jiang, Y.; Ye, M.; Liu, L.; Guo, Z.; Han, Z.; Wang, Z.; Zhou, X. An Ultralocalized Cas13a Assay Enables Universal and Nucleic Acid Amplification-Free Single-Molecule RNA Diagnostics. ACS Nano 2021, 15, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, J.; Wu, G.; Weng, Z.; Song, Y.; Zhang, Y.; Vanegas, J.A.; Avery, L.; Gao, D.Z.; Sun, H.; et al. Amplification-Free Detection of SARS-CoV-2 and Respiratory Syncytial Virus Using CRISPR Cas13a and Graphene Field-Effect Transistors. Angew. Chem. Int. Ed. Engl. 2022, 61, e202203826. [Google Scholar] [PubMed]
- Samanta, D.; Ebrahimi, S.B.; Ramani, N.; Mirkin, C.A. Enhancing CRISPR-Cas-Mediated Detection of Nucleic Acid and Non-nucleic Acid Targets Using Enzyme-Labeled Reporters. J. Am. Chem. Soc. 2022, 144, 16310–16315. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Tian, T.; Jiang, Y.; Xiong, E.; Zhu, D.; Zhou, X. A CRISPR/Cas9 eraser strategy for contamination-free PCR end-point detection. Biotechnol. Bioeng. 2021, 118, 2053–2066. [Google Scholar] [CrossRef] [PubMed]
- Tao, D.; Liu, J.; Nie, X.; Xu, B.; Tran-Thi, T.-N.; Niu, L.; Liu, X.; Ruan, J.; Lan, X.; Peng, G.; et al. Application of CRISPR-Cas12a Enhanced Fluorescence Assay Coupled with Nucleic Acid Amplification for the Sensitive Detection of African Swine Fever Virus. ACS Synth. Biol. 2020, 9, 2339–2350. [Google Scholar] [CrossRef]
- Yang, B.; Shi, Z.; Ma, Y.; Wang, L.; Cao, L.; Luo, J.; Wan, Y.; Song, R.; Yan, Y.; Yuan, K.; et al. LAMP assay coupled with CRISPR/Cas12a system for portable detection of African swine fever virus. Transbound. Emerg. Dis. 2022, 69, e216–e223. [Google Scholar] [CrossRef]
- Wei, N.; Zheng, B.; Niu, J.; Chen, T.; Ye, J.; Si, Y.; Cao, S. Rapid Detection of Genotype II African Swine Fever Virus Using CRISPR Cas13a-Based Lateral Flow Strip. Viruses 2022, 14, 179. [Google Scholar] [CrossRef]
- Zeng, M.; Ke, Y.; Zhuang, Z.; Qin, C.; Li, L.Y.; Sheng, G.; Li, Z.; Meng, H.; Ding, X. Harnessing Multiplex crRNA in the CRISPR/Cas12a System Enables an Amplification-Free DNA Diagnostic Platform for ASFV Detection. Anal. Chem. 2022, 94, 10805–10812. [Google Scholar] [CrossRef]
- Liu, S.; Tao, D.; Liao, Y.; Yang, Y.; Sun, S.; Zhao, Y.; Yang, P.; Tang, Y.; Chen, B.; Liu, Y.; et al. Highly Sensitive CRISPR/Cas12a-Based Fluorescence Detection of Porcine Reproductive and Respiratory Syndrome Virus. ACS Synth. Biol. 2021, 10, 2499–2507. [Google Scholar] [CrossRef]
- Yang, K.; Liang, Y.; Li, Y.; Liu, Q.; Zhang, W.; Yin, D.; Song, X.; Shao, Y.; Tu, J.; Qi, K. Reverse transcription–enzymatic recombinase amplification coupled with CRISPR-Cas12a for rapid detection and differentiation of PEDV wild-type strains and attenuated vaccine strains. Anal. Bioanal. Chem. 2021, 413, 7521–7529. [Google Scholar] [CrossRef]
- Qian, B.; Liao, K.; Zeng, D.; Peng, W.; Wu, X.; Li, J.; Bo, Z.; Hu, Y.; Nan, W.; Wen, Y.; et al. Clustered Regularly Interspaced Short Palindromic Repeat/Cas12a Mediated Multiplexable and Portable Detection Platform for GII Genotype Porcine Epidemic Diarrhoea Virus Rapid Diagnosis. Front. Microbiol. 2022, 13, 920801. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tao, D.; Chen, X.; Shen, L.; Zhu, L.; Xu, B.; Liu, H.; Zhao, S.; Li, X.; Liu, X.; et al. Detection of Four Porcine Enteric Coronaviruses Using CRISPR-Cas12a Combined with Multiplex Reverse Transcriptase Loop-Mediated Isothermal Amplification Assay. Viruses 2022, 14, 833. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xu, L.; Liu, Q.; Cao, Y.; Yang, K.; Song, X.; Shao, Y.; Tu, J.; Qi, K. Enzymatic recombinase amplification coupled with CRISPR-Cas12a for ultrasensitive, rapid, and specific Porcine circovirus 3 detection. Mol. Cell. Probes 2021, 60, 101772. [Google Scholar] [CrossRef]
- Wei, J.; Li, Y.; Cao, Y.; Liu, Q.; Yang, K.; Song, X.; Shao, Y.; Qi, K.; Tu, J. Rapid and Visual Detection of Porcine Parvovirus Using an ERA-CRISPR/Cas12a System Combined with Lateral Flow Dipstick Assay. Front. Cell. Infect. Microbiol. 2022, 12, 879887. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Gong, P.; Zhang, Y.; Wang, Y.; Tao, D.; Fu, L.; Khazalwa, E.M.; Liu, H.; Zhao, S.; Zhang, X.; et al. A one-tube rapid visual CRISPR assay for the field detection of Japanese encephalitis virus. Virus Res. 2022, 319, 198869. [Google Scholar] [CrossRef]
- Wei, N.; Xiong, J.; Ma, J.; Ye, J.; Si, Y.; Cao, S. Development of efficient, sensitive, and specific detection method for Encephalomyocarditis virus based on CRISPR/Cas13a. J. Virol. Methods 2022, 309, 114592. [Google Scholar] [CrossRef]
- Jiang, C.; Tao, D.; Geng, Y.; Yang, H.; Xu, B.; Chen, Y.; Hu, C.; Chen, H.; Xie, S.; Guo, A. Sensitive and Specific Detection of Lumpy Skin Disease Virus in Cattle by CRISPR-Cas12a Fluorescent Assay Coupled with Recombinase Polymerase Amplification. Genes 2022, 13, 734. [Google Scholar] [CrossRef]
- Yao, R.; Xu, Y.; Wang, L.; Wang, D.; Ren, L.; Ren, C.; Li, C.; Li, X.; Ni, W.; He, Y.; et al. CRISPR-Cas13a-Based Detection for Bovine Viral Diarrhea Virus. Front. Vet. Sci. 2021, 8, 603919. [Google Scholar] [CrossRef]
- Chen, X.; Nie, F.; Xiong, Y.; Lin, L.; Shi, M.; Yang, J.; Wang, Y.; Wang, G.; Li, Y.; Huo, D.; et al. Ultra-sensitive and point-of-care detection of Capripoxvirus (CaPV) based on loop-mediated amplification (LAMP) and trans-cleavage activity of CRISPR/Cpf1. Anal. Chim. Acta 2022, 1191, 339330. [Google Scholar] [CrossRef]
- Lei, R.; Li, L.; Wu, P.; Fei, X.; Zhang, Y.; Wang, J.; Zhang, D.; Zhang, Q.; Yang, N.; Wang, X. RPA/CRISPR/Cas12a-Based On-Site and Rapid Nucleic Acid Detection of Toxoplasma gondii in the Environment. ACS Synth. Biol. 2022, 11, 1772–1781. [Google Scholar] [CrossRef]
- Ma, Q.-N.; Wang, M.; Zheng, L.-B.; Lin, Z.-Q.; Ehsan, M.; Xiao, X.-X.; Zhu, X.-Q. RAA-Cas12a-Tg: A Nucleic Acid Detection System for Toxoplasma gondii Based on CRISPR-Cas12a Combined with Recombinase-Aided Amplification (RAA). Microorganisms 2021, 9, 1644. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Y.; Xue, Q.; Zhu, Z.; Zou, M.; Fang, F. A novel rapid visual detection assay for Toxoplasma gondii combining recombinase-aided amplification and lateral flow dipstick coupled with CRISPR-Cas13a fluorescence (RAA-Cas13a-LFD). Parasite 2022, 29, 21. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhang, K.; Wang, Y.; Li, D.; Cui, Z.; Huang, J.; Zhang, S.; Li, X.; Zhang, L. CRISPR/Cas12a-based on-site diagnostics of Cryptosporidium parvum IId-subtype-family from human and cattle fecal samples. Parasites Vectors 2021, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ma, J.; Li, Y.; Kang, L.; Yuan, B.; Li, S.; Chao, J.; Wang, L.; Wang, J.; Su, S.; et al. A general RPA-CRISPR/Cas12a sensing platform for Brucella spp. detection in blood and milk samples. Sens. Actuators B Chem. 2022, 364, 131864. [Google Scholar] [CrossRef]
- Wang, Y.; Ke, Y.; Liu, W.; Sun, Y.; Ding, X. A One-Pot Toolbox Based on Cas12a/crRNA Enables Rapid Foodborne Pathogen Detection at Attomolar Level. ACS Sens. 2020, 5, 1427–1435. [Google Scholar] [CrossRef]
- Fang, T.; Shen, J.; Xue, J.; Jiang, Y.; Guo, D.; Yang, J.; Kong, X.; Xu, X.; Wang, X. Sensitive and rapid detection of Escherichia coli O157:H7 from beef samples based on recombinase aided amplification assisted CRISPR/Cas12a system. J. AOAC Int. 2022, qsac101. [Google Scholar] [CrossRef]
- Zhi, S.; Shen, J.; Li, X.; Jiang, Y.; Xue, J.; Fang, T.; Xu, J.; Wang, X.; Cao, Y.; Yang, D.; et al. Development of Recombinase-Aided Amplification (RAA)-Exo-Probe and RAA-CRISPR/Cas12a Assays for Rapid Detection of Campylobacter jejuni in Food Samples. J. Agric. Food Chem. 2022, 70, 9557–9566. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, T.; Liu, C.; Xu, Q.; Fang, S.; Wu, Y.; Wu, M.; Liu, Q. An ultrasensitive and contamination-free on-site nucleic acid detection platform for Listeria monocytogenes based on the CRISPR-Cas12a system combined with recombinase polymerase amplification. LWT 2021, 152, 112166. [Google Scholar] [CrossRef]
- Xiao, Y.; Ren, H.; Hu, P.; Wang, Y.; Wang, H.; Li, Y.; Feng, K.; Wang, C.; Cao, Q.; Guo, Y.; et al. Ultra-Sensitive and Rapid Detection of Pathogenic Yersinia enterocolitica Based on the CRISPR/Cas12a Nucleic Acid Identification Platform. Foods 2022, 11, 2160. [Google Scholar] [CrossRef]
- Tao, D.; Liu, J.; Li, Q.; Jiang, Y.; Xu, B.; Khazalwa, E.M.; Gong, P.; Xu, J.; Ma, Y.; Ruan, J.; et al. A Simple, Affordable, and Rapid Visual CRISPR-Based Field Test for Sex Determination of Earlier Porcine Embryos and Pork Products. Mol. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, J.; Li, H.-T.; Zhang, T.; Dong, Y.; Deng, S.; Lv, Y.; He, Q.; Deng, R. CRISPR-Cas system meets DNA barcoding: Development of a universal nucleic acid test for food authentication. Sens. Actuators B Chem. 2022, 353, 131138. [Google Scholar] [CrossRef]
- Zhao, G.; Wang, J.; Yao, C.; Xie, P.; Li, X.; Xu, Z.; Xian, Y.; Lei, H.; Shen, X. Alkaline lysis-recombinase polymerase amplification combined with CRISPR/Cas12a assay for the ultrafast visual identification of pork in meat products. Food Chem. 2022, 383, 132318. [Google Scholar] [CrossRef] [PubMed]
- Tao, D.; Xiao, X.; Lan, X.; Xu, B.; Wang, Y.; Khazalwa, E.M.; Pan, W.; Ruan, J.; Jiang, Y.; Liu, X.; et al. An Inexpensive CRISPR-Based Point-of-Care Test for the Identification of Meat Species and Meat Products. Genes 2022, 13, 912. [Google Scholar] [CrossRef]
- Pan, R.; Liu, J.; Wang, P.; Wu, D.; Chen, J.; Wu, Y.; Li, G. Ultrasensitive CRISPR/Cas12a-Driven SERS Biosensor for On-Site Nucleic Acid Detection and Its Application to Milk Authenticity Testing. J. Agric. Food Chem. 2022, 70, 4484–4491. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiong, E.; Tian, T.; Cheng, M.; Lin, W.; Sun, J.; Zhou, X. CASLFA: CRISPR/Cas9-mediated lateral flow nucleic acid assay. bioRxiv 2019. [Google Scholar] [CrossRef]
- Srivastava, S.; Upadhyay, D.J.; Srivastava, A. Next-Generation Molecular Diagnostics Development by CRISPR/Cas Tool: Rapid Detection and Surveillance of Viral Disease Outbreaks. Front. Mol. Biosci. 2020, 7, 582499. [Google Scholar] [CrossRef]
- Welch, N.L.; Zhu, M.; Hua, C.; Weller, J.; Mirhashemi, M.E.; Nguyen, T.G.; Mantena, S.; Bauer, M.R.; Shaw, B.M.; Ackerman, C.M.; et al. Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants. Nat. Med. 2022, 28, 1083–1094. [Google Scholar] [CrossRef]
- Thakku, S.G.; Ackerman, C.M.; Myhrvold, C.; Bhattacharyya, R.P.; Livny, J.; Ma, P.; Gomez, G.I.; Sabeti, P.C.; Blainey, P.C.; Hung, D.T. Multiplexed detection of bacterial nucleic acids using Cas13 in droplet microarrays. PNAS Nexus 2022, 1, pgac021. [Google Scholar] [CrossRef]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Labun, K.; Montague, T.G.; Gagnon, J.A.; Thyme, S.B.; Valen, E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016, 44, W272–W276. [Google Scholar] [CrossRef]
- Pliatsika, V.; Rigoutsos, I. “Off-Spotter”: Very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs. Biol. Direct 2015, 10, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.-H.; Kim, J.-S.; Bae, S. Web-Based CRISPR Toolkits: Cas-OFFinder, Cas-Designer, and Cas-Analyzer. Methods Mol. Biol. 2021, 2162, 23–33. [Google Scholar] [PubMed]
- Liu, H.; Wei, Z.; Dominguez, A.; Li, Y.; Wang, X.; Qi, L.S. CRISPR-ERA: A comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics 2015, 31, 3676–3678. [Google Scholar] [CrossRef] [PubMed]
- Heigwer, F.; Kerr, G.; Boutros, M. E-CRISP: Fast CRISPR target site identification. Nat. Methods 2014, 11, 122–123. [Google Scholar] [CrossRef]
- Zhao, C.; Zheng, X.; Qu, W.; Li, G.; Li, X.; Miao, Y.-L.; Changzhi, Z.; Liu, X.; Li, Z.; Ma, Y.; et al. CRISPR-offinder: A CRISPR guide RNA design and off-target searching tool for user-defined protospacer adjacent motif. Int. J. Biol. Sci. 2017, 13, 1470–1478. [Google Scholar] [CrossRef]
- Zhu, H.; Richmond, E.; Liang, C. CRISPR-RT: A web application for designing CRISPR-C2c2 crRNA with improved target specificity. Bioinformatics 2018, 34, 117–119. [Google Scholar] [CrossRef]
- Zhu, H.; Liang, C. CRISPR-DT: Designing gRNAs for the CRISPR-Cpf1 system with improved target efficiency and specificity. Bioinformatics 2019, 35, 2783–2789. [Google Scholar] [CrossRef]
- McKenna, A.; Shendure, J. FlashFry: A fast and flexible tool for large-scale CRISPR target design. BMC Biol. 2018, 16, 74. [Google Scholar] [CrossRef]
- Wu, C.; Chen, Z.; Li, C.; Hao, Y.; Tang, Y.; Yuan, Y.; Chai, L.; Fan, T.; Yu, J.; Ma, X.; et al. CRISPR-Cas12a-Empowered Electrochemical Biosensor for Rapid and Ultrasensitive Detection of SARS-CoV-2 Delta Variant. Nano-Micro Lett. 2022, 14, 159. [Google Scholar] [CrossRef]
- Hu, T.; Ke, X.; Ou, Y.; Lin, Y. CRISPR/Cas12a-Triggered Chemiluminescence Enhancement Biosensor for Sensitive Detection of Nucleic Acids by Introducing a Tyramide Signal Amplification Strategy. Anal. Chem. 2022, 94, 8506–8513. [Google Scholar] [CrossRef]
- Ke, X.; Ou, Y.; Lin, Y.; Hu, T. Enhanced chemiluminescence imaging sensor for ultrasensitive detection of nucleic acids based on HCR-CRISPR/Cas12a. Biosens. Bioelectron. 2022, 212, 114428. [Google Scholar] [CrossRef] [PubMed]
- Garigliany, M.; Desmecht, D.; Tignon, M.; Cassart, D.; Lesenfant, C.; Paternostre, J.; Volpe, R.; Cay, A.B.; van den Berg, T.; Linden, A. Phylogeographic Analysis of African Swine Fever Virus, Western Europe, 2018. Emerg. Infect. Dis. 2019, 25, 184–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, K.; Sun, W.; Zhao, J.; Yin, Y.; Si, H.; Qu, S.; Lu, W. Development a multiplex RT-PCR assay for simultaneous detection of African swine fever virus, classical swine fever virus and atypical porcine pestivirus. J. Virol. Methods 2021, 287, 114006. [Google Scholar] [CrossRef]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef]
- Chu, J.-Q.; Hu, X.-M.; Kim, M.-C.; Park, C.-S.; Jun, M.-H. Development and validation of a recombinant nucleocapsid protein-based ELISA for detection of the antibody to porcine reproductive and respiratory syndrome virus. J. Microbiol. 2009, 47, 582–588. [Google Scholar] [CrossRef]
- Chai, Z.; Ma, W.; Fu, F.; Lang, Y.; Wang, W.; Tong, G.; Liu, Q.; Cai, X.; Li, X. A SYBR Green-based real-time RT-PCR assay for simple and rapid detection and differentiation of highly pathogenic and classical type 2 porcine reproductive and respiratory syndrome virus circulating in China. Arch. Virol. 2013, 158, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Spear, A.; Faaberg, K.S. Development of a genome copy specific RT-qPCR assay for divergent strains of type 2 porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2015, 218, 1–6. [Google Scholar] [CrossRef]
- Lin, C.-N.; Lin, W.-H.; Hung, L.-N.; Wang, S.-Y.; Chiou, M.-T. Comparison of viremia of type II porcine reproductive and respiratory syndrome virus in naturally infected pigs by zip nucleic acid probe-based real-time PCR. BMC Vet. Res. 2013, 9, 181. [Google Scholar] [CrossRef]
- Huang, Y.-W.; Dickerman, A.W.; Piñeyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.-J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 2013, 4, e00737-13. [Google Scholar] [CrossRef]
- Jung, K.; Saif, L.J. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015, 204, 134–143. [Google Scholar] [CrossRef]
- Li, W.; van Kuppeveld, F.J.M.; He, Q.; Rottier, P.J.M.; Bosch, B. Cellular entry of the porcine epidemic diarrhea virus. Virus. Res. 2016, 226, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, G.; Wang, J.; Man, K.; Yang, Q. Cell attenuated porcine epidemic diarrhea virus strain Zhejiang08 provides effective immune protection attributed to dendritic cell stimulation. Vaccine 2017, 35, 7033–7041. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Li, G.; Stasko, J.; Thomas, J.T.; Stensland, W.R.; Pillatzki, A.E.; Gauger, P.C.; Schwartz, K.J.; Madson, D.; Yoon, K.-J.; et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Marthaler, D.; Wang, Q.; Culhane, M.R.; Rossow, K.D.; Rovira, A.; Collins, J.; Saif, L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014, 20, 1620–1628. [Google Scholar] [CrossRef]
- Godfroid, J.; Scholz, H.; Barbier, T.; Nicolas, C.; Wattiau, P.; Fretin, D.; Whatmore, A.; Cloeckaert, A.; Blasco, J.; Moriyon, I.; et al. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev. Vet. Med. 2011, 102, 118–131. [Google Scholar] [CrossRef]
- Moreno, E. The one hundred year journey of the genus Brucella (Meyer and Shaw 1920). FEMS Microbiol. Rev. 2021, 45, fuaa045. [Google Scholar] [CrossRef]
- Głowacka, P.; Żakowska, D.; Naylor, K.; Niemcewicz, M.; Bielawska-Drózd, A. Brucella—Virulence Factors, Pathogenesis and Treatment. Pol. J. Microbiol. 2018, 67, 151–161. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.-D.; Huang, S.-Y.; Zhu, X.-Q. Diagnosis of toxoplasmosis and typing of Toxoplasma gondii. Parasites Vectors 2015, 8, 292. [Google Scholar] [CrossRef]
- Nissapatorn, V. Toxoplasma gondii and HIV: A never-ending story. Lancet HIV 2017, 4, e146–e147. [Google Scholar] [CrossRef]
- Zhou, P.; Chen, Z.; Li, H.-L.; Zheng, H.; He, S.; Lin, R.-Q.; Zhu, X.-Q. Toxoplasma gondii infection in humans in China. Parasites Vectors 2011, 4, 165. [Google Scholar] [CrossRef]
- Nayeri, T.; Sarvi, S.; Moosazadeh, M.; Daryani, A. Global prevalence of Toxoplasma gondii infection in the aborted fetuses and ruminants that had an abortion: A systematic review and meta-analysis. Vet. Parasitol. 2021, 290, 109370. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Cervantes, R.; Córdova-Izquierdo, A. Sexing sperm of domestic animals. Trop. Anim. Health Prod. 2012, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schinckel, A.P.; Mahan, D.C.; Wiseman, T.G.; Einstein, M.E. Growth of protein, moisture, lipid, and ash of two genetic lines of barrows and gilts from twenty to one hundred twenty-five kilograms of body weight1. J. Anim. Sci. 2008, 86, 460–471. [Google Scholar] [CrossRef]
- Osada, M.; Iwabuchi, H.; Aoki, T.; Sasaki, K.; Ushijima, H.; Ozawa, T. Economic evaluation of artificial insemination of sex-sorted semen on a Brown Swiss dairy farm—A case study. Anim. Sci. J. 2019, 90, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Carabús, A.; Sainz, R.D.; Oltjen, J.W.; Gispert, M.; Font-I-Furnols, M. Growth of total fat and lean and of primal cuts is affected by the sex type. Animal 2017, 11, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Choi, T.J.; Cho, K.H.; Cho, E.S.; Lee, J.J.; Chung, H.J.; Baek, S.Y.; Jeong, Y.D. Effects of Sex and Breed on Meat Quality and Sensory Properties in Three-way Crossbred Pigs Sired by Duroc or by a Synthetic Breed Based on a Korean Native Breed. Korean J. Food Sci. Anim. Resour. 2018, 38, 544–553. [Google Scholar]
- Gunawan, A.; Sahadevan, S.; Neuhoff, C.; Große-Brinkhaus, C.; Gad, A.; Frieden, L.; Tesfaye, D.; Tholen, E.; Looft, C.; Uddin, M.J.; et al. RNA Deep Sequencing Reveals Novel Candidate Genes and Polymorphisms in Boar Testis and Liver Tissues with Divergent Androstenone Levels. PLoS ONE 2013, 8, e63259. [Google Scholar] [CrossRef]
- Barnett, J.; Begen, F.; Howes, S.; Regan, A.; McConnon, A.; Marcu, A.; Rowntree, S.; Verbeke, W. Consumers’ confidence, reflections and response strategies following the horsemeat incident. Food Control 2016, 59, 721–730. [Google Scholar] [CrossRef]
- Lopez-Calleja, I.M.; Gonzalez, I.; Fajardo, V.; Hernández, P.E.; García, T.; Martín, R. Application of an indirect ELISA and a PCR technique for detection of cows’ milk in sheep’s and goats’ milk cheeses. Int. Dairy J. 2007, 17, 87–93. [Google Scholar] [CrossRef]
- Jian, S.-H.; Yeh, P.-J.; Wang, C.-H.; Chen, H.-C.; Chen, S.-F. Analysis of heterocyclic amines in meat products by liquid chromatography—Tandem mass spectrometry. J. Food Drug Anal. 2019, 27, 595–602. [Google Scholar] [CrossRef]
- Liu, F.X.; Cui, J.Q.; Park, H.; Chan, K.W.; Leung, T.; Tang, B.Z.; Yao, S. Isothermal Background-Free Nucleic Acid Quantification by a One-Pot Cas13a Assay Using Droplet Microfluidics. Anal. Chem. 2022, 94, 5883–5892. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).