Spatial and Temporal Expression Characteristics of the HBB Gene Family in Six Different Pig Breeds
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Identification of Hemoglobin-Encoding Genes in Pigs and Mice
2.2. Evolutionary Analysis and Motif Prediction of Human, Mouse and Pig Hemoglobin-Encoding Genes
2.3. Primer Design
2.4. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
2.5. Statistics
3. Results
3.1. Identification of the Pig Hemoglobin-Encoding Gene Family
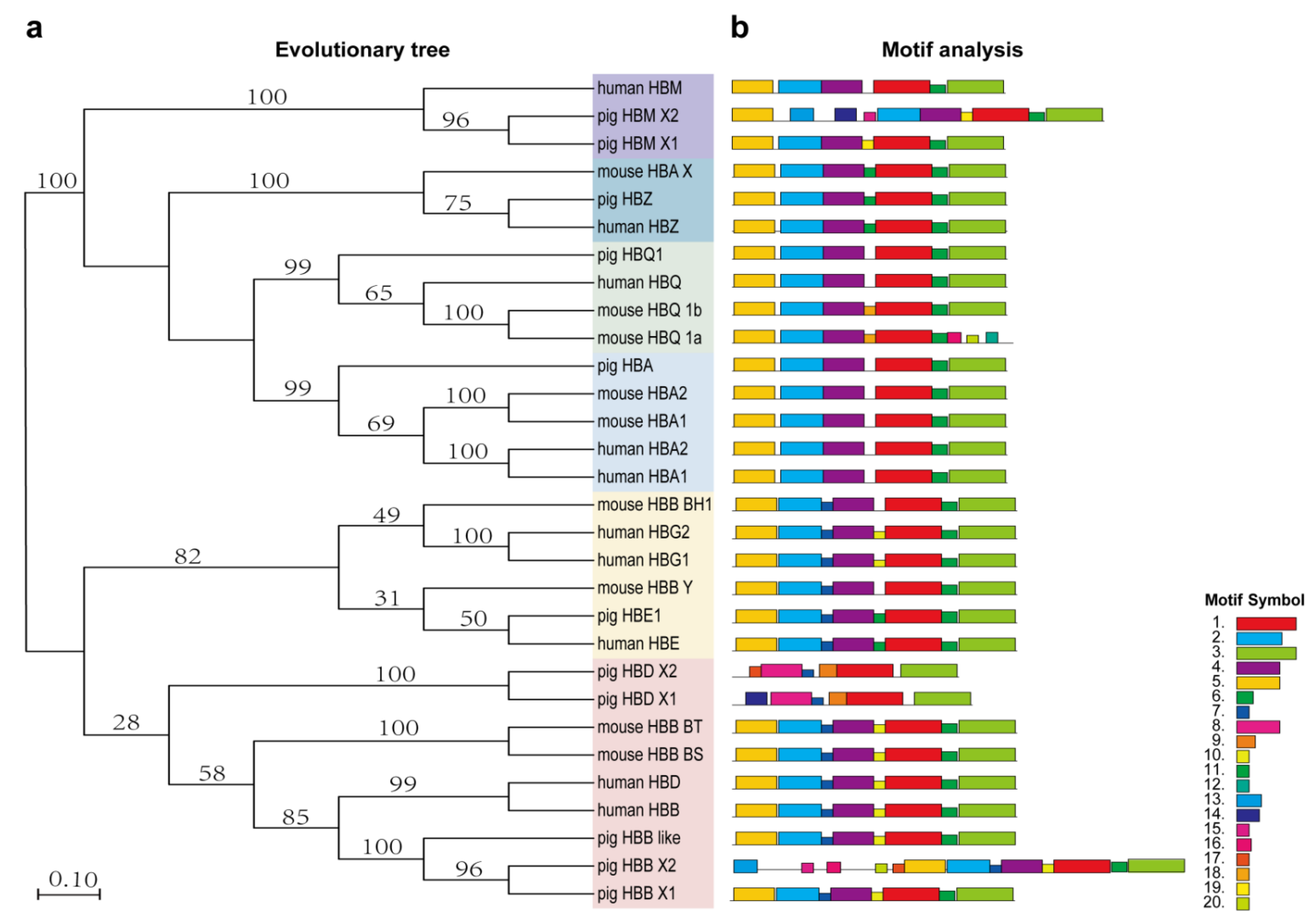
3.2. Evolutionary Analysis of Human, Pig and Mouse Hemoglobin-Encoding Genes
3.3. HBB and HBB-like Gene Sequence Analysis
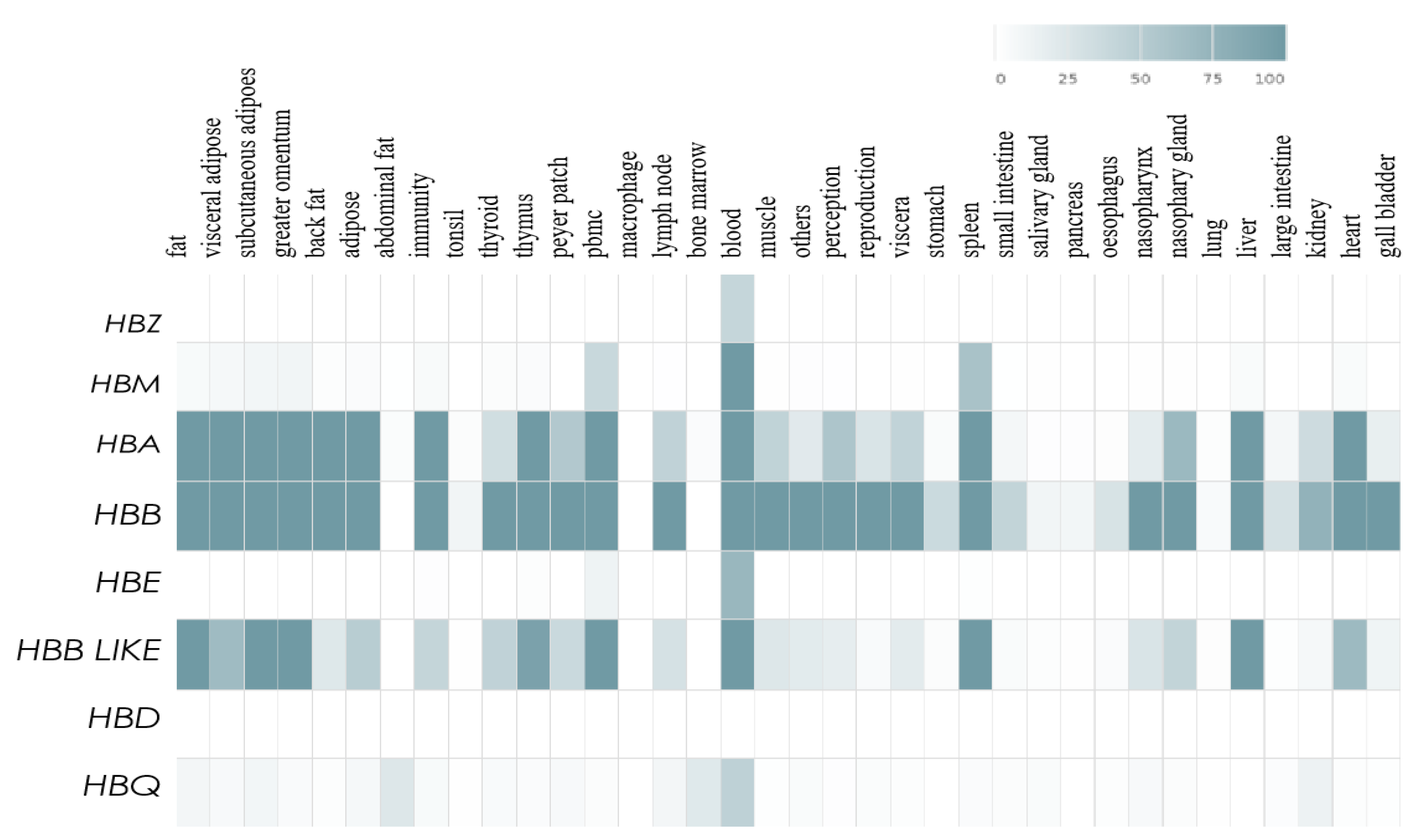
3.4. Expression Levels of the HBB Gene Family in Different Tissues
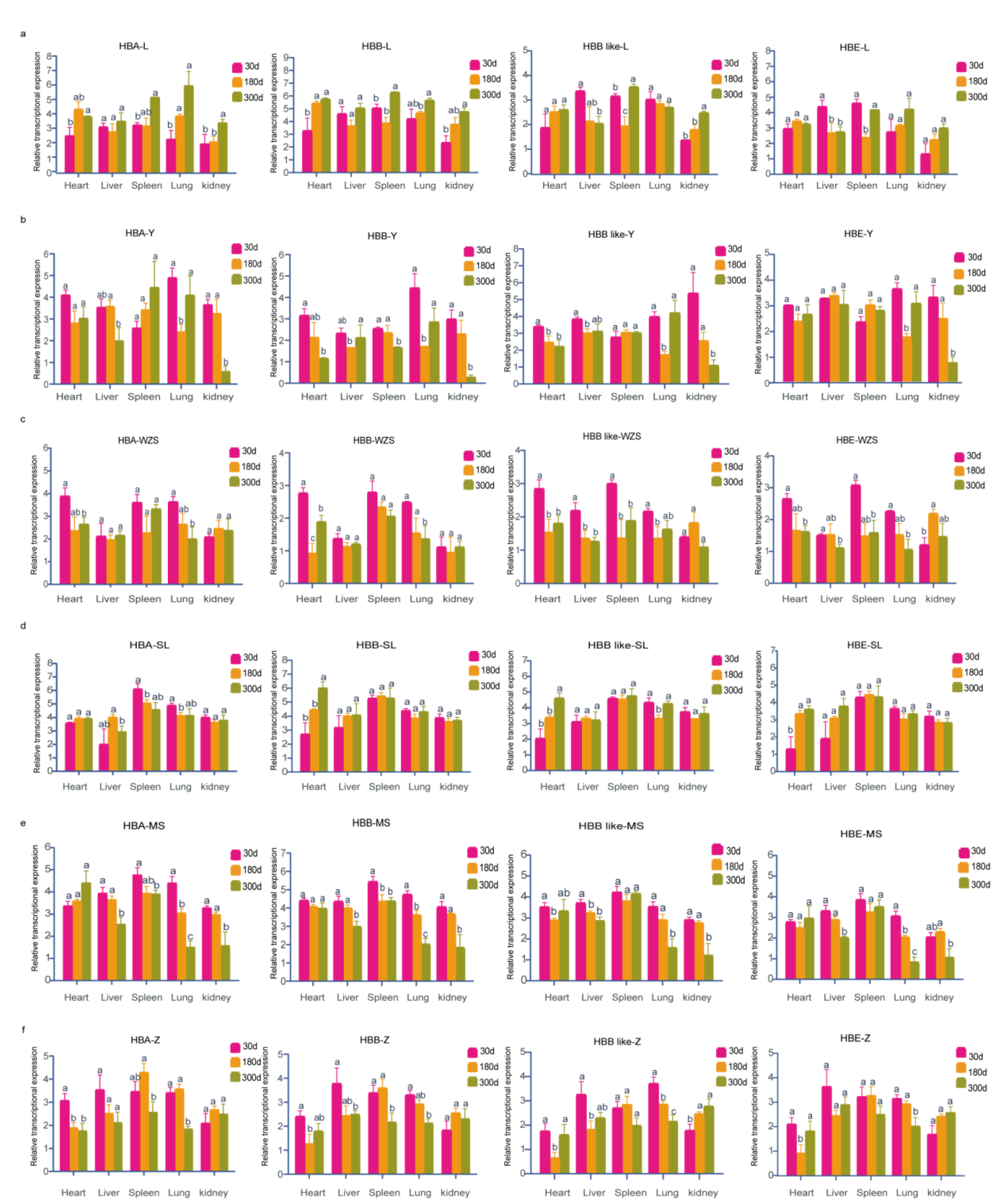
3.5. Spatiotemporal Expression Patterns of Hemoglobin-Encoding Genes in Adult Pigs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, H.; Huang, Q.; Yu, Z.; Zhong, Z. Molecular analysis of alpha- and beta-thalassemia in Meizhou region and comparison of gene mutation spectrum with different regions of southern China. J. Clin. Lab. Anal. 2021, 35, e24105. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wu, X. Epidemiology and treatment of beta thalassemia major in China. Pediatr. Investig. 2019, 4, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Cao, A.; Kan, Y.W. The prevention of thalassemia. Cold Spring Harb. Perspect. Med. 2013, 3, a011775. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cui, Q.; Zhou, W.; Qiu, L.; Han, B. Comparison of gene mutation spectrum of thalassemia in different regions of China and Southeast Asia. Mol. Genet. Genomic Med. 2019, 7, e680. [Google Scholar] [CrossRef]
- Mahdieh, N.; Rabbani, B. Beta thalassemia in 31,734 cases with HBB gene mutations: Pathogenic and structural analysis of the common mutations; Iran as the crossroads of the Middle East. Blood Rev. 2016, 30, 493–508. [Google Scholar] [CrossRef]
- Marengo-Rowe, A.J. Structure-function relations of human hemoglobins. Bayl. Univ. Med. Cent. Proc. 2006, 19, 239–245. [Google Scholar] [CrossRef]
- Zittersteijn, H.A.; Harteveld, C.L.; Klaver-Flores, S.; Lankester, A.C.; Hoeben, R.C.; Staal, F.J.T.; Gonçalves, M.A.F.V. A small key for a heavy door: Genetic therapies for the treatment of hemoglobinopathies. Front. Genome Ed. 2021, 4, 617780. [Google Scholar] [CrossRef]
- Cao, A.; Moi, P. Regulation of the globin genes. Pediatr. Res. 2002, 51, 415–421. [Google Scholar] [CrossRef][Green Version]
- Huehns, E.R.; Flynn, F.V.; Butler, E.A.; Beaven, G.H. Two new haemoglobin variants in a very young human embryo. Nature 1961, 189, 496–497. [Google Scholar] [CrossRef]
- Oneal, P.A.; Gantt, N.M.; Schwartz, J.D.; Bhanu, N.V.; Lee, Y.T.; Moroney, J.W.; Reed, C.H.; Schechter, A.N.; Luban, N.L.; Miller, J.L. Fetal hemoglobin silencing in humans. Blood 2006, 108, 2081–2086. [Google Scholar] [CrossRef]
- Sripichai, O.; Fucharoen, S. Fetal hemoglobin regulation in β-thalassemia: Heterogeneity, modifiers and therapeutic approaches. Expert. Rev. Hematol. 2016, 9, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Peschle, C.; Mavilio, F.; Migliaccio, G.; Migliaccio, A.R.; Russo, G.; Mastroberardino, G.; Marinucci, M. Erythropoietic development and hemoglobin switching in human embryos: Cellular and molecular aspects. Prog. Clin. Biol. Res. 1985, 191, 383–396. [Google Scholar] [PubMed]
- Su, Q.; Chen, S.; Wu, L.; Tian, R.; Yang, X.; Huang, X.; Chen, Y.; Peng, Z.; Chen, J. Severe Thalassemia Caused by Hb Zunyi [β147(HC3)Stop→Gln; HBB: C.442T>C)] on the β-Globin Gene. Hemoglobin 2019, 43, 7–11. [Google Scholar] [CrossRef]
- Lithanatudom, P.; Smith, D.R. Analysis of protein profiling studies of β-thalassemia/Hb E disease. Proteom. Clin. Appl. 2016. [CrossRef] [PubMed]
- Galanello, R.; Origa, R. Beta-thalassemia. Orphanet. J. Rare Dis. 2010, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Azman, N.F.; Abdullah, W.Z.; Hanafi, S.; Diana, R.; Bahar, R.; Johan, M.F.; Zilfalil, B.A.; Hassan, R. Genetic polymorphisms of HbE/beta thalassemia related to clinical presentation: Implications for clinical diversity. Ann. Hematol. 2020, 99, 729–735. [Google Scholar] [CrossRef]
- Munkongdee, T.; Tongsima, S.; Ngamphiw, C.; Wangkumhang, P.; Peerapittayamongkol, C.; Hashim, H.B.; Fucharoen, S.; Svasti, S. Predictive SNPs for β-thalassemia/HbE disease severity. Sci. Rep. 2021, 11, 10352. [Google Scholar] [CrossRef]
- He, X.; Sheng, M.; Xu, M.; Xiong, C.; Ren, Z. Rapid identification of common β-thalassemia mutations in the Chinese population using duplex or triplex amplicon genotyping by high-resolution melting analysis. Genet. Test. Mol. Biomarkers 2010, 14, 851–856. [Google Scholar] [CrossRef]
- Zhang, W.; Cai, W.W.; Zhou, W.P.; Li, H.P.; Li, L.; Yan, W.; Deng, Q.K.; Zhang, Y.P.; Fu, Y.X.; Xu, X.M. Evidence of gene conversion in the evolutionary process of the codon 41/42 (-CTTT) mutation causing beta-thalassemia in southern China. J. Mol. Evol. 2008, 66, 436–445. [Google Scholar] [CrossRef]
- He, S.; Li, J.; Li, D.M.; Yi, S.; Lu, X.; Luo, Y.; Liang, Y.; Feng, C.; Chen, B.; Zheng, C.; et al. Molecular characterization of α- and β-thalassemia in the Yulin region of Southern China. Gene 2018, 655, 61–64. [Google Scholar] [CrossRef]
- Weatherall, D.J. Keynote address: The challenge of thalassemia for the developing countries. Ann. N. Y. Acad. Sci. 2005, 1054, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Weatherall, D.J. Thalassemia as a global health problem: Recent progress toward its control in the developing countries. Ann. N. Y. Acad. Sci. 2010, 1202, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.K.; Hara, H.; Yazer, M. Genetically engineered pigs as a source for clinical red blood cell transfusion. Clin. Lab. Med. 2010, 30, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.K.C.; Ezzelarab, M.; Iwase, H.; Hara, H. Perspectives on the optimal genetically engineered pig in 2018 for initial clinical trials of kidney or heart xenotransplantation. Transplantation 2018, 102, 1974–1982. [Google Scholar] [CrossRef]
- Yamamoto, T.; Bikhet, M.H.; Marques, M.B.; Nguyen, H.Q.; Cui, Y.; Javed, M.; Raza, S.S.; Ayares, D.; Iwase, H.; Cooper, D.K.C.; et al. Initial experimental experience of triple-knockout pig red blood cells as potential sources for transfusion in alloimmunized patients with sickle cell disease. Transfusion 2021, 61, 3104–3118. [Google Scholar] [CrossRef]
- Rogers, C.S. Genetically engineered livestock for biomedical models. Transgenic Res. 2016, 25, 345–359. [Google Scholar] [CrossRef]
- Ou, Z.; Niu, X.; He, W.; Chen, Y.; Song, B.; Xian, Y.; Fan, D.; Tang, D.; Sun, X. The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human β-thalassemia in mice. Sci. Rep. 2016, 6, 32463. [Google Scholar] [CrossRef]
- Yang, Y.; Kang, X.; Hu, S.; Chen, B.; Xie, Y.; Song, B.; Zhang, Q.; Wu, H.; Ou, Z.; Xian, Y.; et al. CRISPR/Cas9-mediated β-globin gene knockout in rabbits recapitulates human β-thalassemia. J. Biol. Chem. 2021, 296, 100464. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic. Acids. Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef]
- Hung, J.H.; Weng, Z. Sequence Alignment and Homology Search with BLAST and ClustalW. Cold Spring Harb. Protoc. 2016, 11, 10–1101. [Google Scholar] [CrossRef]
- Li, D.; Xu, R.; Lv, D.; Zhang, C.; Yang, H.; Zhang, J.; Wen, J.; Li, C.; Tan, X. Identification of the core pollen-specific regulation in the rice OsSUT3 promoter. Int. J. Mol. Sci. 2020, 21, 1909. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, N.F. Treatment strategies for hemoglobin E beta-thalassemia. Blood Rev. 2012, 26, S28–S30. [Google Scholar] [CrossRef]
- Bao, X.; Zhang, X.; Wang, L.; Wang, Z.; Huang, J.; Zhang, Q.; Ye, Y.; Liu, Y.; Chen, D.; Zuo, Y.; et al. Epigenetic inactivation of ERF reactivates γ-globin expression in β-thalassemia. Am. J. Hum. Genet 2021, 108, 709–721. [Google Scholar] [CrossRef]
- Zakaria, N.A.; Islam, M.A.; Abdullah, W.Z.; Bahar, R.; Mohamed Yusoff, A.A.; Abdul Wahab, R.; Shamsuddin, S.; Johan, M.F. Epigenetic insights and potential modifiers as therapeutic targets in β-thalassemia. Biomolecules 2021, 11, 755. [Google Scholar] [CrossRef]
- Muncie, H.L., Jr.; Campbell, J. Alpha and beta thalassemia. Am. Fam. Physician 2009, 80, 339–344. [Google Scholar]
- Yin, X.L.; Wu, Z.K.; He, Y.Y.; Zhou, T.H.; Zhou, Y.L.; Zhang, X.H. Treatment and complications of thalassemia major in Guangxi, Southern China. Pediatr. Blood Cancer 2011, 57, 1174–1178. [Google Scholar] [CrossRef]

| Primers | Sequence (5′–3′) | Product Length, bp |
|---|---|---|
| SUS-Qhbb1-F | aatgtggacgaagttggtggt | 270 |
| SUS-Qhbb1-R | gttgcccaggagcctgaagt | |
| SUS-HBB-like-F | tcggctgctgtcctcggcctgca | 303 |
| SUS-HBB-like-R | gttgcccaggagcctgaagt | |
| SUS-HBE1-F | tcctggtggtctacccttgg | 233 |
| SUS-HBE1-R | gttgcccaggagcctgaagt | |
| SUS-HBA-F | agaggccctggaaagaatgt | 278 |
| SUS-HBA-R | ggttgaaatcatcggggtgg | |
| 18S-F | gtaacccgttgaaccccatt | 151 |
| 18S-R | ccatccaatcggtagtagcg |
| Species | Gene Symbol | Protein ID | Chromosome |
|---|---|---|---|
| Homo sapiens | HBB | ENST00000335295 | 11 |
| HBD | ENST00000292901 | 11 | |
| HBE1 | ENST00000396895 | 11 | |
| HBG1 | ENST00000330597 | 11 | |
| HBG2 | ENST00000336906 | 11 | |
| HBA1 | ENST00000320868 | 16 | |
| HBA2 | ENST00000251595 | 16 | |
| HBM | ENST00000356815 | 16 | |
| HBQ1 | ENST00000199708 | 16 | |
| HBZ | ENST00000252951 | 16 | |
| Sus scrofa | HBB X1 | ENSSSCP00000067521 | 9 |
| HBB X2 | ENSSSCP00015021905 | 9 | |
| HBB-like | ENSSSCP00000015649 | 9 | |
| HBD X1 | ENSSSCP00000059042 | 9 | |
| HBD X2 | ENSSSCP00000045757 | 9 | |
| HBE1 | ENSSSCP00000015648 | 9 | |
| HBA | ENSSSCP00000028944 | 3 | |
| HBM X1 | ENSSSCP00000063903 | 3 | |
| HBM X2 | ENSSSCP00000008516 | 3 | |
| HBQ1 | ENSSSCP00000029523 | 3 | |
| HBZ | ENSSSCP00000008514 | 3 | |
| Mus Musculus | HBA-x | ENSMUSP00000020531 | 11 |
| HBQ 1a | ENSMUSP00000020535 | 11 | |
| HBQ 1b | ENSMUSP00000098936 | 11 | |
| HBA a1 | ENSMUSP00000090897 | 11 | |
| HBA a2 | ENSMUSP00000090895 | 11 | |
| HBB bs | ENSMUSP00000023934 | 7 | |
| HBB y | ENSMUSP00000033229 | 7 | |
| HBB bh1 | ENSMUSP00000064865 | 7 | |
| HBB bt | ENSMUSP00000095794 | 7 | |
| HBB bh2 | ENSMUSP00000102479 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Liu, Z.; Mu, Y.; Huang, L.; Li, K.; Zhang, J. Spatial and Temporal Expression Characteristics of the HBB Gene Family in Six Different Pig Breeds. Genes 2022, 13, 1822. https://doi.org/10.3390/genes13101822
Guo X, Liu Z, Mu Y, Huang L, Li K, Zhang J. Spatial and Temporal Expression Characteristics of the HBB Gene Family in Six Different Pig Breeds. Genes. 2022; 13(10):1822. https://doi.org/10.3390/genes13101822
Chicago/Turabian StyleGuo, Xin, Zhiguo Liu, Yulian Mu, Lei Huang, Kui Li, and Jing Zhang. 2022. "Spatial and Temporal Expression Characteristics of the HBB Gene Family in Six Different Pig Breeds" Genes 13, no. 10: 1822. https://doi.org/10.3390/genes13101822
APA StyleGuo, X., Liu, Z., Mu, Y., Huang, L., Li, K., & Zhang, J. (2022). Spatial and Temporal Expression Characteristics of the HBB Gene Family in Six Different Pig Breeds. Genes, 13(10), 1822. https://doi.org/10.3390/genes13101822






