Abstract
The extreme diversity and complexity of angiosperms is well known. Despite the fact that parasitic plants are angiosperms, little is known about parasitic plant mitogenomic diversity, complexity, and evolution. In this study, we obtained and characterized the mitogenomes of three Cistanche species (holoparasitic plants) from China to compare the repeats, segment duplication and multi-copy protein-coding genes (PCGs), to clarify the phylogenetic and evolution relationship within the Lamiales order, and to identify the mitochondrial plastid insertions (MTPT) in Cistanche mitogenomes. The results showed that the mitogenome sizes of the three Cistanche species ranged from 1,708,661 to 3,978,341 bp. The Cistanche species genome encodes 75–126 genes, including 37–65 PCGs, 31–58 tRNA genes and 3–5 rRNA genes. Compared with other Lamiales and parasitic species, the Cistanche species showed extremely high rates of multi-copy PCGs, ranging from 0.13 to 0.58 percent of the total number of PCGs. In addition, 37–133 Simple Sequence Repeat (SSRs) were found in these three mitogenomes, the majority of which were the mononucleotides Adenine/Thymine. The interspersed repeats contained forward and palindromic repeats. Furthermore, the segment-duplication sequence size ranged from 199,584 to 2,142,551 bp, accounting for 24.9%, 11.7% and 53.9% of the Cistanche deserticola, Cistanche salsa and Cistanche tubulosa mitogenome, respectively. Furthermore, the Ka/Ks analysis suggested that the atp4, ccmB, ccmFc and matR were probably positively selected during Lamiales evolution. The Cistanche plastome suggested the presence of MTPT. Moreover, 6–12 tRNA, 9–15 PCGs fragments and 3 rRNA gene fragments in the Cistanche mitogenomes were found in the MTPT regions. This work reports the Cistanche species mitogenome for the first time, which will be invaluable for study the mitogenome evolution of Orobanchaceae family.
Keywords:
Cistanche; mitogenome; MTPT; repeat; segment duplication; multi-copy PCGs; substitution rate 1. Introduction
The Cistanche species are a group of non-photosynthetic parasitic plants. Cistanche is an Old World genus with about two dozen species. They have been divided into four well supported and geographically differentiated clades: East Asian Clade, Northwest African Clade, Southwest Asian Clade and Widespread Clade [1]. The Cistanche genus belongs to the Orobancheaceae family. The Orobanchaceae family contains about 2000 species (mostly parasitic plants) in 90–115 genera, and it belongs to Lamiales order [2]. Depending on the need of a host to complete its life cycle, Orobancheaceae plants are typically classified as obligate and facultative parasites. In addition, depending on their photosynthesis ability, Orobancheaceae plants are divided into a group with photosynthetic activity (hemiparasites) and a group with no photosynthetic activity (holoparasites) [3]. The Orobanchaceae family is an excellent model system for studying the evolution of parasitism in plants [4].
Previous studies showed that the genome size, genome structure, and gene content of Orobanchaceae (parasitic plants) plastomes vary dramatically compared with other angiosperm plants [5,6]. Furthermore, severe gene loss and pseudogenization within Orobanchaceae species plastomes were found [7]. To date, 46 complete Orobanchaceae plastid genomes have been deposited in GenBank, while only one Orobanchaceae mitogenome has been deposited. The complete mitogenome of an Orobanchaceae species, Castilleja paramensis, has the lowest gene-loss rate (7.69%) among the seven Orobanchaceae species. The low level of gene loss in the mitogenomes suggests that the parasitic plants still have a typical mitochondrial function [8].
The first complete mitogenome from parasitic mistletoes also exhibited significant degradation [9]. In addition, the parasitic mitogenomes about which previous reports were published were highly divergent in many aspects, including genome size, genome structure, gene loss, etc. For example, the Viscum scurruloideum mitogenome was 66 kb in size, with one circular and one linear chromosome [9]. The Gastrodia elata mitogenome was 20 times larger than that of V. scurruloideum, and it contained 12 circular and 7 linear chromosomes [10]. It is unknown, however, whether this observation applies generally to other parasitic plants in terms of genome structures.
Mitochondria are organelles that participate in a variety of metabolic activities linked to energy generation, synthesis and destruction [11]. The most well-known function of mitochondria is oxidative phosphorylation, which uses the proton gradient to produce ATP for metabolic activity [12]. In general, the mitogenome represents mitochondrial genetic information. Compared to those of animals and fungi, plant mitogenomes display many unique features. The sizes of the mitogenomes in angiosperms range from 200 kb to 11 Mb, and differ greatly among species [9,13]. Most angiosperm mitogenomes contain 24 to 41 PCGs and 2 or 3 rRNA genes [14,15,16,17]. The mitogenome expansion is largely fueled by DNA duplications and the insertion of foreign DNA, such as nuclear DNA, plastid-derived DNA (referred to as mitochondrial plastid insertions, MTPTs), and even horizontal gene transfers (HGTs) [15,18,19].
In addition, the synonymous substitution rates are typically quite low in plant mitogenomes and are similar among PCGs. A surprising amount of synonymous rate variation was also seen in some plants’ mitochondrial PCGs [20]. The variability of the above mitochondrial sequences and PCGs has sparked researchers’ interest in the mitogenome. To date, 8592 complete plastomes have been deposited in GenBank, while only 457 complete plant mitogenomes have been deposited. Therefore, it is necessary to study plant mitogenomes.
Most Cistanche species are traditionally used as medicinal herbs. In particular, C. deserticola is known as desert ginseng for its nourishing effects [21]. Several studies reported the chemical components and pharmacological effects of Cistanche species [22,23,24]. Previously, we reported the features of four Cistanche plastomes in China, and we found that the Cistanche plastomes were significantly different from those of other angiosperms [25]. By contrast, the study of the mitogenomes of Cistanche species is still in its infancy.
In this work, for the first time, we sequenced, assembled and characterized three full Cistanche species mitogenomes to: (1) remedy the lack of knowledge of Cistanche mitogenomes; (2) explore the complexity and diversity of Cistanche mitogenomes in terms of genome size, multi-copy PCGs, repeat, segment duplication, phylogenetic relationship and substitution rate; and (3) analyze the MTPT in the Cistanche mitogenomes. The results from this study will provide invaluable information for Cistanche-mitogenome evolution.
2. Results
2.1. Characteristics of the Cistanche Mitogenomes
The three Cistanche mitogenomes contained multiple linear chromosomes (Table 1; Figures S1–S3). The lengths of the C. deserticola, C. salsa and C. tubulosa mitogenomes were 1,860,774 bp, 1,708,661 bp and 3,978,341 bp, respectively (Table 1). Compared with C. deserticola and C. salsa, the C. tubulosa mitogenome was significantly larger. The overall GC contents of the C. deserticola, C. salsa and C. tubulosa mitogenomes were 44.59%, 44.52% and 44.57%, respectively (Table 1).

Table 1.
Basic information on Cistanche species mitogenomes. In the row, Chromosomes, the number means the total number of chromosomes, while L denotes linear. Se-dup means segment duplication.
We annotated the three Cistanche species mitogenomes. The functional and structural classification of the annotated genes are shown in Table 2. According to our annotation, the C. deserticola mitogenome contained 82 genes, including 37 PCGs, 39 tRNA and 5 rRNA. The C. salsa mitogenome contained 75 genes, including 41 PCGs, 31 tRNA and 3 rRNA. The C. tubulosa mitogenome contained 126 genes, including 65 PCGs, 58 tRNA and 4 rRNA (Table 1). The functional genes were divided into ten classes, according to American College of Medical Genetics and Genomics (ACMG): Complex I (nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7 and nad9); complex II (sdh3 and sdh4); complex III (cob); complex IV (cox1, cox2 and cox3); complex V (atp1, atp4, atp6, atp8 and atp9); cytochromec biogenesis (ccmB, ccmC, ccmFc and ccmFn); intron maturase (matR); secY independent transport (mttB); ribosomal protein large subunit (rpl2, rpl5, rpl10 and rpl16), and ribosomal protein small subunit (rps3, rps4, rps10, rps12, rps13 and rps14) (Table 2).

Table 2.
Protein-coding genes in Cistanche species mitogenomes. ●: no intron containing gene present; Ө: trans-spliced gene present; ○: cis-spliced gene present; Ψ: pseudogene present; ×: gene absent.
It is known that mitochondrial PCGs are variable in genetic structure. The functional genes were divided into three classes: non-intron-containing, trans-spliced and cis-spliced gene. Most PCGs belonging to the same functional categories shared similar genetic structures. We found that four, seven and eight PCGs were trans-spliced genes in C. deserticola (ccmFc, cox1, nad4 and nad7), C. salsa (atp1, ccmFc, cob, nad7, rpl2, rps3 and rps10) and C. tubulosa (ccmFc, cox1, nad4, nad5, nad7, rpl10, rps3 and rps12), respectively (Table 2). In addition, we found that three, four and two PCGs were cis-spliced genes in C. deserticola (nad1, nad2 and nad5), C. salsa (nad1, nad2, nad4 and nad5) and C. tubulosa (nad1 and nad2), respectively (Table 2). Notably, the plastid PCGs (rpl16 and rps14) were likely to be two pseudogenes (Table 2).
2.2. Comparison of Multi-Copy Protein-Coding Genes (PCGs) in the Three Cistanche Species and Eight Other Lamiales and Six Parasitic Species Mitogenomes
We compared the mitogenome size, GC content and PCGs copy of the Cistanche species and other Lamiales species with published mitogeomes. The published Lamiales species included Boea hygrometrica, Mimulus guttatus, Ajuga reptans, Salvia miltiorrhiza, Hesperelaea palmeri, Castilleja paramensis, Utricularia reniformis and Rotheca serrate (Table S2). Their GC contents ranged from 43.27% to 45.5%, which were fairly similar (Table 1 and Table S2). However, the sizes of these mitogenomes were extremely variable (Figure 1, Table 1 and Table S2). The C. tubulosa mitogenome (3,978,341 bp) was the largest; its size was 11.3 times larger than the smallest mitogenome (A. reptans, 352,069 bp) (Table 1 and Table S2). The Cistanche mitogenomes were the largest among all the mitogenomes found in the Lamiales order. To determine whether there were any correlations between the PCG copy numbers and the mitogenome size, the PCGs copy numbers from those 11 mitogenomes were compared. The duplication of PCGs was observed in all the Lamiales mitogenomes. The degree of duplication was especially high in the Cistanche genus (Figure 1). The mitochondrial PCGs were divided into two categories, core genes and variable genes, according to a previous study [9]. Among the Cistanche mitogenomes, the proportion of duplicated core genes ranged from 13% to 58%, in the following order: C. tubulosa (58%), C. salsa (25%) and C. deserticola (13%) (Figure 1 and Table S4). The proportion of duplicated variable genes ranged from 0–35%, in the following order: C. tubulosa (35%), C. deserticola (6%) and C. salsa (0%) (Figure 1 and Table S4). Among other Lamiales species, the duplication of core genes was only present in the H. palmeri and U. reniformis mitogenomes (Figure 1). Furthermore, the duplication of variable genes was present in the E. guttata, H. palmeri and U. reniformis mitogenomes (Figure 1).
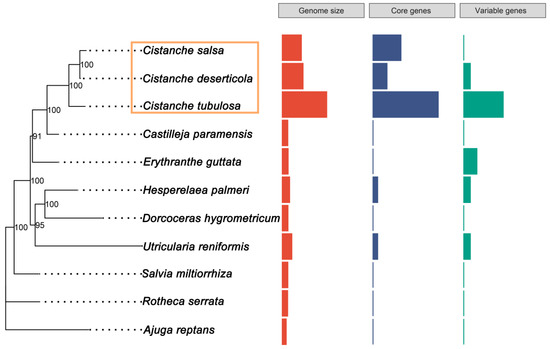
Figure 1.
Genome size and protein-coding-gene contents among 11 Lamiales species mitogenomes.
We also compared the size, GC content and number of PCG copies of the mitogenomes from the Cistanche species and several parasitic plants, including C. paramensis, Cynomorium coccineum, Epirxanthes elongate, Lophophytum mirabile, Viscum album and V. scurruloideum (Table S5). Their GC contents ranged from 43.52% to 47.4%, which were fairly similar (Table 1 and Table S5). However, the sizes of these mitogenomes varied greatly (Figure S4, Table 1 and Table S5). The smallest mitogenome was from V. scurruloideum (65,873 bp). The Cistanche mitogenomes remained the largest among the parasitic plants (Figure S4, Table 1 and Table S5). It is worth noting that the mitogenome size of C. coccineum, a holoparasitic plant, was also over 1 Mb (Table S5).
In addition to C. paramesis, V. scurruloideum and V. album, the duplication of PCGs was also present in the mitogenomes of the parasitic plants (Figure S4 and Table S6). Among the Cistanche mitogenomes, the proportion of duplicated core genes ranged from 13% to 0.63%, in the following order: C. tubulosa (63%), C. salsa (25%) and C. deserticola (13%) (Figure S4 and Table S6). Furthermore, the proportion of duplicated variable genes ranged from 0–35%, in the following order: C. tubulosa (35%), C. deserticola (6%) and C. deserticola (0) (Figure S4 and Table S6). Overall, C. tubulosa and C. coccineum had the highest proportions of duplicated core genes and variable genes (Figure S4 and Table S6).
2.3. Identification of MTPTs
To identify the MTPTs in the Cistanche mitogenomes, we compared the Cistanche mitogenome sequences with their plastome sequences. For C. deserticola, 158 high-scoring segment pairs (HSPs) were found. These 158 fragments were 35,165 bp in length in total, accounting for 1.89% of the mitogenome length (Figure S5). The fragment length ranged from 33 to 1156 bp. The annotated results showed that they were all plastid genes, including 11 complete tRNA genes: trnV-GAC (2), trnN-GUU, trnM-CAU, trnW-CCA (2), trnS-GGA (2), trnS-GCU (2) and trnS-GGA. Furthermore, nine fragments were annotated as partial plastid PCGs: rpl2 (170 bp), ycf2 (276, 387, 216, 61, 36, 61 bp), rps14 (199 bp) and rps4 (193 bp). The remaining fragments were identified as plastid ribosome RNAs (rrn 5, 16S and 23) (Table S7).
For C. salsa, 128 HSPs were identified. These 128 fragments were 28,963 bp in length in total, accounting for 1.64% of the mitogenome length (Figure S6). The fragment length ranged from 33 to 1155 bp. The annotated results showed that they were all plastid genes, including 12 complete tRNA genes: trnS-GGA (5), trnN-GUU (2), trnD-GUC (2), trnW-CCA, trnM-CAU and trnV-GAC. Furthermore, nine fragments were annotated as partial plastid PCGs: rps4 (193 bp), rps14 (199,193 bp) and ycf2 (216, 170, 61, 387, 276, 396 bp). The remaining fragments were identified as plastid ribosome RNAs (rrn 5, 16S and 23) (Table S8).
For C. tubulosa, 139 HSPs were found. These 139 fragments were 26, 911 bp in length in total, accounting for 0.68% of the mitogenome length (Figure S7). The fragment length ranged from 35 to 1156 bp. The annotated results showed that they were all plastid genes, including six complete tRNA genes: trnD-GUC (2), trnN-GUU, trnD-GUC, trnN-GUU and trnR-ACG. Furthermore, 15 fragments were annotated as partial plastid PCGs: ycf2 (128, 81, 110, 81, 257, 110, 110, 128, 77 bp), ycf1 (580, 580 bp), accD (243, 31, 243 bp) and rpl2 (138 bp). The remaining fragments were identified as plastid ribosome RNA (rrn 5, 16S and 23) (Table S9).
2.4. Repeats and Segment Duplication Analysis
The types and numbers of repeats varied among the three mitogenomes. SSRs are sequences composed of repeats with motifs 1 to 6 bp in length. Among the Cistanche mitogenomes, the number of SSRs ranged from 37–133 in the following order: C. tubulosa (133), C. deserticola (44) and C. salsa (37). Polyadenine or polythymine repeat types were the most prevalent mononucleotide SSRs (Figure 2A and Table S10). This result was in agreement with the fact that the AT content (55.41–55.43%) was higher than the GC content (44.57–44.59%) in the Cistanche mitogenomes (Table 1).
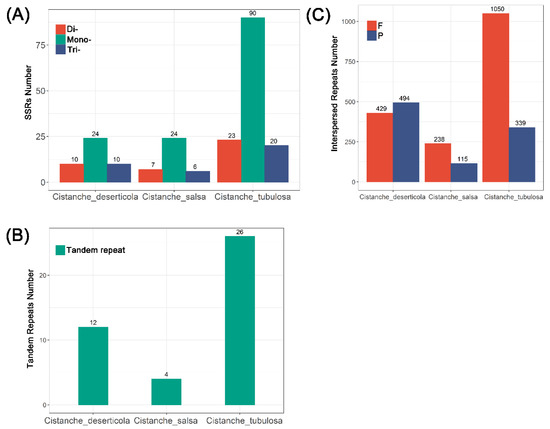
Figure 2.
Simple sequence repeats (SSRs), interspersed repeats and tandem repeats in 3 Cistanche species mitogenomes. (A) Comparison of SSRs among the three mitogenomes. Each color column represents a different SSR-repeat type. (B) Comparison of tandem repeats among the three mitogenomes. (C) Comparison of interspersed repeats in the three mitogenomes. Each color column represents a different interspersed-repeat type. The number of repeats in each category is shown on the top of the corresponding columns.
Next, we detected the interspersed repeats by REPuter. The interspersed repeats were divided into four types: forward, palindrome, reverse and complement repeats. In the Cistanche mitogenomes, the forward and palindromic repeats were the main types of interspersed repeats (Figure 2B and Tables S11–S13). Only 353 interspersed repeats were detected in the C. salsa mitogenome (Table S12), and more than 1300 in the C. tubulosa (Table S13).
In addition to the SSRs and interspersed repeats, we also detected tandem repeats >30 bp in length and similarities of >90%. The number of interspersed repeats ranged from 4 to 26 in the Cistanche mitogenomes (Figure 2C). The number of repeat units ranged from 1.8 to 2.5 copies per tandem repeat, and the repeat sizes ranged from 21 to 127 bp (Tables S14–S16).
The segment-duplication-identification results showed that the segment sequences ranged from 199,584 bp to 2,142,551 bp in length (Table 1), accounting for 24.9%, 11.7% and 53.9% of the lengths of the C. deserticola, C. salsa and C. tubulosa mitogenomes, respectively (Figure 3). In the C. deserticola mitogenome, 39 alignments were identified. The lengths of the alignments ranged from 5078 bp to 38,025 bp (Table S17). By contrast, only 14 alignments were identified in the C. salsa mitogenome. The lengths of the alignments ranged from 5385 bp to 23,085 bp (Table S18). It is worth noting that 168 alignments were found in the C. tubulosa mitogenome. The lengths of the alignments ranged from 5169 bp to 64,106 bp (Table S19). These repeat sand segment duplications might have promoted genome rearrangement and contributed to the variations in genome size.
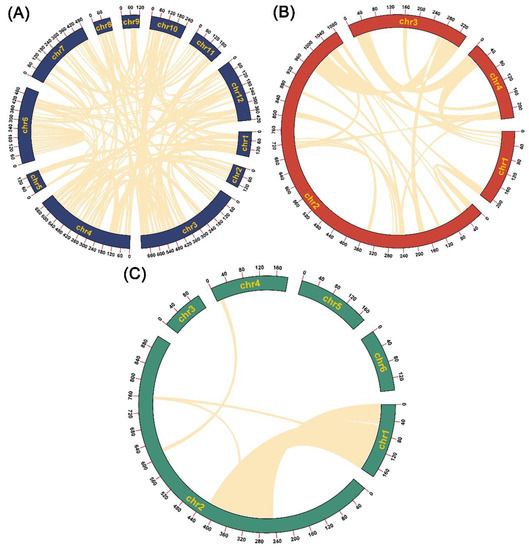
Figure 3.
The mitogenomic-segment-duplication distributions of the 3 Cistanche species. (A) Cistanche tubulosa. (B) Cistanche deserticola. (C) Cistanche salsa. The outermost circle marks the position of the mitogenome. The bright yellow arcs indicate segment duplication.
2.5. Phylogenetic Analysis by Mitogenome Sequences
The phylogeny was reconstructed using shared mitochondrial PCGs from 11 Lamiales mitogenomes using the maximum-likelihood (ML) method. The sister genus of Cistanche was Castilleja, with a bootstrap score (BS) of 100 (Figure 4). These two genera belong to the Orobanchaceae family. The species of Cistanche were distributed in two main clades. The first clade (BS: 100) was formed by C. deserticola and C. salsa, with the same mitogenome size. The second clade contained C. tubulosa with a BS of 100. These two clades were subsequently clustered together (BS: 100) (Figure 4). The bootstrap scores were high for all the branches, indicating the high degree of reliability of the phylogenetic tree. Moreover, the phylogenetic relationship of the Cistanche species constructed using the mitogenomes was congruent with that using the plastid genome, as shown in our previous studies.

Figure 4.
Phylogenetic relationships of Cistanche species with 8 other Lamiales species. The tree was constructed based on the amino-acid sequences of 49 mitochondrial protein-coding genes, including atp1, atp4, atp6, atp8, atp9, atpB, atpE, ccmB, ccmC, ccmFc, ccmFn, cob, cox1, cox2, cox3, cytB, ItrA, matR, mttB, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, petG, petL, rbcL, rpl2, rpl5, rpl10, rpl16, rpl23, rpl36, rps1, rps3, rps4, rps7, rps10, rps11, rps12, rps13, rps14, rps19, sdh3 and sdh4. Species in red box were three Cistanche species.
2.6. The Substitution Rate of Mitochondrial PCGs
The shared mitochondrial PCGs were used to estimate the nucleotide substitution rate of the mitochondrial PCGs in Lamiales. For each of the 28 PCGs, the pairwise Ka/Ks ratios were calculated. We found that the Ka/Ks rations of the four PCGs were over 1.0 in most of the species (Figure 5 and Table S20). These four PCGs were atp4, ccmB, ccmFc and matR, suggesting potential positive selection. However, most of the mitochondrial PCGs showed low Ka/Ks ratios, indicating possible purifying selection. In particular, the Ka/Ks ration of atp9, cox1, cox3 and nad4L showed a relatively low value (Figures S8–S31).
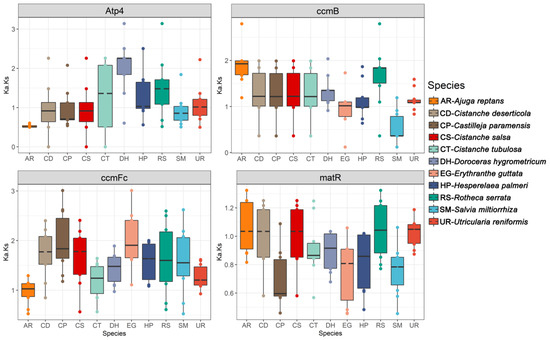
Figure 5.
Boxplots of pairwise Ka/Ks values among each retained mitochondrial protein-coding gene within the 11 Lamiales species.
3. Discussion
3.1. Genome Expansion in C. tubulosa
A substantial portion of plant mitogenomes may be made up of small repetitive sequences [26]. For example, low-complexity repetitive DNA made up 5–10% of the sequences in the Citrullus and Cucurbita mitogenomes [27]. Furthermore, similar ratios of low-complexity repetitive DNA sequences were observed in other plants [9,28]. In our study, C. tubulosa, C. salsa and C. deserticola had repeat sequences that were 174,618 bp, 32,375 bp and 149,945 bp long, accounting for 4.4%, 1.89% and 8.06% of the mitogenome sizes, respectively. These results were consistent with the results of previous studies, which showed that repeat sequences can cause changes in mitogenome size [9,26,27,28].
C. deserticola possessed the largest proportion of repetition sequences among the three Cistanche mitogenomes in our study. However, the mitogenome size of C. derserticola was close to that of C. salsa, which means that other factors may also play important roles in mitogenome size in addition to small repetitive sequence. Segment duplication and multi-copy protein-coding genes were two other factors causing mitogenome expansion.
In the C. tubulosa mitogenome, the sequences of the duplicated segments were 2,142,511 bp in length, accounting for 53.9% of the whole mitogenome size. Similarly, Sloan reported that 4.6-megabyte repeats were identified in the mitogenome of Silene conica, accounting for 40.8% of the whole mitogenome [13]. Furthermore, multi-copy protein-coding genes might result in mitogenome expansion. In the C. tubulosa mitogenome, the proportions of duplicated core genes and variable genes were 58% and 35%, respectively. For typical large mitogenomes (Cucumis species), some protein-coding genes (rps19) are also presented twice in both Citrullus and Cucurbita [14]. In C. tubulosa, complex Ⅰ (nad4, nad4L, nad6 and nad7), complex Ⅳ (cox1 and cox2), complex Ⅴ (atp4, atp8 and atp9), Cytochromec biogenesis (ccmFc and ccmFn), Ribosomal protein small subunit (rps3, rps4 and rps14), Intron maturase (matR) and SecY independent transport (mttB) protein-coding gene all had multiple copies. We speculated that this was related to the fact that holoparasitic plants do not conduct photosynthesis. In addition, environmental stress might up-regulate the expression of some related genes. Previous studies suggested that the plant mitochondrial electron-transport chain could improve plant performance under stressful environmental conditions [29]. Unlike C. deserticola and C. salsa, C. tubulosa experience salt stress and cold stress rather than drought stress [30,31,32]. This might lead to the duplication of genes in C. tubulosa. In summary, small repeats, segment duplication, and multi-copy genes were the main causes of the mitogenome expansion.
3.2. The Presence of MTPTs
In angiosperm plants, MTPTs are almost always present [33]. In our study, we found 158 C. deserticola, 128 C. salsa and 139 C. tubulosa MTPT sequences. They were 35,165 bp, 28,963 bp and 26,911 bp in length, accounting for 1.89%, 1.64% and 0.68% of the C. deserticola, C. salsa and C. tubulosa mitogenomes, respectively. Cheng et al. suggested that 26.87-kilobyte MTPT fragments were found in Suaeda glauca, accounting for 5.18% of the mitogenome [34]. In addition, the MTPTs discovered in Salix suchowensis account for 11.3% (17.5 kb) of the plastome and 2.8% (18.1 kb) of the mitogenome [35]. Interestingly, the proportion of MTPTs in the Cistanche mitogenomes was relatively low. In addition, our results showed that, in the MTPT sequences, the plastid PCGs and ribosome RNA were partial sequences. By contrast, the plastid tRNA genes had complete sequences in the MTPT fragments. The partial loss of plastid PCGs and ribosomal RNAs suggest that they might no longer function in the MTPT sequences. This supports the theory that fragments of DNA from plastomes usually become nonfunctional pseudogenes, while some tRNA genes still perform normal functions [36].
4. Materials and Methods
4.1. Sampling, DNA Extraction, and Genome Sequencing
Fresh samples of C. deserticola, C. salsa and C. tubulosa were collected from the Alxa League (Inner Mongolia Autonomous Region), Tacheng City (Xinjiang Uygur Autonomous Region), Hotan Prefecture (Xinjiang Uygur Autonomous Region), China (Table S1). The samples were identified by Professor Yulin Lin and stored at the Herbarium of the Chinese Academy of Medical Science and Peking Union Medicinal College (under specimens registry numbers CMPB13484, CMPB13485 and CMPB13487). Total DNA extraction was carried out using a plant genomic DNA extraction kit (Tiangen Biotech, Beijing, China). Utilizing NEBNext® library building kit [37], a DNA library with an insert size of 400 bp was constructed. Subsequently, Illumina HiSeq4000 sequencing platform was used for sequencing. The sequencing produced 5.44, 5.16 and 4.34 G of raw data, respectively (Table S1). Trimmomatic was used to filter the raw data to obtain the clean data [38]. In total, 4.88 G, 4.62 G and 3.92 G clean data were obtained, respectively. The plant samples were also used for Oxford Nanopore sequencing. Library construction, quality detection and sequencing were conducted following the manufacturer’s standard protocol. Consequently, 78.96 G, 39.19 G and 59.52 G of raw data were obtained, and 48 G, 31.96 G and 54.56 G remained after filtering and qualification (Table S1).
4.2. Mitogenome Assembly and Annotation
Eight Lamiales mitogenomes were downloaded as references from NCBI (Table S2). We initially enriched the mitogenome-related clean reads from Oxford Nanopore data using USEARCH [39]. The filtered Nanopore reads were assembled into contigs using Nextdenovo v2.4.0 (available online: https://github.com/Nextomics/NextDenovo (20 December 2020)) with the default parameters. The obtained contigs were then used as references, named Cistanche Structure Contigs. Illumina paired-end reads were mapped back to the Cistanche Structure Contigs using Minimap2 [40] and SAMtools [41]. We extracted the filtered Illumina paired-end reads and assembled them into contigs using SPAdes v. 3.10.1 [42]. By comparing the assembly of short-reads and long-reads using Minimap2 [40], we preliminarily determined which contig was the putative mitochondrial molecule. The assembly contigs obtained above were corrected with the Illumina paired-end reads using NextPolish1.3.1 [43]. Draft mitochondrial contigs were processed further following the steps below. Firstly, we compared the sequences with those in GenBank using BLASTn program to determine whether they were mitochondrial reads [44]. Second, we annotated the mitogenomes using MITOFY to determine whether the sequences contained mitochondrial genes [14].
4.3. Identification of Mitochondrial Plastid DNAs (MTPTs)
Cistanche mitogenomes were compared with C. deserticola (MN614127), C. slsa (MN614128) and C. tubulosa (MN614129) plastomes to identify MTPTs using BLASTn [44]. The BLASTn parameters was selected based on those reported previously [45]. Furthermore, TBtools was used to visualize the BLASTn results [46]. With the aid of Cistanche plastomes as a reference, the identified transferred DNA segments were annotated using BLASTn [44].
4.4. Analysis of Simple Sequence Repeats (SSRs), Tandem Repeats, Interspersed Repeats, and Segment Duplication
Online website MISA (Available online: http://webblast.ipk-gatersleben.de/misa/ (15 January 2021)) was used to identify the SSRs in mitochondrial genome. These SSRs included mono-, di-, tri-, tetra-, penta-, and hexanucleotides with minimum numbers of 10, 6, 5, 5, 5 and 5, respectively. With the default parameters, Tandem Repeat Finder [47] was used to identify tandem repeats. In addition, REPuter was used to identify forward, reverse, palindromic and complementary repeat sequences [48]. The minimum repeat size was set to 30 bp and the identity of the repeat units was ≥90%. Segment duplications were identified by comparing the mitochondrial genome to itself using BLASTN with the parameter setting e-value = 1 × 10−5. All alignments with length > 5000 bp and score > 90% were considered segment duplication for calculations of segment-duplication number. TBtools was used to visualize the BLASTn results [46].
4.5. Phylogenetic Analyses and Estimation of Nucleotide-Substitution Rates
For phylogenetic analyses, the DNA sequences of shared mitochondrial genome PCGs from 11 Lamiales species, including the Cistanche species in this work (Table S2), were used in the construction. The mitogenomes of other 8 Lamiales species were downloaded from GenBank Organelle Genome Resource database. PhyloSuite (v1.2.1) was used to extract shared mitochondrial PCGs from Lamiales species [49]. MAFFT (v7.450) was used to align the corresponding amino-acid sequences [50]. The aligned amino-acid sequences were concatenated and used to construct the phylogenetic trees through the maximum-likelihood (ML) method, using Solanum lycopersicum (MF034193) and Nicotiana tabacum (NC_006581.1) as outgroups. The bootstrap analysis was performed with 1000 replicates. We used the yn00 program in PAML v 4.9 [51] to calculate the nonsynonymous substitution rate (dN) and synonymous substitution rate (dS) for PCGs with the F3 × 4 codon model.
5. Conclusions
In conclusion, this study provides a first insight into the structural diversity and complexity of Cistanche mitogenomes. Our results answered the three scientific questions that were posed in the introduction. First, the complete mitogenomes of C. deserticola, C. salsa and C. tubulosa were successfully assembled, which was a significant accomplishment in the study of Cistanche mitogenomes. Second, the C. tubulosa was close to 4 Mb in size, indicating a significant expansion. Furthermore, the C. tubulosa mitogenomes differed significantly from those of the C. deserticola and C. salsa in terms of numbers of duplicated PCGs and segment duplication. Three Cistanche species were formed into one clade, close to the species of Orobanchaceae. Additionally, the topology of the Lamiales in the present study was highly similar to that in the APG IV system. Third, MTPT sequences were identified in three Cistanche species mitogenomes, with partial PCGs and ribosome RNA fragments, and complete tRNA from the plastomes. The results of this study therefore revealed many fascinating aspects of mitogenome diversity and complexity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13101791/s1. Figure S1: Mitochondrial genome map of Cistanche deserticola. The outside circle shows the GC content. The inside circle represents the protein coding gene, tRNA and rRNA; Figure S2: Mitochondrial genome map of Cistanche salsa. The outside circle shows the GC content. The inside circle represents the protein coding gene, tRNA and rRNA; Figure S3: Mitochondrial genome map of Cistanche tubulosa. The outside circle shows the GC content. The inside circle represents the protein coding gene, tRNA and rRNA; Figure S4: Genome size and protein-coding gene content of 9 parasitic plants mitochondrial genomes; Figure S5: The collinearity analysis among C. deserticola mitochondrial genome and plastid genome; Figure S6: The collinearity analysis among C. salsa mitochondrial genome and plastid genome; Figure S7: The collinearity analysis among C. tubulosa mitochondrial genome and plastid genome; Figure S8: Boxplots of pairwise Ka/Ks values in Atp1 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S9: Boxplots of pairwise Ka/Ks values in Atp6 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S10: Boxplots of pairwise Ka/Ks values in Atp8 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S11: Boxplots of pairwise Ka/Ks values in Atp9 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S12: Boxplots of pairwise Ka/Ks values in ccmC gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S13: Boxplots of pairwise Ka/Ks values in ccmFn gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S14: Boxplots of pairwise Ka/Ks values in cob gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S15: Boxplots of pairwise Ka/Ks values in cox1 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S16: Boxplots of pairwise Ka/Ks values in cox2 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S17: Boxplots of pairwise Ka/Ks values in cox3 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S18: Boxplots of pairwise Ka/Ks values in mttB gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S19: Boxplots of pairwise Ka/Ks values in nad1 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S20: Boxplots of pairwise Ka/Ks values in nad2 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S21: Boxplots of pairwise Ka/Ks values in nad3 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S22: Boxplots of pairwise Ka/Ks values in nad4 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S23: Boxplots of pairwise Ka/Ks values in nad4L gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S24: Boxplots of pairwise Ka/Ks values in nad5 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S25: Boxplots of pairwise Ka/Ks values in nad6 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S26: Boxplots of pairwise Ka/Ks values in nad7 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S27: Boxplots of pairwise Ka/Ks values in rpl5 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S 28: Boxplots of pairwise Ka/Ks values in rpl10 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S29: Boxplots of pairwise Ka/Ks values in rps12 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S30: Boxplots of pairwise Ka/Ks values in rps13 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Figure S31: Boxplots of pairwise Ka/Ks values in sdh4 gene within 11 Lamiales species. AR—Ajuga reptans; CD—Cistanche deserticola; CP—Castilleja paramensis; CS—Cistanche salsa; CT—Cistanche tubulosa; DH—Doroceras hygrometricum; EG—Erythranthe guttata; HP—Hesperelaea palmeri; RS—Rotheca serrate; SM—Salvai miltiorrhiza; UR—Utricularia reniformis; Table S1: Sampling and sequencing data information; Table S2: Mitogenome information of 8 Lamiales species and 2 outgroups; Table S3: Mitochondrial genome information of Chenopodiaceae species used in HGT events prediction; Table S4: Comparison of mitochondrial protein coding genes copy with 11 Lamiales mitogenomes; Table S5: Mitogenome information of parasitic plants; Table S6: Comparison of mitochondrial protein coding genes copy with 9 parasitic plant mitogenomes; Table S7: List of potential MTPTs of C. deserticola; Table S8: List of potential MTPTs of C. salsa; Table S9: List of potential MTPTs of C. tubulosa; Table S10 Types and numbers of SSRs in the Cistanche mitogenome; Table S11: Interspersed repeat sequences identified in C. deserticola mitogenome; Table S12: Interspersed repeat sequences identified in C. salsa mitogenome; Table S13: Interspersed repeat sequences identified in C. tubulosa mitogenome; Table S14: Tandem repeat sequences identified in the mitogenome of C. deserticola;Table S15: Tandem repeat sequences identified in the mitogenome of C. salsa; Table S16: Tandem repeat sequences identified in the mitogenome of C. tubulosa; Table S17: Segment duplication sequences in C. deserticola mitogenome; Table S18: Segment duplication sequences in C. salsa mitogenome; Table S19: Segment duplication sequences in C. tubulosa mitogenome; Table S20: Pairwise Ka/Ks ratios in different mitochondrial genes of 11 Lamiales plants.
Author Contributions
L.H., C.L. and H.C. conceived the project; W.X. performed the experiments; Y.M. and H.C. analyzed data; all authors discussed the results; Y.M. and H.C. wrote the original draft and prepared the figures. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Science Foundation of China Funds (no. 81872966), National Natural Science Foundation of China (no. 82073960), the Open Fund of State Key Laboratory of Southwestern Chinese Medicine Resources (no. SCMR20210), National Science and Technology Fundamental Resources Investigation Program of China (2018FY100701) and CAMS Innovation Fund for Medical Sciences (2021-12M-1-022, 2021-I2M-1-071).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The assembled chloroplast genomes of C. deserticola, C. salsa and C. tubulosa were deposited in GenBank with the accession numbers ON890398–ON890419.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Ataei, N.; Schneeweiss, G.M.; Garcia, M.A.; Krug, M.; Lehnert, M.; Valizadeh, J.; Quandt, D. A multilocus phylogeny of the non-photosynthetic parasitic plant Cistanche (Orobanchaceae) refutes current taxonomy and identifies four major morphologically distinct clades. Mol. Phylogenet. Evol. 2020, 151, 106898. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.S.; Park, S. Complete plastid and mitochondrial genomes of Aeginetia indica reveal intracellular gene transfer (IGT), horizontal gene transfer (HGT), and cytoplasmic male sterility (CMS). Int. J. Mol. Sci. 2021, 22, 6143. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, T.C.; Qin, Q.; Ren, Z.; Zhao, J.; Takahiro, Y.; Masami, H.; Crabbe, M.; Li, J.; Yang, Z. Complete chloroplast genome sequence of holoparasite Cistanche desertiscola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS ONE 2013, 8, e58747. [Google Scholar] [CrossRef]
- Li, X.; Feng, T.; Randle, C.; Schneeweiss, G.M. Phylogenetic Relationships in Orobanchaceae Inferred From Low-Copy Nuclear Genes: Consolidation of Major Clades and Identification of a Novel Position of the Non-photosynthetic Orobanche Clade Sister to All other Parasitic Orobanchaceae. Front. Plant Sci. 2019, 10, 902. [Google Scholar] [CrossRef] [PubMed]
- Gruzdev, E.V.; Kadnikov, V.V.; Beletsky, A.V.; Mardanov, A.V.; Ravin, N.V. Extensive plastome reduction and loss of photosynthesis genes in Diphelypaea coccinea, a holoparasitic plant of the family Orobanchaceae. PeerJ 2019, 7, e7830. [Google Scholar] [CrossRef]
- Wicke, S.; Müller, K.F.; de Pamphilis, C.W.; Quandt, D.; Wickett, N.J.; Zhang, Y.; Renner, S.S.; Schneeweiss, G.M. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 2013, 25, 3711–3725. [Google Scholar] [CrossRef] [PubMed]
- Frailey, D.C.; Chaluvadi, S.R.; Vaughn, J.N.; Coatney, C.G.; Bennetzen, J.L. Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant Biol. 2018, 18, 30. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Zhu, A.; Kozaczek, M.; Shah, N.; Pabón-Mora, N.; González, F.; Mower, J.P. Limited mitogenomic degradation in response to a parasitic lifestyle in Orobanchaceae. Sci. Rep. 2016, 6, 36285. [Google Scholar] [CrossRef] [PubMed]
- Skippington, E.; Barkman, T.J.; Rice, D.W.; Palmer, J.D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. USA 2015, 112, E3515–E3524. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Jin, X.H.; Liu, J.; Zhao, X.; Zhou, J.H.; Wang, X.; Wang, D.Y.; Lai, C.J.S.; Xu, W.; Huang, J.W.; et al. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 2018, 9, 1615–1626. [Google Scholar] [CrossRef]
- Shtolz, N.; Dan, M. The mitochondrial genome-on selective constraints and signatures at the organism, cell, and single mitochondrion levels. Front. Ecol. Evol. 2019, 7, 342. [Google Scholar] [CrossRef]
- Stechmann, A.; Hamblin, K.; Pérez-Brocal, V.; Gaston, D.; Richmond, G.; Giezen, M.; Clark, C.; Roger, A.J. Organelles in blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr. Biol. 2008, 18, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef]
- Alverson, A.J.; Wei, X.X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef]
- Kan, S.L.; Shen, T.T.; Gong, P.; Ran, J.H.; Wang, X.Q. The complete mitochondrial genome of Taxus cuspidata (Taxaceae): Eight protein-coding genes have transferred to the nuclear genome. BMC Evol. Biol. 2020, 20, 10. [Google Scholar] [CrossRef]
- Pinard, D.; Myburg, A.A.; Mizrachi, E. The plastid and mitochondrial genomes of Eucalyptus grandis. BMC Genom. 2019, 20, 132. [Google Scholar] [CrossRef]
- Roulet, M.E.; Garcia, L.E.; Gandini, C.L.; Sato, H.; Sanchez-Puerta, M.V. Multichromosomal structure and foreign tracts in the Ombrophytum subterraneum (Balanophoraceae) mitochondrial genome. Plant Mol. Biol. 2020, 103, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Sloan, D.B.; Alverson, A.J. Plant mitochondrial genome diversity: The genomics revolution. In Plant Genome Diversity; Wendel, J.F., Greilhuber, J., Dolezel, J., Leitch, I.J., Eds.; Springer: Vienna, Austria, 2012; Volume 1, pp. 123–144. [Google Scholar]
- Park, S.; Grewe, F.; Zhu, A.; Ruhlman, T.A.; Sabir, J.; Mower, J.P.; Jansen, R.K. Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. New Phytol. 2015, 208, 570–583. [Google Scholar] [CrossRef]
- Liu, F.; Fan, W.S.; Yang, J.B.; Xiang, C.L.; Mower, J.P.; Li, D.Z.; Zhu, A.D. Episodic and guanine-cytosine-biased bursts of intragenomic and interspecific synonymous divergence in Ajugoideae (Lamiaceae) mitogenomes. New Phytol. 2020, 228, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, X.; Xie, W. Cistanche deserticola Y. C. Ma, “Desert Ginseng”: A Review. Am. J. Chin. Med. 2012, 40, 1123–1141. [Google Scholar] [CrossRef] [PubMed]
- Bougandoura, A.; D'Abrosca, B.; Ameddah, S.; Scognamiglio, M.; Mekkiou, R.; Fiorentino, A.; Benayache, S.; Benayache, F. Chemical constituents and in vitro anti-inflammatory activity of Cistanche violacea Desf. (Orobanchaceae) extract. Fitoterapia 2016, 109, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Miao, M.; Bai, M.; Wei, Z. Phenylethanoid glycosides of Cistanche on menopausal syndrome model in mice. Saudi Pharm. J. 2017, 25, 537–547. [Google Scholar] [CrossRef]
- Wang, D.; Wang, H.; Li, G. The antidepressant and cognitive improvement activities of the traditional Chinese herb Cistanche. Evid. Based Complement. Altern. Med. 2017, 2017, 3925903. [Google Scholar] [CrossRef]
- Xu, W.Q.; Chen, H.M.; Tian, L.X.; Jiang, M.; Yang, Q.Q.; Wang, L.Q.; Ahmad, B.; Huang, L.F. Extensive gene loss in the plastome of holoparasitic plant Cistanche tubulosa (Orobanchaceae). Mitochondrial DNA B Resour. 2020, 5, 2679–2681. [Google Scholar] [CrossRef]
- Andre, C.; Levy, A.; Walbot, V. Small repeated sequences and the structure of plant mitochondrial genomes. Trends Genet. 1992, 8, 128–132. [Google Scholar] [CrossRef]
- Ward, B.L.; Anderson, R.S.; Bendich, A.J. The mitochondrial genome is large and variable in a family of plants (cucurbitaceae). Cell 1981, 25, 793–803. [Google Scholar] [CrossRef]
- Guo, W.H.; Grewe, F.; Fan, W.S.; Young, G.J.; Knoop, V.; Palmer, J.D.; Mower, J.P. Ginkgo and welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Biol. Evol. 2016, 33, 1448–1460. [Google Scholar] [CrossRef]
- Moller, I. Plant mitochondrial and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Du, Z.; Pei, J.; Huang, L. Chemical Diversity and Prediction of Potential Cultivation Areas of Cistanche Herbs. Sci. Rep. 2019, 9, 19737. [Google Scholar] [CrossRef]
- Song, Y.; Zeng, K.; Jiang, Y.; Tu, P. Cistanches Herba, from an endangered species to a big brand of Chinese medicine. Med. Res. Rev. 2021, 41, 1539–1577. [Google Scholar] [CrossRef] [PubMed]
- Thorogood, C.J.; Leon, C.J.; Lei, D.; Aldughayman, M.; Hawkins, J.A. Desert hyacinths: An obscure solution to a global problem? Plants People Planet 2021, 3, 302–307. [Google Scholar] [CrossRef]
- Wang, X.-C.; Chen, H.; Yang, D.; Liu, C. Diversity of mitochondrial plastid DNAs (MTPTs) in seed plants. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2017, 29, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; He, X.; Priyadarshani, S.; Wang, Y.; Qin, Y. Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca. BMC Genom. 2020, 22, 167. [Google Scholar] [CrossRef]
- Ye, N.; Wang, X.; Li, J.; Bi, C.; Xu, Y.; Wu, D.; Ye, Q. Assembly and comparative analysis of complete mitochondrial genome sequence of an economic plant Salix suchowensis. PeerJ 2017, 5, e3148. [Google Scholar] [CrossRef]
- Kitazaki, K.; Kubo, T.; Kagami, H.; Matsumoto, T.; Fujita, A.; Matsuhira, H.; Matsunaga, M.; Mikami, T. A horizontally transferred tRNACys gene in the sugar beet mitochondrial genome: Evidence that the gene is present in diverse angiosperms and its transcript is aminoacylated. Plant J. 2011, 68, 267–272. [Google Scholar] [CrossRef]
- Emerman, A.B.; Bowman, S.K.; Barry, A.; Henig, N.; Patel, K.M.; Gardner, A.F.; Hendrickson, C.L. NEBNext Direct: A novel, rapid, hybridization-based approach for the capture and library conversion of genomic regions of interest. Curr. Protoc. Mol. Biol. 2017, 119, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Marc, L.; Bjoern, U. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data, P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fan, J.; Sun, Z.; Liu, S. NextPolish: A fast and efficient genome polishing tool for long-read assembly. Bioinformatics 2020, 36, 2253–2255. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, Y.; Shan, Y.; Pei, X.; Yong, S.; Liu, C.; Yu, J. Assembly of the complete mitochondrial genome of an endemic plant, Scutellaria tsinyunensis, revealed the existence of two conformations generated by a repeat-mediated recombination. Planta 2021, 254, 36. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- John, R.; Li, S.; Mar, A.K.; Standley, D.M.; Kazutaka, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).