Genome-Wide Survey and Analysis of Microsatellites in Waterlily, and Potential for Polymorphic Marker Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source of Genomic Sequences
2.2. Identification of Microsatellite and Primer Design
2.3. SSR Location, GO and KEEG Enrichment
2.4. Plant Materials and DNA Extraction
2.5. Validation of SSR Primer Pairs by PCR Amplification
2.6. Data Analysis
3. Results
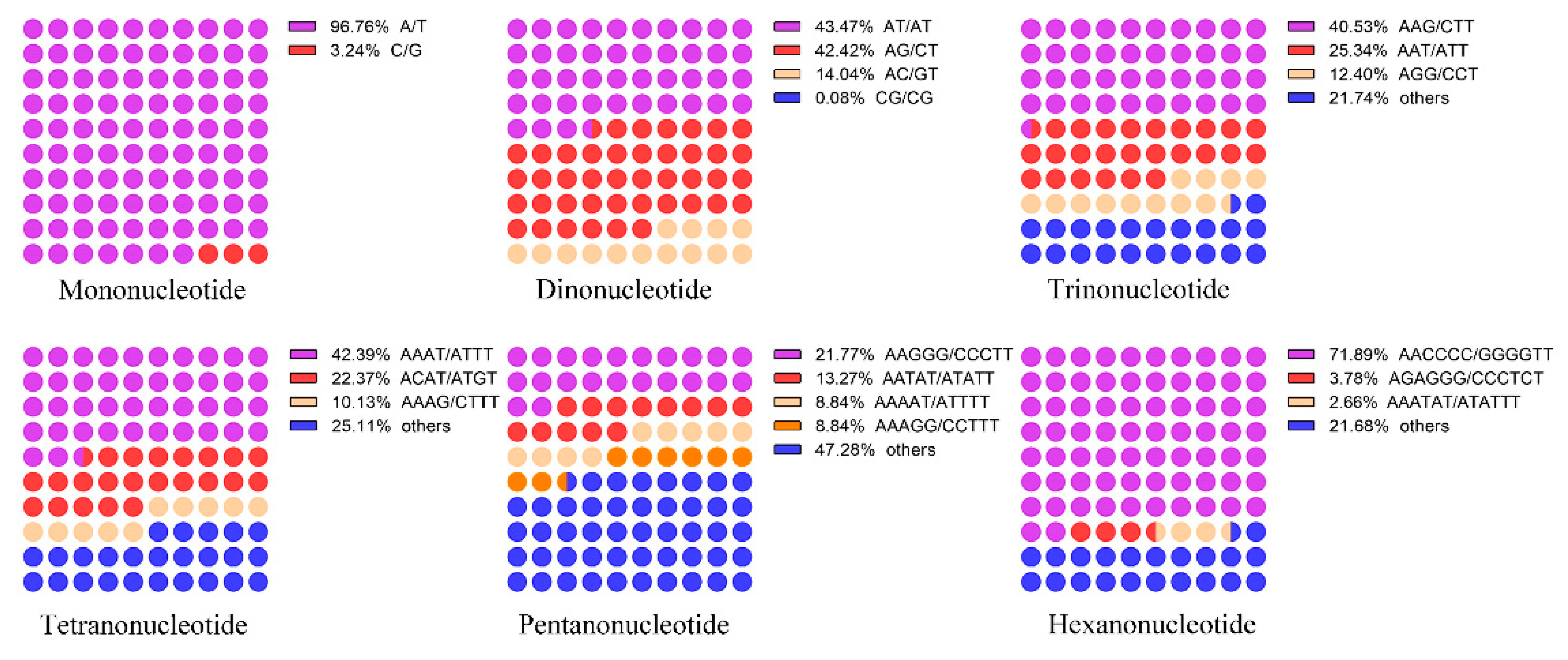
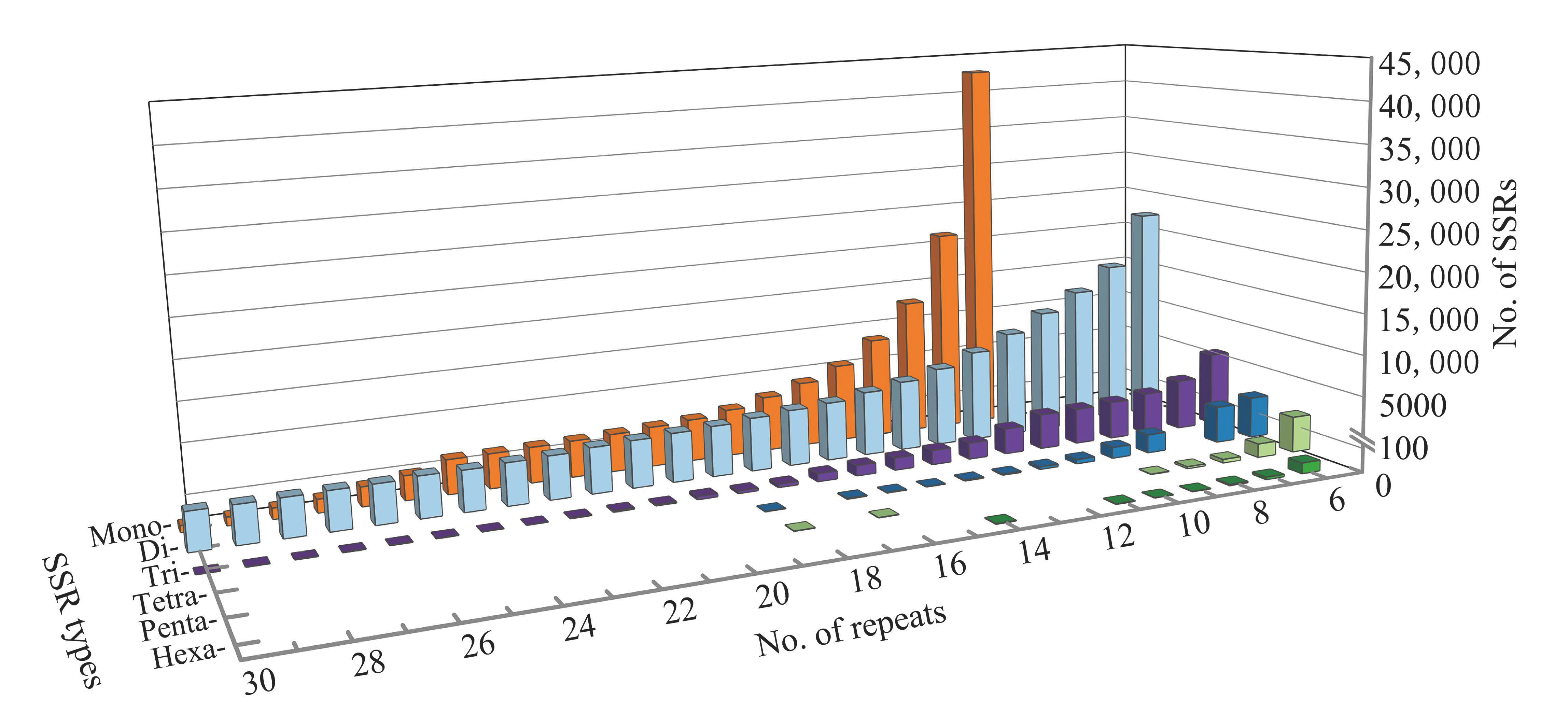
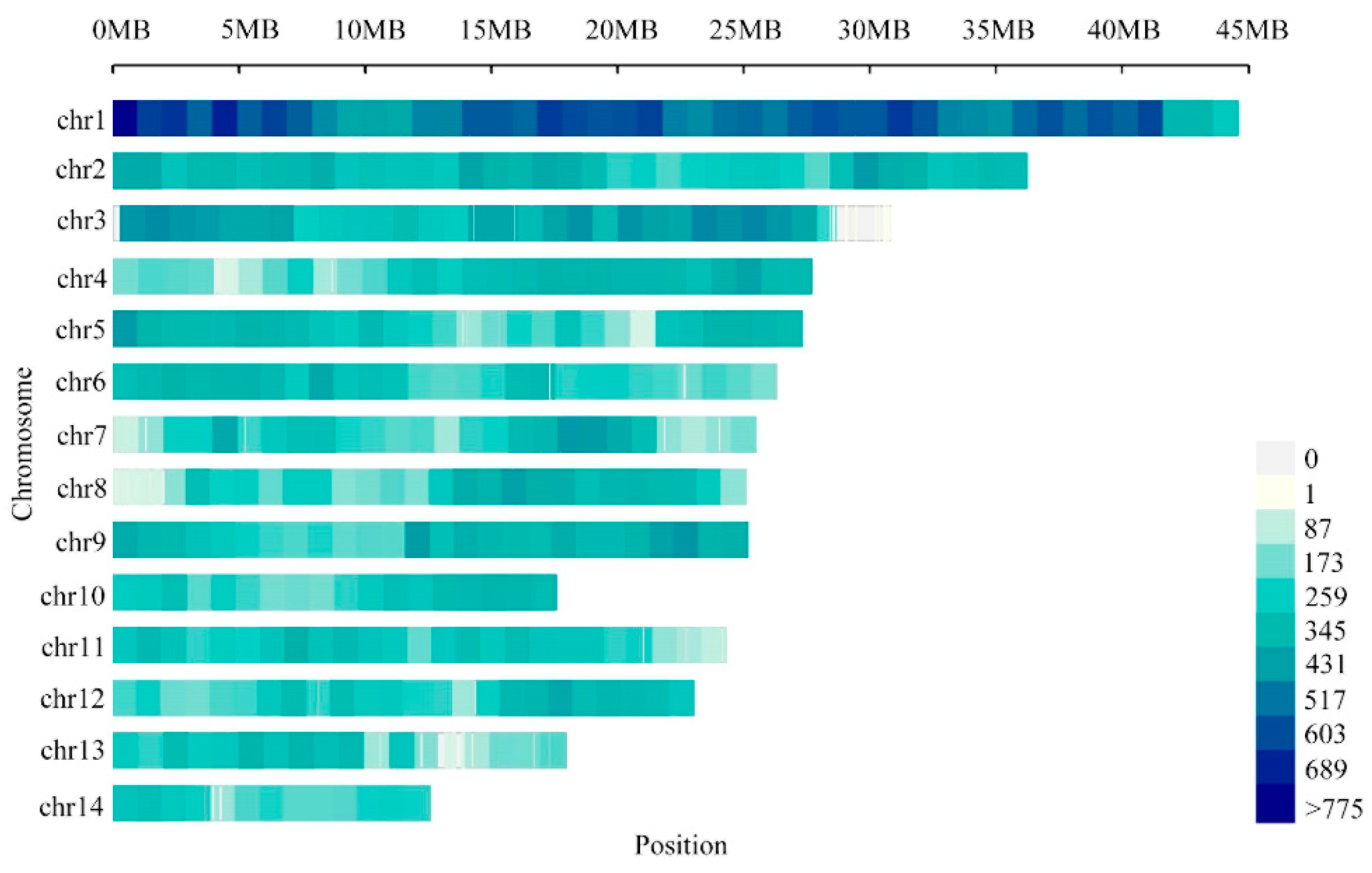
3.1. SSR Motifs Content in the N. colorata Genome
3.2. Characterization of SSR Motifs in the N. colorata Genome
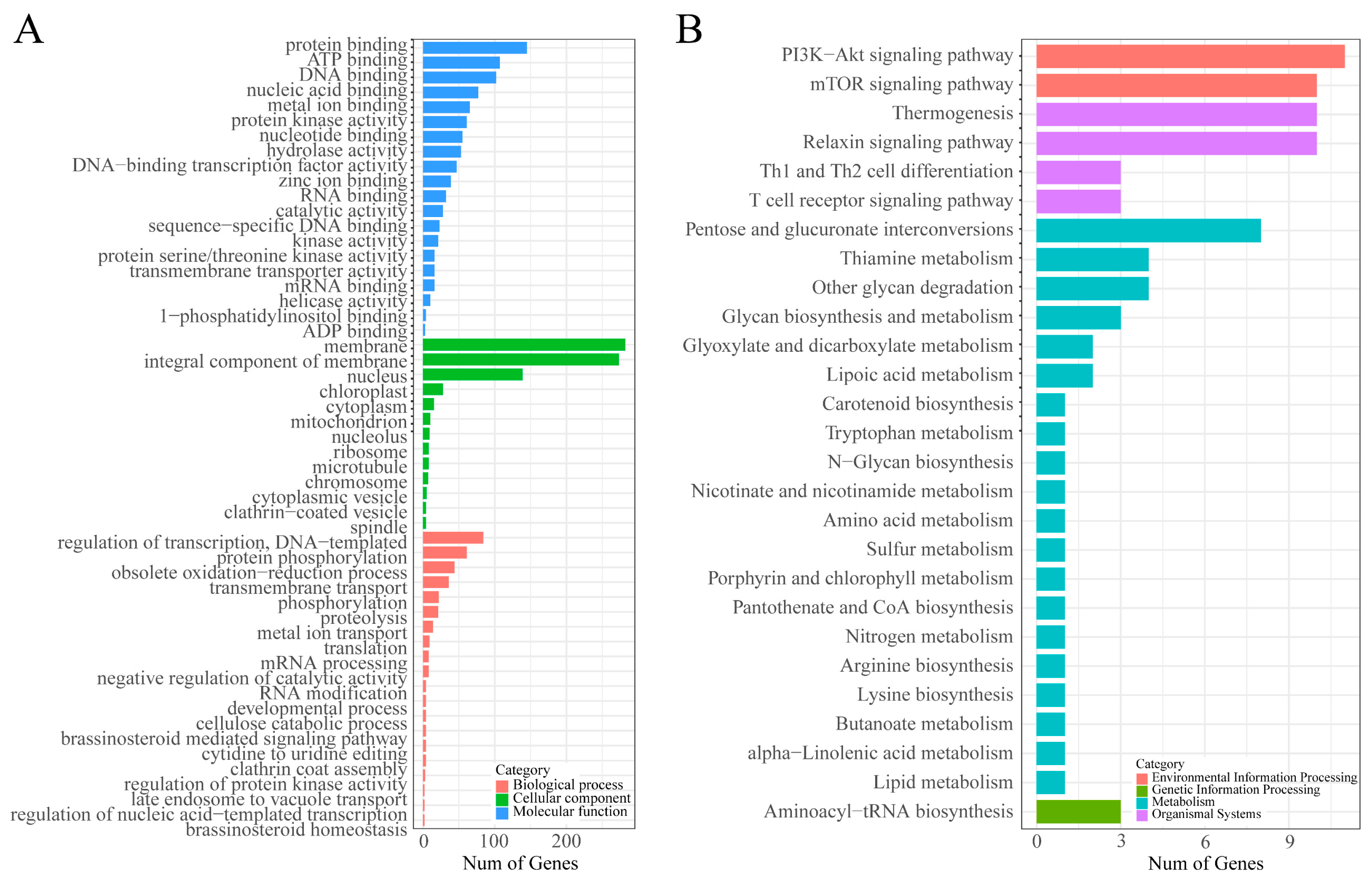
3.3. GO and KEGG Enrichment Analysis of CDSs Containing Microsatellite in the N. colorata Genome
3.4. Genome-Wide SSR Marker Development
3.5. Validation Analysis of SSR Primers
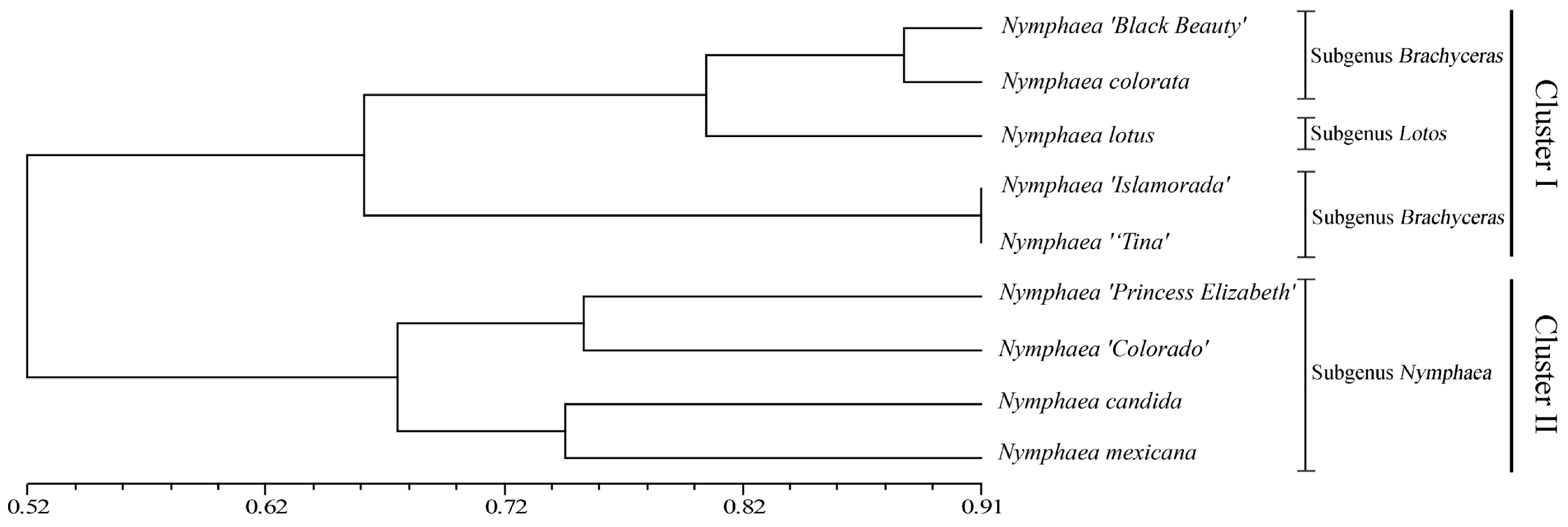
3.6. Cluster Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slocum, P.D. Waterlilies and Lotuses: Species, Cultivars, and New Hybrids; Timber Press: Portland, OR, USA, 2005; pp. 1–7. ISBN 9780881926842. [Google Scholar]
- Yoo, M.J.; Bell, C.D.; Soltis, P.S.; Soltis, D.E. Divergence times and historical biogeography of Nymphaeales. Syst. Bot. 2005, 30, 693–704. [Google Scholar] [CrossRef]
- Huang, G.Z.; Deng, H.Q.; Li, Z.X.; Li, G. The Waterlilies; Chinese Forestry Publishing House: Beijing, China, 2008; pp. 1–15. ISBN 9787503853326. [Google Scholar]
- Tang, H.B.; Zhang, L.S.; Chen, F.; Zhang, X.T.; Chen, F.; Ma, H.; Peer, Y.V. Nymphaea colorata (Blue-Petal Water Lily). Trends Genet. 2020, 36, 718–719. [Google Scholar] [CrossRef] [PubMed]
- Koga, K.; Kadono, K.; Setoguchi, H. The genetic structure of populations of the vulnerable aquatic macrophyte Ranunculus nipponicus (Ranunculaceae). J. Plant Res. 2007, 120, 167–174. [Google Scholar] [CrossRef]
- Hu, J.H.; Pan, L.; Liu, H.G.; Wang, S.Z.; Wu, Z.H.; Ke, W.D.; Ding, Y. Comparative analysis of genetic diversity in sacred lotus (Nelumbo nucifera Gaertn.) using AFLP and SSR markers. Mol. Biol. Rep. 2012, 39, 3637–3647. [Google Scholar] [CrossRef]
- Kumar, H.; Priya, P.; Singh, N.; Kumar, M.; Choudhary, B.K.; Kumar, L.; Singh, I.S.; Kumar, N. RAPD and ISSR marker-based comparative evaluation of genetic diversity among Indian germplasms of Euryale ferox: An aquatic food plant. Appl. Biochem. Biotechnol. 2016, 180, 1345–1360. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, J.; Jiang, M.; Wang, W.; Jiang, J.; Li, Y.; Yang, L.; Zhou, X. Development of genome-wide SSR markers in rapeseed by next generation sequencing. Gene 2021, 798, 145798. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.G.; Singh, N.V.; Parashuram, S.; Bohra, A.; Mundewadikar, D.M.; Sangnure, V.R.; Babu, K.D.; Sharma, J. Genome wide identification, characterization and validation of novel miRNA-based SSR markers in pomegranate (Punica granatum L.). Physiol. Mol. Biol. Plants 2020, 26, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xing, M.; Song, L.; Yan, J.; Lu, W.; Zeng, A. Genome-wide analysis of simple sequence repeats in Cabbage (Brassica oleracea L.). Front. Plant. Sci. 2021, 12, 726084. [Google Scholar] [CrossRef]
- Hou, S.; Ren, X.; Yang, Y.; Wang, D.; Du, W.; Wang, X.; Li, H.; Han, Y.; Liu, L.; Sun, Z. Genome-wide development of polymorphic microsatellite markers and association analysis of major agronomic traits in core germplasm resources of Tartary Buckwheat. Front. Plant Sci. 2022, 13, 819008. [Google Scholar] [CrossRef]
- Martina, M.; Acquadro, A.; Barchi, L.; Gulino, D.; Brusco, F.; Rabaglio, M.; Portis, F.; Portis, E.; Lanteri, S. Genome-wide survey and development of the first microsatellite markers database (AnCorDB) in Anemone coronaria L. Int. J. Mol. Sci. 2022, 23, 3126. [Google Scholar] [CrossRef]
- Chaveerach, A.; Tanee, T.; Sudmoon, R. Molecular identification and barcodes for the genus Nymphaea. Acta Biol. Hung. 2011, 62, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Jeremy, D.; Suman, K.; Sayawada, R.R.; Pramod, T. Sequence characteristics and phylogenetic implications of the nrDNA internal transcribed spacers (ITS) in the genus Nymphaea with focus on some Indian representatives. Plant Syst. Evol. 2012, 298, 93–108. [Google Scholar] [CrossRef]
- Parveen, S.; Singh, N.; Adit, A.; Kumaria, S.; Tandon, R.; Agarwal, M.; Jagannath, A.; Goel, S. Contrasting reproductive strategies of two Nymphaea species affect existing natural genetic diversity as assessed by microsatellite markers: Implications for conservation and wetlands restoration. Front. Plant Sci. 2022, 13, 773572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.S.; Chen, F.; Zhang, X.T.; Li, Z.; Tang, H.B. The water lily genome and the early evolution of flowering plants. Nature 2020, 577, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Zuo, L.H.; Zhang, S.; Zhang, J.; Liu, Y.; Yu, X.; Yang, M.; Wang, J. Primer development and functional classification of EST-SSR markers in Ulmus species. Tree Genet. Genomes 2020, 16, 74–87. [Google Scholar] [CrossRef]
- Li, J.M.; Li, S.Q.; Kong, L.J.; Wang, L.H.; Wei, A.Z.; Liu, Y.L. Genome survey of Zanthoxylum bungeanum and development of genomic-SSR markers in congeneric species. Biosci. Rep. 2020, 40, BSR20201101. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer 3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.J.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nuclei. Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schffer, A.A.; Zhang, J.; Zhang, Z.; Webb, M.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, G.; Chen, A.; Li, J.J.; Huang, S.Q.; Tang, H.J.; Chang, L.; Zhao, L.N.; Li, D.F. Genome-wide development of simple sequence repeats database for flax (Linum usitatissimum L.) and its use for genetic diversity assessment. Genet. Resour. Crop Evol. 2020, 67, 865–874. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T.B.J.; Yeh, Z.H.; Mao, J.X. POPGENE Version 1.31: Microsoft Window-Based Freeware for Population Genetic Analysis; University of Alberta: Edmonton, AB, USA, 1997. [Google Scholar]
- Han, B.; Wang, C.B.; Tang, Z.H.; Ren, Y.K.; Li, Y.; Zhang, D.Y.; Dong, Y.H.; Zhao, X.H. Genome-wide analysis of microsatellite markers based on sequenced database in Chinese spring wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0141540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dkhar, J.; Kumaria, S.; Rao, S.R.; Tandon, P. Molecular phylogenetics and taxonomic reassessment of four Indian representatives of the genus Nymphaea. Aquat. Bot. 2010, 93, 135–139. [Google Scholar] [CrossRef] [Green Version]
- Poczai, P.; Mátyás, K.K.; Szabó, I.; Varga, I.; Hyvönen, J.; Cernák, I.; Gorji, A.M.; Decsi, K.; Taller, J. Genetic variability of thermal Nymphaea (Nymphaeaceae) populations based on ISSR markers: Implications on relationships, hybridization, and conservation. Plant Mol. Biol. Rep. 2011, 29, 906–918. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.D.; Yang, W.R.; Zhang, Q.X.; Cheng, T.R.; Pan, H.T.; Xu, Z.D.; Zhang, J.; Chen, C.G. Genome-wide characterization and linkage mapping of simple sequence repeats in Mei (Prunus mume Sieb. et Zucc.). PLoS ONE 2013, 8, e59562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, P.G.; Singh, N.V.; Sharma, J.; Bohra, A.; Raghavendra, K.P.; Mane, R.; Mundewadikar, D.M.; Babu, K.D.; Sharma, J. Comprehensive characterization and validation of chromosome-specific highly polymorphic SSR markers from pomegranate (Punica granatum L.) cv. Tunisia Genome. Front. Plant Sci. 2021, 12, 337. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, B.; Hui, L.; Chang, Y.S.; Han, Y.Y.; Li, J.; Wei, G.C.; Zhao, S.; Khan, M.A.; Zhou, Y.; et al. Identification, characterization, and utilization of genome-wide simple sequence repeats to identify a QTL for acidity in apple. BMC Genom. 2012, 1, 537. [Google Scholar] [CrossRef] [Green Version]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef]
- Yang, M.; Han, Y.; VanBuren, R.; Ming, R.; Xu, L.; Han, Y.; Liu, Y. Genetic linkage maps for Asian and American lotus constructed using novel SSR markers derived from the genome of sequenced cultivar. BMC Genom. 2012, 13, 653. [Google Scholar] [CrossRef]
- Cavagnaro, P.F.; Senalik, D.A.; Yang, L.M.; Simon, P.W.; Harkins, T.T.; Kodira, C.D.; Huang, S.W.; Weng, Y.Q. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom. 2010, 11, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.; Hong, Y.B.; Li, S.X.; Liu, H.; Li, H.F.; Zhang, J.N.; Lan, H.F.; Liu, H.Y.; Liu, X.Y.; Wen, S.J.; et al. Genome-wide identification of microsatellite markers from cultivated peanut (Arachis hypogaea L.). BMC Genom. 2019, 20, 799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.Y.; Li, J.Y.; Qin, G.H. Genome-wide distribution of simple sequence repeats in pomegranate and their application to the analysis of genetic diversity. Tree Genet. Genomes 2020, 16, 36. [Google Scholar] [CrossRef]
- Li, S.F.; Zhang, G.J.; Li, X.; Wang, L.J.; Yuan, J.H.; Deng, C.L.; Gao, W.J. Genome-wide identification and validation of simple sequence repeats (SSRs) from Asparagus officinalis. Mol. Cell. Probes 2016, 30, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.M. The contribution of slippage-like processes to genome evolution. J. Mol. Evol. 1995, 41, 1038–1047. [Google Scholar] [CrossRef]
- Mokhtar, M.M.; Adawy, S.S.; El-Assal, S.D.; Hussein, E.H. Genic and intergenic SSR database generation, SNPs determination and pathway annotations, in date palm (Phoenix dactylifera L.). PLoS ONE 2016, 11, e0159268. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.T.; Zhang, Y.J.; Qiao, L.; Chen, B. Comparative analyses of simple sequence repeats (SSRs) in 23 mosquito species genomes: Identification, characterization and distribution (Diptera: Culicidae). Insect Sci. 2019, 26, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, S.; Wills, C.; Metzgar, D. Analysis of simple sequence repeats in mammalian cell cycle genes. Recent. Adv. DNA Gene Seq. 2014, 8, 20–29. [Google Scholar] [CrossRef]
- Liu, W.C.; Xu, Y.T.; Li, Z.K.; Fan, J.; Yang, Y. Genome-wide mining of microsatellites in king cobra (Ophiophagus hannah) and cross-species development of tetranucleotide SSR markers in Chinese cobra (Naja atra). Mol. Biol. Rep. 2019, 46, 6087–6098. [Google Scholar] [CrossRef]
- Stone, J. Plant protein kinase families and signal transduction. Plant Physiol. 1995, 108, 451–457. [Google Scholar] [CrossRef]
- Navari-Izzo, F.; Quartacci, M.F.; Sgherri, C. Lipoic acid: A unique antioxidant in the detoxification of activated oxygen species. Plant Physiol. Biochem. 2002, 40, 463–470. [Google Scholar] [CrossRef]
- Zheng, X.F.; You, Y.N.; Diao, Y.; Zheng, X.W.; Xie, K.Q.; Zhou, M.Q.; Hu, Z.L.; Wang, Y.W. Development and characterization of genic-SSR markers from different Asia lotus (Nelumbo nucifera) types by RNA-seq. Genet. Mol. Res. 2015, 14, 11171–11184. [Google Scholar] [CrossRef]
- Fu, Y.R.; Liu, F.L.; Li, S.; Tian, D.K.; Dong, L.; Chen, Y.C.; Su, Y. Genetic diversity of wild Asian lotus (Nelumbo nucifera) from northern China. Hortic. Plant J. 2021, 7, 13. [Google Scholar] [CrossRef]
- Nurainee, S.; Seiji, T.; Nakao, K.; Samak, K. Molecular phylogeny and postharvest morphology of petals in two major Nelumbo nucifera cultivars in Thailand. Agric. Nat. Resour. 2018, 52, 45–52. [Google Scholar] [CrossRef]
- Rai, M.K.; Phulwaria, M.; Shekhawat, N.S. Transferability of simple sequence repeat (SSR) markers developed in guava (Psidium guajava L.) to four Myrtaceae species. Mol. Biol. Rep. 2013, 40, 5067–5071. [Google Scholar] [CrossRef] [PubMed]
- Endo, C.; Yamamoto, N.; Kobayashi, M.; Nakamura, Y.; Yokoyama, K.; Kurusu, T.; Yano, K.; Tada, Y. Development of simple sequence repeat markers in the halophytic turf grass Sporobolus virginicus and transferable genotyping across multiple grass genera/species/genotypes. Euphytica 2017, 213, 56. [Google Scholar] [CrossRef]
- Tuler, A.C.; Carrijo, T.T.; Nóia, L.R.; Ferreira, A.; Peixoto, A.L.; da Silva Ferreira, M.F. SSR markers: A tool for species identification in Psidium (Myrtaceae). Mol. Biol. Rep. 2015, 42, 1501–1513. [Google Scholar] [CrossRef]

| Locus | Core Motif | Chr. | Primer Sequences (5′–3′) | Na | Ne | I | Ho | He | Fis | PIC |
|---|---|---|---|---|---|---|---|---|---|---|
| Ny-2.1 | (T)13 | 2 | F: AGAGCTGAGATTGGTTTGAAGC R: TCAGCGATTTCTTCTTGGGAT | 2.000 | 1.800 | 0.637 | 0.000 | 0.471 | 1.000 | 0.444 |
| Ny-2.2 | (CT)10 | 2 | F: TTTGTGGTGCGACTTCTTGC R: GTTAACAGCAGCCTTCACCG | 3.000 | 1.409 | 0.557 | 0.333 | 0.307 | −0.149 | 0.290 |
| Ny-3.2 | (A)12 | 3 | F: CTATGGACAACACATGCCGC R: CACGAGCAACAAGACCAGTA | 2.000 | 1.800 | 0.637 | 0.000 | 0.471 | 1.000 | 0.444 |
| Ny-4.2 | (GA)33 | 4 | F: CACGGCGAGGGGACAATATA R: CCGCCAATTCCACCATTCAT | 3.000 | 1.906 | 0.787 | 0.333 | 0.503 | 0.299 | 0.475 |
| Ny-5.1 | (A)13 | 5 | F: AAAGAACTATGCGGACCTGC R: CAAGCTCAGGACATGGTTCG | 2.000 | 2.000 | 0.693 | 0.111 | 0.529 | 0.778 | 0.500 |
| Ny-5.2 | (TC)20(TA)25 | 5 | F: CCGCAGTTAGTGTCACATGG R: CGCGTTCTCCTTTGCCAATA | 3.000 | 2.656 | 1.037 | 0.222 | 0.660 | 0.644 | 0.624 |
| Ny-6.2 | (T)10…(AT)10 | 6 | F: AATCAATGCTTCCATGGCCG R: TCATGTCCGGGATTCTAGGC | 2.000 | 1.976 | 0.687 | 0.000 | 0.523 | 1.000 | 0.494 |
| Ny-9.1 | (TA)17(GA)14 | 9 | F: ACCAAGGACTGCGAGTGTAT R: ATTTGAGTTGAGGGTTGCCG | 2.000 | 1.670 | 0.591 | 0.556 | 0.425 | −0.385 | 0.401 |
| Ny-10.1 | (AG)8… (AG)10 | 10 | F: CCCAGCATCGTAAATGACCG R: TTGGAGGAGGAGGAGATTGC | 3.000 | 2.418 | 0.981 | 0.667 | 0.621 | −0.137 | 0.586 |
| Ny-11.1 | (AG)6…(T)11 | 11 | F: AGCGTCACAACACTCCACTA R: AGGATTAGATGGGGCTCTGC | 2.000 | 1.906 | 0.668 | 0.333 | 0.503 | 0.299 | 0.475 |
| Ny-12.1 | (TC)10(TA)13 | 12 | F: AGGAGAAAACAGAGTGGGGC R: AGCATGCATGTATTCCCCAT | 2.000 | 1.800 | 0.637 | 0.222 | 0.471 | 0.500 | 0.444 |
| Ny-13.1 | (CT)6(CA)9 | 13 | F: CAGATGCAAGGATGGGAAGC R: GCAATGGGGATGATGAAGGC | 2.000 | 2.000 | 0.693 | 0.556 | 0.529 | −0.111 | 0.500 |
| Ny-13.2 | (TC)20(TA)12 | 13 | F: GCCTACCCATGTCCTCTGAT R: CCCTGTTCTGTTTGTGTTGC | 2.000 | 1.976 | 0.687 | 0.000 | 0.523 | 1.000 | 0.494 |
| Mean | - | - | - | 2.308 | 1.947 | 0.715 | 0.256 | 0.503 | 0.441 | 0.475 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Yang, M.; Guo, J.; Liu, J.; Chu, G.; Xu, Y. Genome-Wide Survey and Analysis of Microsatellites in Waterlily, and Potential for Polymorphic Marker Development. Genes 2022, 13, 1782. https://doi.org/10.3390/genes13101782
Huang X, Yang M, Guo J, Liu J, Chu G, Xu Y. Genome-Wide Survey and Analysis of Microsatellites in Waterlily, and Potential for Polymorphic Marker Development. Genes. 2022; 13(10):1782. https://doi.org/10.3390/genes13101782
Chicago/Turabian StyleHuang, Xiang, Meihua Yang, Jiaxing Guo, Jiachen Liu, Guangming Chu, and Yingchun Xu. 2022. "Genome-Wide Survey and Analysis of Microsatellites in Waterlily, and Potential for Polymorphic Marker Development" Genes 13, no. 10: 1782. https://doi.org/10.3390/genes13101782
APA StyleHuang, X., Yang, M., Guo, J., Liu, J., Chu, G., & Xu, Y. (2022). Genome-Wide Survey and Analysis of Microsatellites in Waterlily, and Potential for Polymorphic Marker Development. Genes, 13(10), 1782. https://doi.org/10.3390/genes13101782




