Characterisation of the Complete Mitochondrial Genome of Critically Endangered Mustela lutreola (Carnivora: Mustelidae) and Its Phylogenetic and Conservation Implications
Abstract
1. Introduction
2. Results
2.1. Mitochondrial Genome Structure, Organization and Composition
2.2. Repetitive and Palindromic Sequences
2.3. Protein-Coding Genes and Codon Usage
2.4. Transfer RNA Genes
2.5. Ribosomal RNA Genes
2.6. Non-Coding Regions
2.7. Mitochondrial DNA Sequence Heterogeneity
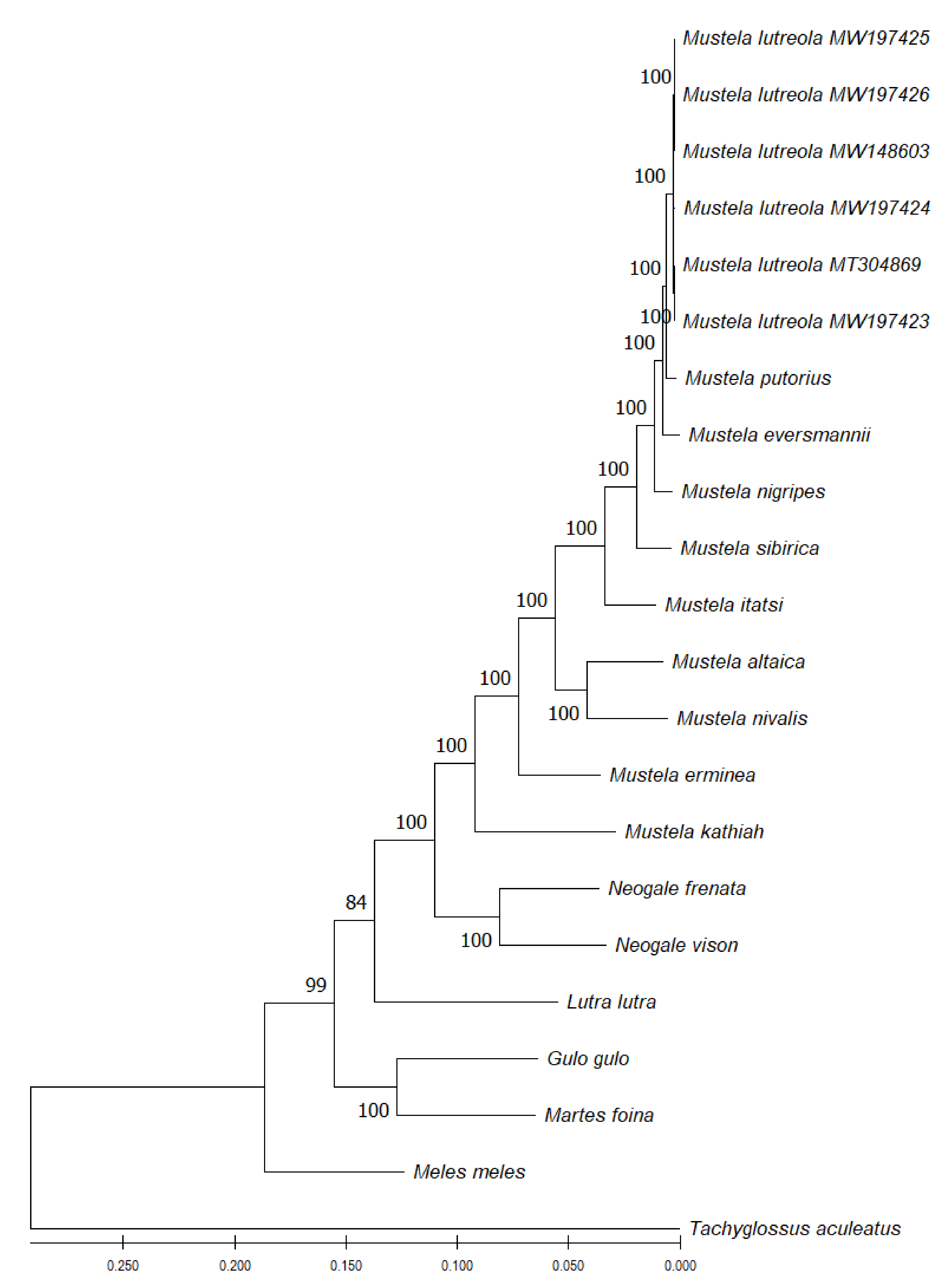
2.8. Interspecies Comparative and Phylogenetic Analyses of the European Mink Mitogenome
3. Discussion
3.1. General Features of the European Mink Mitochondrial Genome
3.2. mtDNA Sequence Heterogeneity
3.3. Phylogenetic Considerations
4. Materials and Methods
4.1. Sample Collection and Mitochondrial DNA Extraction
4.2. Mitochondrial Genome Sequencing and Assembly
4.3. Gene Annotation and Sequence Analysis
4.4. Analysis of Sequence Heterogeneity, Interspecies Comparison and Phylogenetic Inference
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Youngman, P.M. Mustela lutreola. Mamm. Species 1990, 362, 1–3. [Google Scholar] [CrossRef]
- Heptner, V.G.; Naumov, N.P.; Yurgenson, P.B.; Sludskii, A.A.; Chirkova, A.F.; Bannikov, A.G. Part 1b: Carnivora. Mammals of the Soviet Union, 1st ed.; Smithsonian Institution Libraries and The National Science Foundation Press: New Delhi, India, 2001; Volume 2, pp. 1078–1106. [Google Scholar]
- Sidorovich, V. Mustelids in Belarus: Evolutionary Ecology, Demography and Interspecific Relationships; Zolotoi Uley: Minsk, Belarus, 1997; pp. 1–263. [Google Scholar]
- Sidorovich, V.; MacDonald, D.W. Density dynamics and changes in habitat use by the European mink and other native mustelids in connection with the American mink expansion in Belarus. Neth. J. Zool. 2001, 51, 107–126. [Google Scholar] [CrossRef]
- Maran, T.; Skumatov, D.; Gomez, A.; Põdra, M.; Abramov, A.V.; Dinets, V. Mustela lutreola. The IUCN Red List of Threatened Species. 2016. E.T14018A45199861. Available online: https://www.iucnredlist.org/species/14018/45199861 (accessed on 1 February 2021).
- Maran, T. European mink: Setting of goal for conservation and Estonian case study. Galemys 2003, 15, 1–11. [Google Scholar]
- Harrington, L.A.; Põdra, M.; Gómez, A.; Maran, T. Raising awareness of the plight of the critically endangered European mink in Spain is not miscommunication: A response to Melero. Biodivers. Conserv. 2018, 27, 269–271. [Google Scholar] [CrossRef]
- Maran, T.; Põdra, M.; Harrington, L.A.; Macdonald, D.W. European mink: Restoration attempts for a species on the brink of extinction. In Biology and Conservation of Musteloids; Macdonald, D.W., Newman, C., Harrington, L.A., Eds.; Oxford University Press: Oxford, UK, 2017. [Google Scholar] [CrossRef]
- Maran, T.; Fienieg, E.; Schad, K. Long-Term Management Plan for European Mink (Mustela lutreola) European Endangered Species Programme (EEP). 2017. Available online: http://www.nouvelle-aquitaine.developpement-durable.gouv.fr/IMG/pdf/2017_european_mink_long_term_management_plan.pdf (accessed on 1 February 2021).
- Kiseleva, N.V. The Current State of the European Mink in Russia. Available online: https://lutreoladotorg.files.wordpress.com/2018/04/the-current-state-of-the-european-mink-in-russia-kiseleva-2017.pdf (accessed on 1 February 2021).
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, UK, 2010; pp. 1–644. [Google Scholar]
- Melbourne, B.A.; Hastings, A. Extinction risk depends strongly on factors contributing to stochasticity. Nature 2008, 454, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Fauvergue, X.; Vercken, E.; Malausa, T.; Hufbauer, R. The biology of small, introduced populations, with special reference to biological control. Evol. Appl. 2012, 5, 424–443. [Google Scholar] [CrossRef]
- Skorupski, J. Fifty Years of Research on European Mink Mustela lutreola L., 1761 Genetics: Where Are We Now in Studies on One of the Most Endangered Mammals? Genes 2020, 11, 1332. [Google Scholar] [CrossRef] [PubMed]
- Skorupski, J.; Panicz, R.; Śmietana, P.; Napora-Rutkowski, Ł.; Soroka, M.; Kolek, L.; Budniak, M.; Wąsowicz, B.; Keszka, S.; Kempf, M.; et al. Conservation Genetics in Poland—Theory and Practice, 1st ed.; University of Szczecin: Szczecin, Poland, 2017; Available online: https://www.researchgate.net/profile/Jakub-Skorupski-2/publication/320187990_Conservation_genetics_in_Poland_-_theory_and_practice/links/59d39980aca2721f436cd734/Conservation-genetics-in-Poland-theory-and-practice.pdf (accessed on 1 February 2021).
- Michaux, J.; Libois, R.; Davison, A.; Chevret, P.; Rosoux, R. Is the western population of the European mink, (Mustela lutreola), a distinct Management Unit for conservation? Biol. Conserv. 2004, 115, 357–367. [Google Scholar] [CrossRef]
- Michaux, J.R.; Hardy, O.J.; Justy, F.; Fournier, P.; Kranz, A.; Cabria, M.; Davison, A.; Rosoux, R.; Libois, R. Conservation genetics and population history of the threatened European mink Mustela lutreola, with an emphasis on the west European population. Mol. Ecol. 2005, 14, 2373–2388. [Google Scholar] [CrossRef]
- Witzenberger, K.A.; Hochkirch, A. Ex situ conservation genetics: A review of molecular studies on the genetic consequences of captive breeding programmes for endangered animal species. Biodivers. Conserv. 2011, 20, 1843–1861. [Google Scholar] [CrossRef]
- Attard, C.; Moller, L.; Sasaki, M.; Hammer, M.; Bice, C.; Brauer, C.; Carvalho, D.; Harris, J.; Beheregaray, L. A novel holistic framework for genetic-based captive-breeding and reintroduction programs. Conserv. Biol. 2016, 30, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Steiner, C.C.; Putnam, A.S.; Hoeck, P.E.; Ryder, O.A. Conservation Genomics of Threatened Animal Species. Annu. Rev. Anim. Biosci. 2013, 1, 261–281. [Google Scholar] [CrossRef]
- Themudo, G.E.; Rufino, A.C.; Campos, P.F. Complete mitochondrial DNA sequence of the endangered giant sable antelope (Hippotragus niger variani): Insights into conservation and taxonomy. Mol. Phylogenet. Evol. 2015, 83, 242–249. [Google Scholar] [CrossRef]
- Supple, M.A.; Shapiro, B. Conservation of biodiversity in the genomics era. Genome Biol. 2018, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resources Genbank. Available online: http://www.ncbi.nlm.nih.gov/genbank/ (accessed on 19 September 2021).
- Davison, A.; Griffiths, H.I.; Brookes, R.C.; Maran, T.; Macdonald, D.W.; Sidorovich, V.E.; Kitchener, A.C.; Irizar, I.; Villate, I.; Gonzalez-Esteban, J.; et al. Mitochondrial DNA and palaeontological evidence for the origins of endangered European mink, Mustela lutreola. Anim. Conserv. 2000, 3, 345–355. [Google Scholar] [CrossRef]
- Hosoda, T.; Suzuki, H.; Harada, M.; Tsuchiya, K.; Han, S.-H.; Zhang, Y.-P.; Kryukov, A.P.; Lin, L.-K. Evolutionary trends of the mitochondrial lineage differentiation in species of genera Martes and Mustela. Genes. Genet. Syst. 2000, 75, 259–267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sato, J.J.; Hosoda, T.; Wolsan, M.; Tsuchiya, K.; Yamamoto, M.; Suzuki, H. Phylogenetic Relationships and Divergence Times among Mustelids (Mammalia: Carnivora) Based on Nucleotide Sequences of the Nuclear Interphotoreceptor Retinoid Binding Protein and Mitochondrial CytochromebGenes. Zool. Sci. 2003, 20, 243–264. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.J.; Finarelli, J.A.; Zehr, S.; Hsu, J.; Nedbal, M.A. Molecular Phylogeny of the Carnivora (Mammalia): Assessing the Impact of Increased Sampling on Resolving Enigmatic Relationships. Syst. Biol. 2005, 54, 317–337. [Google Scholar] [CrossRef]
- Kurose, N.; Abramov, A.V.; Masuda, R. Molecular phylogeny and taxonomy of the genus Mustela (Mustelidae, Carnivora), inferred from mitochondrial DNA sequences: New perspectives on phylogenetic status of the back-striped weasel and American mink. Mammal Study 2008, 33, 25–33. [Google Scholar] [CrossRef]
- Korablev, M.P.; Korablev, P.N.; Korablev, N.P.; Tumanov, I.L. Polymorphism of the endangered European mink (Mustela lutreola, Carnivora, Mustelidae) population in the central forest reserve and neighboring areas. Biol. Bull. 2014, 41, 620–628. [Google Scholar] [CrossRef]
- Wilson, D.E.; Reeder, D.-A.M. (Eds.) Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2005; pp. 1–142. [Google Scholar]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-L.; Feng, R.-Q.; Li, M.; Guo, Z.-L.; Zhang, L.-J.; Luo, F.-Z.; Cao, Y.; Yuan, M.-L. The Complete Mitogenome of Pyrrhocoris tibialis (Hemiptera: Pyrrhocoridae) and Phylogenetic Implications. Genes 2019, 10, 820. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Hall, B.D. Codon selection in yeast. J. Biol. Chem. 1982, 257, 3026–3031. [Google Scholar] [CrossRef]
- Schultz, D.T.; Eizenga, J.; Corbett-Detig, R.B.; Francis, W.R.; Christianson, L.M.; Haddock, S.H. Conserved novel ORFs in the mitochondrial genome of the ctenophore Beroe forskalii. PeerJ. 2020, 8, e8356. [Google Scholar] [CrossRef]
- Jang, K.H.; Ryu, S.H.; Hwang, U.W. Mitochondrial genome of the eurasian otter Lutra lutra (Mammalia, Carnivora, Mustelidae). Genes Genom. 2009, 31, 19–27. [Google Scholar] [CrossRef]
- Hixson, J.; Wong, T.W.; A Clayton, D. Both the conserved stem-loop and divergent 5′-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J. Biol. Chem. 1986, 261, 2384–2390. [Google Scholar] [CrossRef]
- Sbisà, E.; Tanzariello, F.; Reyes, A.; Pesole, G.; Saccone, C. Mammalian mitochondrial D-loop region structural analysis: Identification of new conserved sequences and their functional and evolutionary implications. Gene 1997, 205, 125–140. [Google Scholar] [CrossRef]
- Zhang, H.H.; Xu, C.Z.; Ma, J.Z. Structure of the mtDNA control region and phylogeny of the Mustelidae species. Acta. Ecol. Sin. 2009, 29, 3585–3592. [Google Scholar]
- Ki, J.-S.; Hwang, D.-S.; Park, T.-J.; Han, S.-H.; Lee, J.-S. A comparative analysis of the complete mitochondrial genome of the Eurasian otter Lutra lutra (Carnivora; Mustelidae). Mol. Biol. Rep. 2009, 37, 1943–1955. [Google Scholar] [CrossRef]
- Doda, J.N.; Wright, C.T.; Clayton, D.A. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl. Acad. Sci. USA 1981, 78, 6116–6120. [Google Scholar] [CrossRef]
- Ketmaier, V.; Bernardini, C. Structure of the Mitochondrial Control Region of the Eurasian Otter (Lutra lutra; Carnivora, Mustelidae): Patterns of Genetic Heterogeneity and Implications for Conservation of the Species in Italy. J. Hered. 2005, 96, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Haro, M.; Viñas, J.; Mañas, F.; Batet, A.; Ruiz-Olmo, J.; Pla, C. Genetic variability in the complete mitochondrial control region of the Eurasian otter (Lutra lutra) in the Iberian Peninsula. Biol. J. Linn. Soc. 2005, 86, 397–403. [Google Scholar] [CrossRef]
- Walberg, M.W.; Clayton, D.A. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981, 9, 5411–5421. [Google Scholar] [CrossRef] [PubMed]
- Douzery, E.; Randi, E. The mitochondrial control region of Cervidae: Evolutionary patterns and phylogenetic content. Mol. Biol. Evol. 1997, 14, 1154–1166. [Google Scholar] [CrossRef]
- Falkenberg, M. Mitochondrial DNA replication in mammalian cells: Overview of the pathway. Essays Biochem. 2018, 62, 287–296. [Google Scholar] [CrossRef]
- Meyer, S.; Weiss, G.; von Haeseler, A. Pattern of Nucleotide Substitution and Rate Heterogeneity in the Hypervariable Regions I and II of Human mtDNA. Genetics 1999, 152, 1103–1110. [Google Scholar] [CrossRef]
- Zhao, R.-B.; Zhou, C.-Y.; Lu, Z.-X.; Hu, P.; Liu, J.-Q.; Tan, W.-W.; Yang, T.-H. The complete mitochondrial genome of black-footed ferret, Mustela nigripes (Mustela, Mustelinae). Mitochondrial DNA Part A 2014, 27, 1595–1596. [Google Scholar] [CrossRef]
- Gao, W.; Lu, Z.; Liang, Y.; Ren, Z.-M. Complete mitochondrial genome of Mustela sibirica (Carnivora: Mustelidae), a protected and endangered species in China. Mitochondrial DNA Part B 2020, 5, 1081–1083. [Google Scholar] [CrossRef]
- Shalabi, M.A.; Abramov, A.V.; Kosintsev, P.A.; Lin, L.-K.; Han, S.-H.; Watanabe, S.; Yamazaki, K.; Kaneko, Y.; Masuda, R. Comparative phylogeography of the endemic Japanese weasel (Mustela itatsi) and the continental Siberian weasel (Mustela sibirica) revealed by complete mitochondrial genome sequences. Biol. J. Linn. Soc. 2017, 120, 333–348. [Google Scholar] [CrossRef]
- Huang, J.; Yang, B.; Yan, C.; Yang, C.; Tu, F.; Zhang, X.; Yue, B. Phylogenetic analysis of the Mustela altaica (Carnivora: Mustelidae) based on complete mitochondrial genome. Mitochondrial DNA 2013, 25, 255–256. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.J.; Kim, H.R.; Cho, J.Y.; Park, Y.C. Complete mitochondrial genome of the least weasel Mustela nivalis (Mustelidae) in Korea. Mitochondrial DNA Part B 2017, 2, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Peng, D.; Liu, J.; Luan, P.; Liang, L.; Lee, H.; Lee, M.; Ryder, O.A.; Zhang, Y. On the phylogeny of Mustelidae subfamilies: Analysis of seventeen nuclear non-coding loci and mitochondrial complete genomes. BMC Evol. Biol. 2011, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Arnason, U.; Gullberg, A.; Janke, A.; Kullberg, M. Mitogenomic analyses of caniform relationships. Mol. Phylogenet. Evol. 2007, 45, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, T.; Nikaido, M.; Kohno, N.; Fukumoto, Y.; Okada, N.; Hasegawa, M. Molecular phylogenetic study on the origin and evolution of Mustelidae. Gene 2007, 396, 1–12. [Google Scholar] [CrossRef]
- Kim, K.S.; Lee, S.E.; Jeong, H.W.; Ha, J.H. The Complete Nucleotide Sequence of the Domestic Dog (Canis familiaris) Mitochondrial Genome. Mol. Phylogenet. Evol. 1998, 10, 210–220. [Google Scholar] [CrossRef]
- Sun, W.-L.; Zhong, W.; Bao, K.; Liu, H.-L.; Yang, Y.; Wang, Z.; Li, G.-Y. The complete mitochondrial genome of silver fox (Caniformia: Canidae). Mitochondrial DNA Part A 2015, 27, 3348–3350. [Google Scholar] [CrossRef] [PubMed]
- Delisle, I.; Strobeck, C. Conserved Primers for Rapid Sequencing of the Complete Mitochondrial Genome from Carnivores, Applied to Three Species of Bears. Mol. Biol. Evol. 2002, 19, 357–361. [Google Scholar] [CrossRef]
- Peng, R.; Zeng, B.; Meng, X.; Yue, B.; Zhang, Z.; Zou, F. The complete mitochondrial genome and phylogenetic analysis of the giant panda (Ailuropoda melanoleuca). Gene 2007, 397, 76–83. [Google Scholar] [CrossRef]
- Davison, A.; Birks, J.; Griffiths, H.; Kitchener, A.; Biggins, D.; Butlin, R. Hybridization and the phylogenetic relationship between polecats and domestic ferrets in Britain. Biol. Conserv. 1999, 87, 155–161. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal Mitochondrial DNA: Structure and Evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Sun, W.-L.; Wang, S.-J.; Wang, Z.; Liu, H.-L.; Zhong, W.; Yang, Y.-H.; Li, G.-Y. The complete mitochondrial genome sequence of Neovison vison (Carnivora: Mustelidae). Mitochondrial DNA Part A 2016, 27, 1840–1841. [Google Scholar] [CrossRef] [PubMed]
- Lamelas, L.; Aleix-Mata, G.; Rovatsos, M.; Marchal, J.A.; Palomeque, T.; Lorite, P.; Sánchez, A. Complete Mitochondrial Genome of Three Species of the Genus Microtus (Arvicolinae, Rodentia). Animals 2020, 10, 2130. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Tursun, M.; Zhou, S.; Zhang, Y.; Dai, H. Complete mitochondrial genome sequence of Lepus yarkandensis Günther, 1875 (Lagomorpha, Leporidae): Characterization and phylogenetic analysis. ZooKeys 2021, 1012, 135–150. [Google Scholar] [CrossRef]
- Brown, W.M. The mitochondrial genome of animals. In Molecular Evolutionary Genetics; Macintyre, R., Ed.; Plenum Press: New York, NY, USA, 1985; pp. 95–130. [Google Scholar]
- Harrison, R.G. Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol. Evol. 1989, 4, 6–11. [Google Scholar] [CrossRef]
- Zhong, H.-M.; Zhang, H.-H.; Sha, W.-L.; Zhang, C.-D.; Chen, Y.-C. Complete Mitochondrial Genome of the Red Fox (Vuples vuples) and Phylogenetic Analysis with Other Canid Species. Zool. Res. 2010, 31, 122–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, G.; Yang, X.; Zhang, H.; Sun, G.; Zhao, C.; Dou, H. The complete mitochondrial genome sequence of Mustela eversmannii (Carnivora: Mustelidae). Mitochondrial DNA Part A 2015, 27, 3657–3658. [Google Scholar] [CrossRef]
- Yu, M.; Xu, H.; Li, D.; Wu, J.; Wen, A.; Xie, M.; Wang, Q.; Zhu, G.; Ni, Q.; Zhang, M.; et al. The complete mitochondrial genome sequence and phylogenetic analysis of yellow weasel (Mustela sibirica). Mitochondrial DNA Part B 2019, 4, 3698–3699. [Google Scholar] [CrossRef] [PubMed]
- Saccone, C.; Gissi, C.; Reyes, A.; Larizza, A.; Sbisà, E.; Pesole, G. Mitochondrial DNA in metazoa: Degree of freedom in a frozen event. Gene 2001, 286, 3–12. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Emami-Khoyi, A.; Hartley, D.A.; Ross, J.G.; Murphy, E.C.; Paterson, A.M.; Cruickshank, R.H.; Else, T.-A. Complete mitochondrial genome of the stoat (Mustela erminea) and New Zealand fur seal (Arctocephalus forsteri) and their significance for mammalian phylogeny. Mitochondrial DNA Part A 2016, 27, 4597–4599. [Google Scholar] [CrossRef]
- Pereira, S.L. Mitochondrial genome organization and vertebrate phylogenetics. Genet. Mol. Biol. 2000, 23, 745–752. [Google Scholar] [CrossRef]
- Hoelzel, A.R.; Lopez, J.; A Dover, G.; O’Brien, S.J. Rapid evolution of a heteroplasmic repetitive sequence in the mitochondrial DNA control region of carnivores. J. Mol. Evol. 1994, 39, 191–199. [Google Scholar]
- Zhang, D.-X.; Szymura, J.M.; Hewitt, G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 1995, 40, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Lunt, D.H.; Whipple, L.E.; Hyman, B.C. Mitochondrial DNA variable number tandem repeats (VNTRs): Utility and problems in molecular ecology. Mol. Ecol. 1998, 7, 1441–1455. [Google Scholar] [CrossRef] [PubMed]
- Savolainen, P.; Arvestad, L.; Lundeberg, J. mtDNA Tandem Repeats in Domestic Dogs and Wolves: Mutation Mechanism Studied by Analysis of the Sequence of Imperfect Repeats. Mol. Biol. Evol. 2000, 17, 474–488. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, Y.; Chu, K.H. Broader pattern of tandem repeats in the mitochondrial control region of Perciformes. Chin. J. Oceanol. Limnol. 2010, 28, 785–794. [Google Scholar] [CrossRef]
- Broughton, R.E.; Dowling, T.E. Evolutionary dynamics of tandem repeats in the mitochondrial DNA control region of the minnow Cyprinella spiloptera. Mol. Biol. Evol. 1997, 14, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.K.; Boore, J.L.; Moritz, C. Multiple Origins and Rapid Evolution of Duplicated Mitochondrial Genes in Parthenogenetic Geckos (Heteronotia binoei; Squamata, Gekkonidae). Mol. Biol. Evol. 2007, 24, 2775–2786. [Google Scholar] [CrossRef]
- Lavrov, D.V. Rapid Proliferation of Repetitive Palindromic Elements in mtDNA of the Endemic Baikalian Sponge Lubomirskia baicalensis. Mol. Biol. Evol. 2009, 27, 757–760. [Google Scholar] [CrossRef]
- Cabañas, N.; Becerra, A.; Romero, D.; Govezensky, T.; Espinosa-Aguirre, J.J.; Camacho-Carranza, R. Repetitive DNA profile of the amphibian mitogenome. BMC Bioinform. 2020, 21, 197. [Google Scholar] [CrossRef]
- Formenti, G.; Rhie, A.; Balacco, J.; Haase, B.; Mountcastle, J.; Fedrigo, O.; Brown, S.; Capodiferro, M.; Al-Ajli, F.O.; Ambrosini, R.; et al. Complete vertebrate mitogenomes reveal widespread gene duplications and repeats. bioRxiv 2020, 6, 177956. [Google Scholar] [CrossRef]
- Roques, S.; Godoy, J.A.; Negro, J.; Hiraldo, F. Organization and Variation of the Mitochondrial Control Region in Two Vulture Species, Gypaetus barbatus and Neophron percnopterus. J. Hered. 2004, 95, 332–337. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, T.; Lu, W. Complete mitochondrial DNA sequence of the endangered fish (Bahaba taipingensis): Mitogenome characterization and phylogenetic implications. ZooKeys 2015, 546, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Čechová, J.; Lýsek, J.; Bartas, M.; Brázda, V. Complex analyses of inverted repeats in mitochondrial genomes revealed their importance and variability. Bioinformatics 2017, 34, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Saiwaki, T.; Shiga, K.; Fukuyama, R.; Tsutsumi, Y.; Fushiki, S. A unique junctional palindromic sequence in mitochondrial DNA from a patient with progressive external ophthalmoplegia. Mol. Pathol. 2000, 53, 333–335. [Google Scholar] [CrossRef] [PubMed]
- Solano, A.; Gámez, J.; Carod, F.J.; Pineda, M.; Playán, A.; López-Gallardo, E.; Andreu, A.L.; Montoya, J. Characterisation of repeat and palindrome elements in patients harbouring single deletions of mitochondrial DNA. J. Med. Genet. 2003, 40, e86. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef]
- Fearnley, I.M.; Walker, J. Initiation codons in mammalian mitochondria: Differences in genetic code in the organelle. Biochemistry 1987, 26, 8247–8251. [Google Scholar] [CrossRef]
- Watanabe, K. Unique features of animal mitochondrial translation systems-The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases-Proc. Jpn. Acad. Ser. B 2010, 86, 11–39. [Google Scholar] [CrossRef]
- Cantatore, P.; Roberti, M.; Pesole, G.; Ludovico, A.; Milella, F.; Gadaletal, M.N.; Saccone, C. Evolutionary analysis of cytochrome b sequences in some perciformes: Evidence for a slower rate of evolution than in mammals. J. Mol. Evol. 1994, 39, 589–597. [Google Scholar] [CrossRef]
- Hwang, D.-S.; Ki, J.-S.; Jeong, D.-H.; Kim, B.-H.; Lee, B.-K.; Han, S.-H.; Lee, J.-S. A comprehensive analysis of three Asiatic black bear mitochondrial genomes (subspecies ussuricus, formosanus and mupinensis), with emphasis on the complete mtDNA sequence of Ursus thibetanus ussuricus (Ursidae). DNA Sequence 2008, 19, 418–429. [Google Scholar] [CrossRef]
- Yu, H.; Li, Q. Complete mitochondrial DNA sequence of Crassostrea nippona: Comparative and phylogenomic studies on seven commercial Crassostrea species. Mol. Biol. Rep. 2011, 39, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Elhaik, E.; Landan, G.; Graur, D. Can GC Content at Third-Codon Positions Be Used as a Proxy for Isochore Composition? Mol. Biol. Evol. 2009, 26, 1829–1833. [Google Scholar] [CrossRef]
- Uddin, A.; Choudhury, M.N.; Chakraborty, S. Codon usage bias and phylogenetic analysis of mitochondrial ND1 gene in pisces, aves, and mammals. Mitochondrial DNA Part A 2016, 29, 36–48. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986, 24, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Salvato, P.; Simonato, M.; Battisti, A.; Negrisolo, E. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genom. 2008, 9, 331. [Google Scholar] [CrossRef]
- Hao, J.; Sun, Q.; Zhao, H.; Sun, X.; Gai, Y.; Yang, Q. The Complete Mitochondrial Genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and Its Phylogenetic Implication. Comp. Funct. Genom. 2012, 2012, 328049. [Google Scholar] [CrossRef]
- Xu, X.-Z.; Liu, Q.-P.; Fan, L.-J.; Cui, X.-F.; Zhou, X.-P. Analysis of synonymous codon usage and evolution of begomoviruses. J. Zhejiang Univ. Sci. B 2008, 9, 667–674. [Google Scholar] [CrossRef]
- Guo, X.; Bao, J.; Fan, L. Evidence of selectively driven codon usage in rice: Implications for GC content evolution of Gramineaegenes. FEBS Lett. 2007, 581, 1015–1021. [Google Scholar] [CrossRef]
- Breton, S.; Milani, L.; Ghiselli, F.; Guerra, D.; Stewart, D.; Passamonti, M. A resourceful genome: Updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 2014, 30, 555–564. [Google Scholar] [CrossRef]
- Faure, E.; Delaye, L.; Tribolo, S.; Levasseur, A.; Seligmann, H.; Barthélémy, R.-M. Probable presence of an ubiquitous cryptic mitochondrial gene on the antisense strand of the cytochrome oxidase I gene. Biol. Direct 2011, 6, 56. [Google Scholar] [CrossRef]
- Lee, C.; Yen, K.; Cohen, P. Humanin: A harbinger of mitochondrial-derived peptides? Trends Endocrinol. Metab. 2013, 24, 222–228. [Google Scholar] [CrossRef]
- Sun, S.; Hui, M.; Wang, M.; Sha, Z. The complete mitochondrial genome of the alvinocaridid shrimp Shinkaicaris leurokolos (Decapoda, Caridea): Insight into the mitochondrial genetic basis of deep-sea hydrothermal vent adaptation in the shrimp. Comp. Biochem. Physiol. Part D Genom. Proteom. 2018, 25, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Capt, C.; Passamonti, M.; Breton, S. The human mitochondrial genome may code for more than 13 proteins. Mitochondrial DNA Part A 2015, 27, 3098–3101. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.-R.; Chen, Y.; Wu, X.; Hu, J.-C.; Peng, Z.-S.; Yang, J.; Tang, Z.-X.; Zhou, C.-Q.; Li, Y.-M.; Yang, S.-K.; et al. A complete mitochondrial genome sequence of Asian black bear Sichuan subspecies (Ursus thibetanus mupinensis). Int. J. Biol. Sci. 2007, 3, 85–90. [Google Scholar] [CrossRef]
- Gautheret, D.; Konings, D.; Gutell, R.G. U base pairing motifs in ribosomal RNA. RNA 1995, 1, 807–814. [Google Scholar]
- Yokobori, S.-I.; Pääbo, S. tRNA editing in metazoans. Nat. Cell Biol. 1995, 377, 490. [Google Scholar] [CrossRef]
- Smith, D.; Yarus, M. Transfer RNA structure and coding specificity: I. Evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J. Mol. Biol. 1989, 206, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Hardt, W.-D.; Schlegl, J.; Erdmann, V.A.; Hartmann, R.K. Role of the D arm and the anticodon arm in tRNA recognition by eubacterial and eukaryotic RNase P enzymes. Biochemistry 1993, 32, 13046–13053. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Braband, A.; Schierwater, B.; Stadler, P.F. Genetic aspects of mitochondrial genome evolution. Mol. Phylogenet. Evol. 2013, 69, 328–338. [Google Scholar] [CrossRef]
- Dong, Y.; Zhou, L.; Li, B.; Zhao, G. The complete mitochondrial genome of the Black-headed Gull Chroicocephalus ridibundus (Charadriiformes: Laridae). Mitochondrial DNA Part A 2014, 27, 1991–1992. [Google Scholar] [CrossRef]
- Li, B.; Zhou, L.; Liu, G.; Gu, C. Complete mitochondrial genome of Naumann’s thrush Turdus naumanni (Passeriformes: Turdidae). Mitochondrial DNA Part A 2014, 27, 1117–1118. [Google Scholar] [CrossRef]
- Springer, M.S.; Douzery, E. Secondary structure and patterns of evolution among mammalian mitochondrial 12S rRNA molecules. J. Mol. Evol. 1996, 43, 357–373. [Google Scholar] [CrossRef]
- Burk, A.; Douzery, E.J.P.; Springer, M.S. The Secondary Structure of Mammalian Mitochondrial 16S rRNA Molecules: Refinements Based on a Comparative Phylogenetic Approach. J. Mamm. Evol. 2002, 9, 225–252. [Google Scholar] [CrossRef]
- Abhyankar, A.; Park, H.-B.; Tonolo, G.; Luthman, H. Comparative Sequence Analysis of the Non-Protein-Coding Mitochondrial DNA of Inbred Rat Strains. PLoS ONE 2009, 4, e8148. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L.; Olsen, G.J.; Waddell, P.J.; Hillis, D.M. Phylogenetic inference. In Molecular Systematics, 2nd ed.; Hillis, D.M., Moritz, C., Mable, B.K., Eds.; Sinauer Assoc: Sunderland, MA, USA, 1996; pp. 407–514. [Google Scholar]
- Scarpulla, R.C. Transcriptional Paradigms in Mammalian Mitochondrial Biogenesis and Function. Physiol. Rev. 2008, 88, 611–638. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Okumura, H.; Lee, Y.-S.; Kim, K.-S.; Min, M.-S.; Lee, H. Organization and variation of the mitochondrial DNA control region in five Caprinae species. Genes Genom. 2010, 32, 335–344. [Google Scholar] [CrossRef]
- Gemmell, N.J.; Western, P.; Watson, J.M.; Graves, J. Evolution of the mammalian mitochondrial control region-comparisons of control region sequences between monotreme and therian mammals. Mol. Biol. Evol. 1996, 13, 798–808. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saccone, C.; Pesole, G.; Sbisá, E. The main regulatory region of mammalian mitochondrial DNA: Structure-function model and evolutionary pattern. J. Mol. Evol. 1991, 33, 83–91. [Google Scholar] [CrossRef]
- Ledje, C.; Arnason, U. Phylogenetic analyses of complete cytochrome b genes of the order Carnivora with particular emphasis on the Caniformia. J. Mol. Evol. 1996, 42, 135–144. [Google Scholar] [CrossRef]
- Brown, G.G.; Gadaleta, G.; Pepe, G.; Saccone, C.; Sbisà, E. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. J. Mol. Biol. 1986, 192, 503–511. [Google Scholar] [CrossRef]
- Saccone, C.; Attimonelli, M.; Sbisà, E. Structural elements highly preserved during the evolution of the D-loop-containing region in vertebrate mitochondrial DNA. J. Mol. Evol. 1987, 26, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Clayton, D.A. Nuclear gadgets in mitochondrial DNA replication and transcription. Trends Biochem. Sci. 1991, 16, 107–111. [Google Scholar] [CrossRef]
- Shadel, G.S.; Clayton, D.A. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997, 66, 409–435. [Google Scholar] [CrossRef]
- Randi, E.; Lucchini, V. Organization and Evolution of the Mitochondrial DNA Control Region in the Avian Genus Alectoris. J. Mol. Evol. 1998, 47, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Kowal, K.; Tkaczyk, A.; Ząbek, T.; Pierzchała, M.; Ślaska, B. Comparative Analysis of CpG Sites and Islands Distributed in Mitochondrial DNA of Model Organisms. Animals 2020, 10, 665. [Google Scholar] [CrossRef]
- Bellizzi, D.; D’Aquila, P.; Scafone, T.; Giordano, M.; Riso, V.; Riccio, A.; Passarino, G. The Control Region of Mitochondrial DNA Shows an Unusual CpG and Non-CpG Methylation Pattern. DNA Res. 2013, 20, 537–547. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.; Wang, X. How to breakthrough mitochondrial DNA methylation-associated networks. Cell Biol. Toxicol. 2020, 36, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.F.C. Mitochondrial metabolism and DNA methylation: A review of the interaction between two genomes. Clin. Epigenet. 2020, 12, 182. [Google Scholar] [CrossRef]
- Parker, S.C.J.; Margulies, E.H.; Tullius, T.D. The Relationship between Fine Scale DNA Structure, GC content, and Functional elements in 1% of the human genome. Genome Inform. 2008, 20, 199–211. [Google Scholar] [CrossRef]
- Fumagalli, L.; Taberlet, P.; Favre, L.; Hausser, J. Origin and evolution of homologous repeated sequences in the mitochondrial DNA control region of shrews. Mol. Biol. Evol. 1996, 13, 31–46. [Google Scholar] [CrossRef]
- Jang, K.H.; Hwang, U.W. Complete mitochondrial genome of Korean yellow-throated marten, Martes flavigula (Carnivora, Mustelidae). Mitochondrial DNA 2014, 27, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, G.; Chapman, A.M. Length and sequence variation in evening bat D-loop mtDNA. Genetics 1991, 128, 607–617. [Google Scholar] [CrossRef]
- Stanton, D.J.; Daehler, L.L.; Moritz, C.C.; Brown, W.M. Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics 1994, 137, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C.; Brown, W.M. Tandem duplications in animal mitochondrial DNAs: Variation in incidence and gene content among lizards. Proc. Natl. Acad. Sci. USA 1987, 84, 7183–7187. [Google Scholar] [CrossRef] [PubMed]
- Zevering, C.E.; Moritz, C.; Heideman, A.; Sturm, R. Parallel origins of duplications and the formation of pseudogenes in mitochondrial DNA from parthenogenetic lizards (Heteronotia binoei; Gekkonidae). J. Mol. Evol. 1991, 33, 431–441. [Google Scholar] [CrossRef]
- Mundy, N.I.; Winchell, C.S.; Woodruff, D.S. Tandem Repeats and Heteroplasmy in the Mitochondrial DNA Control Region of the Loggerhead Shrike (Lanius ludovicianus). J. Hered. 1996, 87, 21–26. [Google Scholar] [CrossRef]
- Casane, D.; Dennebouy, N.; De Rochambeau, H.; Mounolou, J.C.; Monnerot, M. Nonneutral evolution of tandem repeats in the mitochondrial DNA control region of lagomorphs. Mol. Biol. Evol. 1997, 14, 779–789. [Google Scholar] [CrossRef]
- Hernandez, M.A.; Amezcua, A.; Gutiérrez-Corchero, F.; Campos, F. Identification of Lanius species and subspecies using tandem repeats in the mitochondrial DNA control region. Ibis 2003, 146, 227–230. [Google Scholar] [CrossRef]
- Wang, T.Y.; Tzeng, C.S.; Teng, H.Y.; Chang, T. Phylogeography and identification of a 187-bp-long duplication within the mitochondrial control region of Formosania lacustre (Teleostei: Balitoridae). Zool. Stud. 2007, 46, 569–582. [Google Scholar]
- Omote, K.; Nishida, C.; Dick, M.H.; Masuda, R. Limited phylogenetic distribution of a long tandem-repeat cluster in the mitochondrial control region in Bubo (Aves, Strigidae) and cluster variation in Blakiston’s fish owl (Bubo blakistoni). Mol. Phylogenet. Evol. 2013, 66, 889–897. [Google Scholar] [CrossRef]
- Padhi, A. Geographic variation within a tandemly repeated mitochondrial DNA D-loop region of a North American freshwater fish, Pylodictis olivaris. Gene 2014, 538, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, G.S.; Mayer, F.; Kerth, G.; Petri, B. Evolution of repeated sequence arrays in the D-loop region of bat mitochondrial DNA. Genetics 1997, 146, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Stoneking, M. Hypervariable Sites in the mtDNA Control Region Are Mutational Hotspots. Am. J. Hum. Genet. 2000, 67, 1029–1032. [Google Scholar] [CrossRef]
- Eichmann, C.; Parson, W. Molecular characterization of the canine mitochondrial DNA control region for forensic applications. Int. J. Leg. Med. 2007, 121, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Cabria, M.T.; Gonzalez, E.G.; Gomez-Moliner, B.J.; Michaux, J.R.; Skumatov, D.; Kranz, A.; Fournier, P.; Palazon, S.; Zardoya, R. Patterns of genetic variation in the endangered European mink (Mustela lutreola L., 1761). BMC Evol. Biol. 2015, 15, 141. [Google Scholar] [CrossRef]
- Saccone, C.; De Giorgi, C.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Marmi, J.; López-Giráldez, J.F.; Domingo-Roura, X. Phylogeny, evolutionary history and taxonomy of the Mustelidae based on sequences of the cytochrome b gene and a complex repetitive flanking region. Zool. Scr. 2004, 33, 481–499. [Google Scholar] [CrossRef]
- Fulton, T.L.; Strobeck, C. Molecular phylogeny of the Arctoidea (Carnivora): Effect of missing data on supertree and supermatrix analyses of multiple gene data sets. Mol. Phylogenet. Evol. 2006, 41, 165–181. [Google Scholar] [CrossRef]
- Abramov, A.V.; Meschersky, I.G.; Aniskin, V.M.; Rozhnov, V. The mountain weasel Mustela kathiah (Carnivora: Mustelidae): Molecular and Karyological data. Biol. Bull. 2013, 40, 52–60. [Google Scholar] [CrossRef]
- Yu, L.; Li, Y.-W.; Ryder, O.A.; Zhang, Y.-P. Analysis of complete mitochondrial genome sequences increases phylogenetic re-solution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol. Biol. 2007, 7, 198. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.-C.; Roos, C.; Inoue-Murayama, M.; Inoue, E.; Shih, C.-C.; Pei, K.J.-C.; Vigilant, L. Mitochondrial Genome Sequences Effectively Reveal the Phylogeny of Hylobates Gibbons. PLoS ONE 2010, 5, e14419. [Google Scholar] [CrossRef]
- Duchene, S.; Archer, F.I.; Vilstrup, J.; Caballero, S.; Morin, P.A. Mitogenome Phylogenetics: The Impact of Using Single Regions and Partitioning Schemes on Topology, Substitution Rate and Divergence Time Estimation. PLoS ONE 2011, 6, e27138. [Google Scholar] [CrossRef]
- Ma, H.; Ma, C.; Li, C.; Lu, J.; Zou, X.; Gong, Y.; Wang, W.; Chen, W.; Ma, L.; Xia, L. First mitochondrial genome for the red crab (Charybdis feriata) with implication of phylogenomics and population genetics. Sci. Rep. 2015, 5, 11524. [Google Scholar] [CrossRef]
- Miya, M.; Nishida, M. The mitogenomic contributions to molecular phylogenetics and evolution of fishes: A 15-year retrospect. Ichthyol. Res. 2014, 62, 29–71. [Google Scholar] [CrossRef]
- Zhu, S.; Gao, Y.; Liu, H.; Zhang, S.; Bai, X.; Zhang, M. Phylogenetic relationship of Wolverine Gulo gulo in Mustelidae revealed by complete mitochondrial genome. Mitochondrial DNA Part A 2015, 27, 2937–2938. [Google Scholar] [CrossRef]
- Sullivan, K.A.M.; Platt, R.N.; Bradley, R.D.; Ray, D.A. Whole mitochondrial genomes provide increased resolution and indicate paraphyly in deer mice. BMC Zool. 2017, 2, 11. [Google Scholar] [CrossRef]
- Jia, X.-N.; Xu, S.-X.; Bai, J.; Wang, Y.-F.; Nie, Z.-H.; Zhu, C.-C.; Wang, Y.; Cai, Y.-X.; Zou, J.-X.; Zhou, X.-M. The complete mitochondrial genome of Somanniathelphusa boyangensis and phylogenetic analysis of Genus Somanniathelphusa (Crustacea: Decapoda: Parathelphusidae). PLoS ONE 2018, 13, e0192601. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.M.; McDowell, J.R.; Bennett, M.; Graves, J.E.; Ovenden, J.R. Analysis of whole mitochondrial genome sequences increases phylogenetic resolution of istiophorid billfishes. Bull. Mar. Sci. 2018, 94, 73–84. [Google Scholar] [CrossRef]
- Ibiş, O. Whole mitochondrial genome sequence and phylogenetic relationships of Williams’s jerboa (Scarturus williamsi) from Turkey. PeerJ 2020, 8, e9569. [Google Scholar] [CrossRef] [PubMed]
- Bentley, D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006, 16, 545–552. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT-The BLAST-Like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2007, 24, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Gardiner-Garden, M.; Frommer, M. CpG Islands in vertebrate genomes. J. Mol. Biol. 1987, 196, 261–282. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.; Kocher, T. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Humphrey-Dixon, E.L.; Sharp, R.; Schuckers, M.; Lock, R. Comparative genome analysis suggests characteristics of yeast inverted repeats that are important for transcriptional activity. Genome 2011, 54, 934–942. [Google Scholar] [CrossRef]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Haronikova, L.; Coufal, J.; Stastny, J. Palindrome analyser—A new web-based server for predicting and evaluating inverted repeats in nucleotide sequences. Biochem. Biophys. Res. Commun. 2016, 478, 1739–1745. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Honer Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics, 1st ed.; Columbia University Press: Manhattan, NY, USA, 1987. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Söding, J.S.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Wan, Q.-H.; Wu, H.; Fujihara, T.; Fang, S.-G. Which genetic marker for which conservation genetics issue? Electrophoresis 2004, 25, 2165–2176. [Google Scholar] [CrossRef]
- Islam, M.N.; Sultana, S.; Alam, J. Sequencing and annotation of the complete mitochondrial genome of a threatened labeonine fish, Cirrhinus reba. Genom. Informat. 2020, 18, e32. [Google Scholar] [CrossRef] [PubMed]
- Riddle, A.E.; Pilgrim, K.L.; Mills, L.S.; McKelvey, K.S.; Ruggiero, L.F. Identification of mustelids using mitochondrial DNA and non-invasive sampling. Conserv. Genet. 2003, 4, 241–243. [Google Scholar] [CrossRef]
- Gómez-Moliner, B.J.; Cabria, M.T.; Rubines, J.; Garin, I.; Madeira, M.J.; Elejalde, A.; Aihartza, J.; Fournier, P.; Palazón, S. PCR-RFLP identification of mustelid species: European mink (Mustela lutreola), American mink (M. vison) and polecat (M. putorius) by analysis of excremental DNA. J. Zool. 2004, 262, 311–316. [Google Scholar] [CrossRef]
- Adams, C.I.; Knapp, M.; Gemmell, N.J.; Jeunen, G.-J.; Bunce, M.; Lamare, M.D.; Taylor, H.R. Beyond Biodiversity: Can Environmental DNA (eDNA) Cut It as a Population Genetics Tool? Genes 2019, 10, 192. [Google Scholar] [CrossRef]
- Chagas, A.T.D.A.; Ludwig, S.; Pimentel, J.D.S.M.; de Abreu, N.L.; Nunez-Rodriguez, D.L.; Leal, H.G.; Kalapothakis, E. Use of complete mitochondrial genome sequences to identify barcoding markers for groups with low genetic distance. Mitochondrial DNA Part A 2020, 31, 139–146. [Google Scholar] [CrossRef]
- Jensen, M.R.; Sigsgaard, E.E.; Liu, S.; Manica, A.; Bach, S.S.; Hansen, M.M.; Møller, P.R.; Thomsen, P.F. Genome-scale target capture of mitochondrial and nuclear environmental DNA from water samples. Mol. Ecol. Resour. 2020, 21, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Nabholz, B.; Glémin, S.; Hurst, G.D.D. Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Mol. Ecol. 2009, 18, 4541–4550. [Google Scholar] [CrossRef]
- Harley, E.H.; de Waal, M.; Murray, S.; O’Ryan, C. Comparison of whole mitochondrial genome sequences of northern and southern white rhinoceroses (Ceratotherium simum): The conservation consequences of species definitions. Conserv. Genet. 2016, 17, 1285–1291. [Google Scholar] [CrossRef]
- Ryder, O.A. Species conservation and systematics: The dilemma of subspecies. Trends Ecol. Evol. 1986, 1, 9–10. [Google Scholar] [CrossRef]
- US Fish and Wildlife Service; National Fisheries Service. Policy Regarding the Recognition of Distinct Vertebrate Population Segments under the Endangered Species Act Federal Register; U.S. Government Publishing Office: Washington, DC, USA, 1996; pp. 4721–4725. Available online: https://www.federalregister.gov/documents/1996/02/07/96-2639/policy-regarding-the-recognition-of-distinct-vertebrate-population-segments-under-the-endangered (accessed on 1 February 2021).
- Fraser, D.; Bernatchez, L. Adaptive evolutionary conservation: Towards a unified concept for defining conservation units. Mol. Ecol. 2001, 10, 2741–2752. [Google Scholar] [CrossRef]
- Guerrero, J.; Gallo-Reynoso, J.P.; Biek, R. Mitochondrial DNA diversity, genetic structure, and demographic history of the Neotropical otter (Lontra longicaudis) in Mexico. J. Mammal. 2015, 96, 1162–1173. [Google Scholar] [CrossRef]
- Amstislavsky, S.; Lindeberg, H.; Aalto, J.; Kennedy, M.W. Conservation of European mink (Mustela lutreola): Focus on re-production and reproductive technologies. Reprod. Domest. Anim. 2008, 43, 502–513. [Google Scholar] [CrossRef] [PubMed]
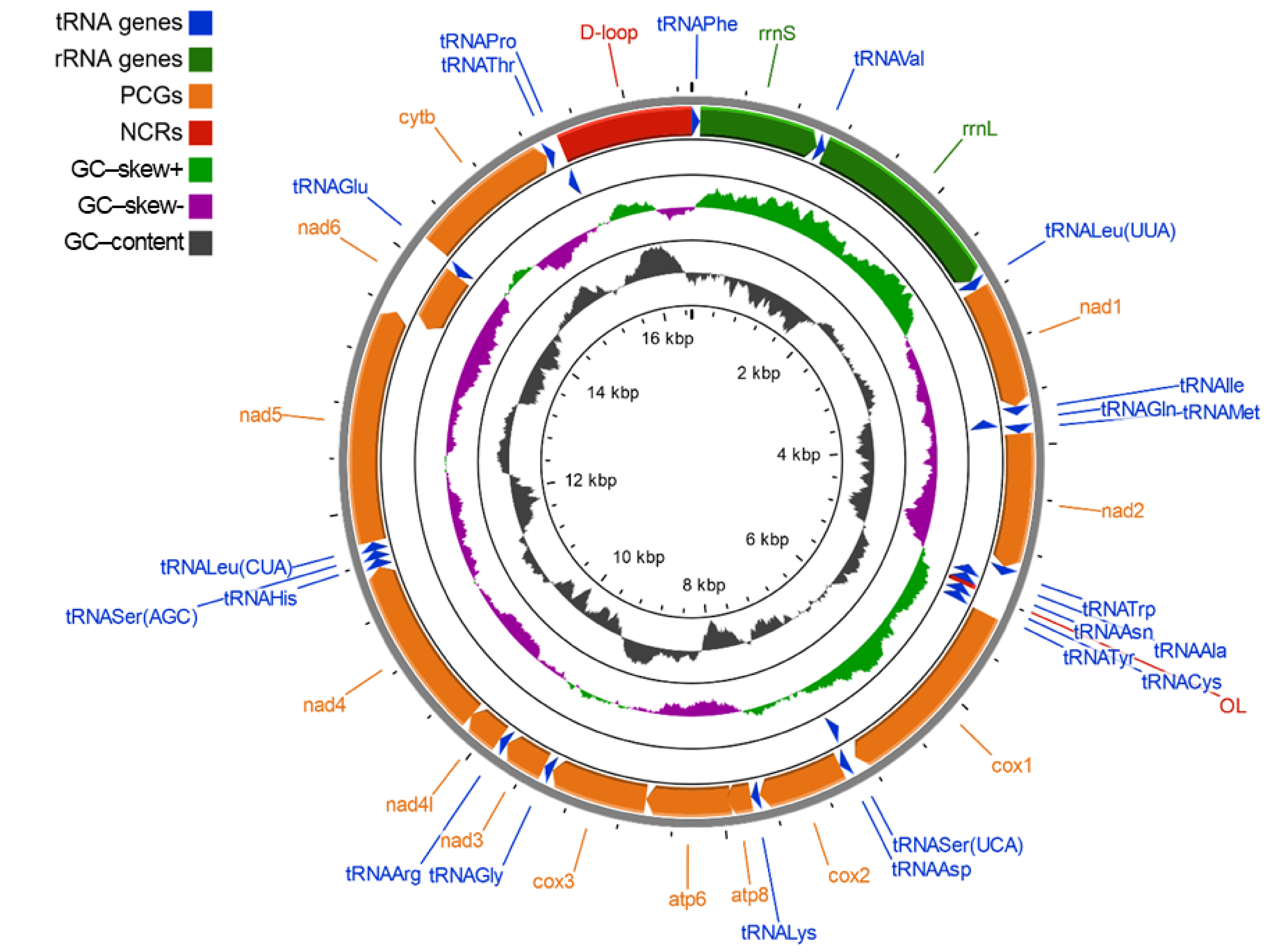

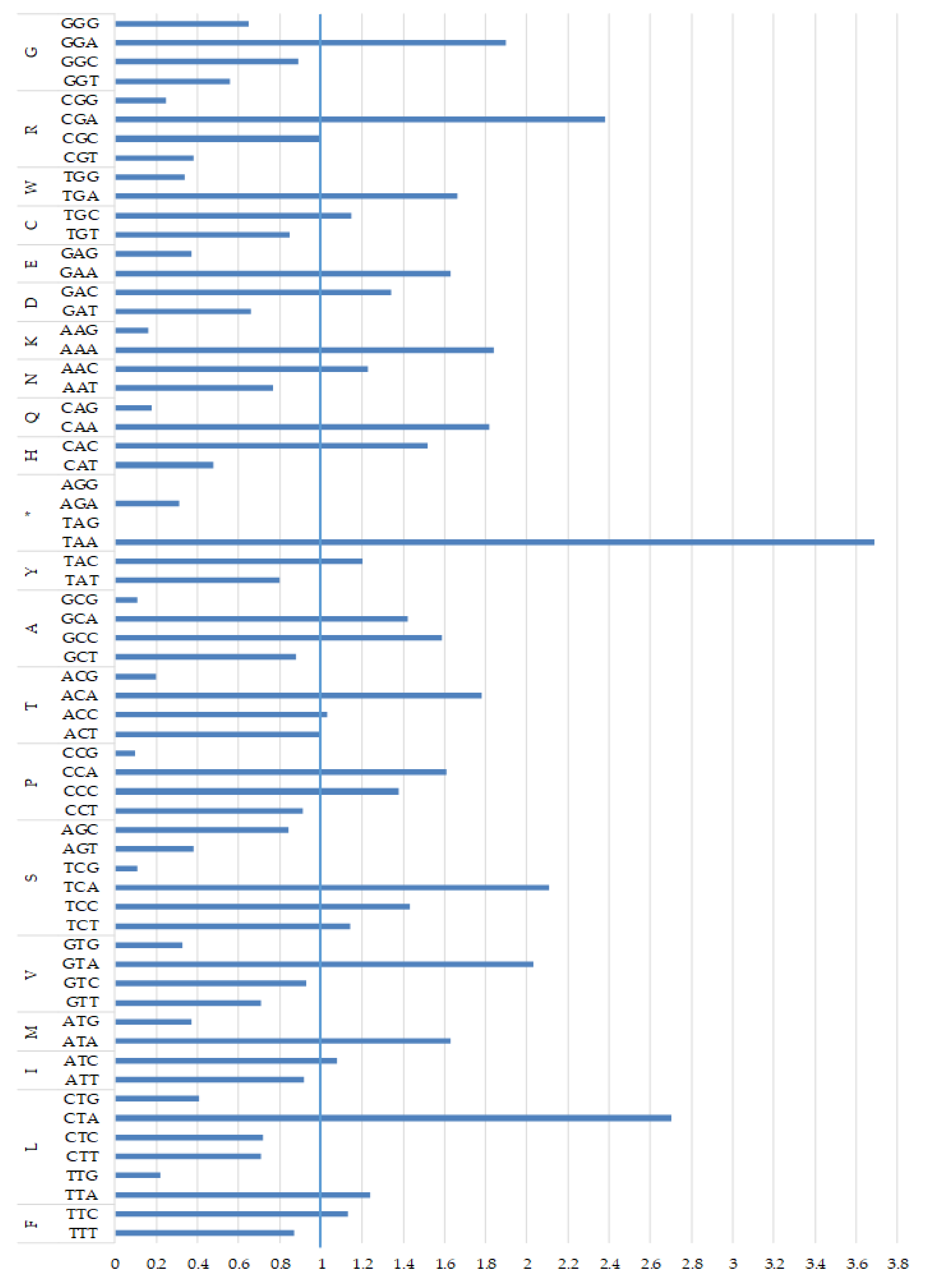
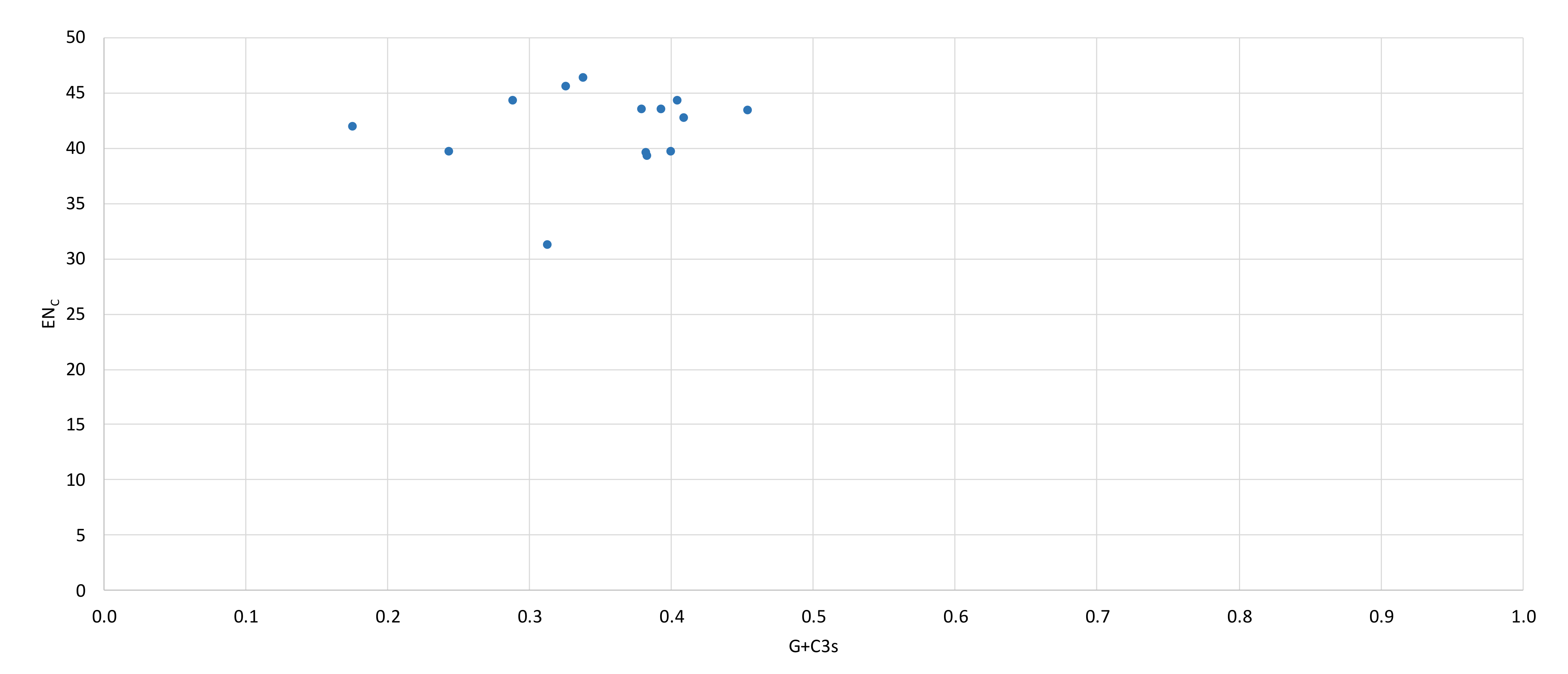
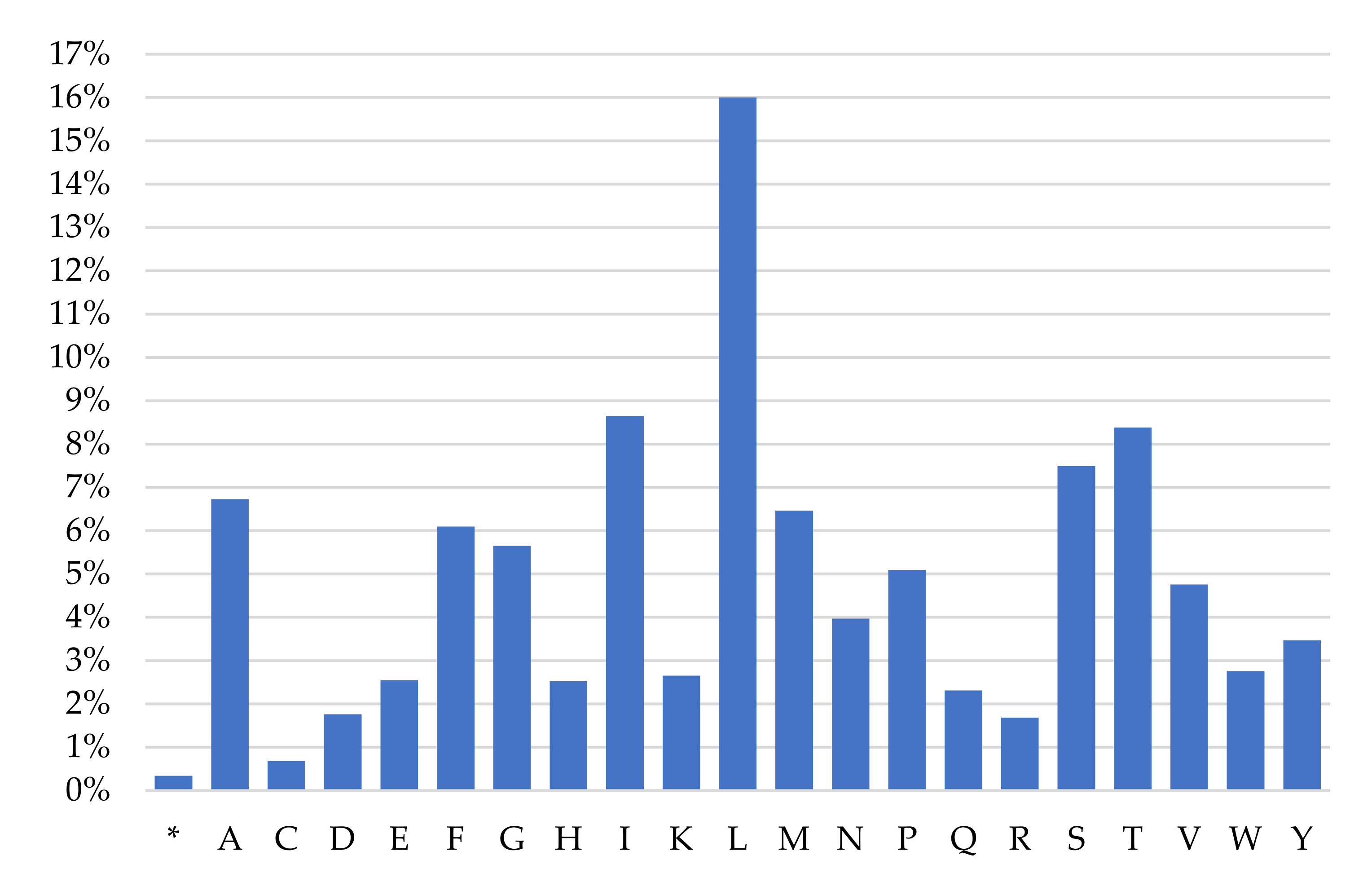

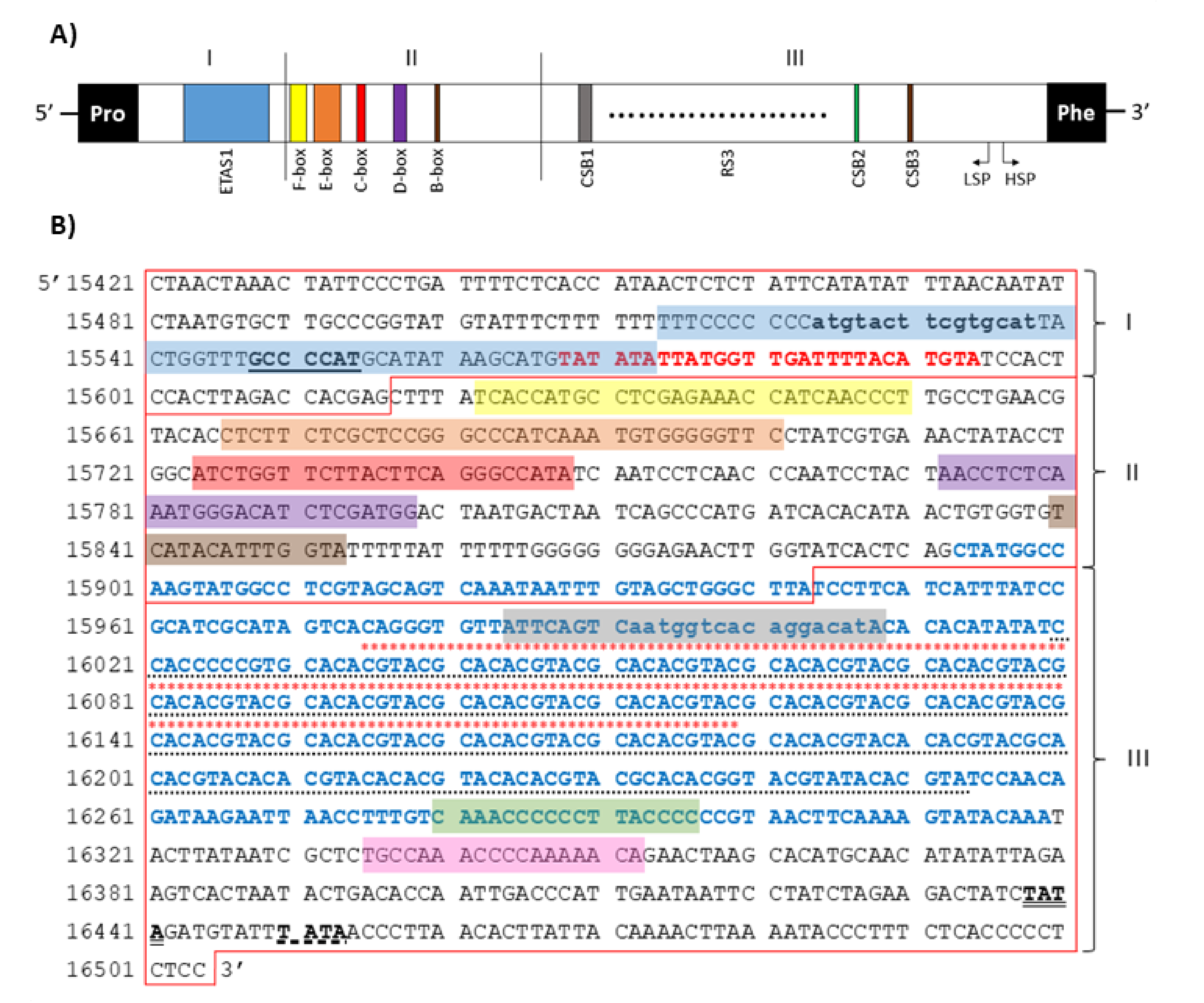
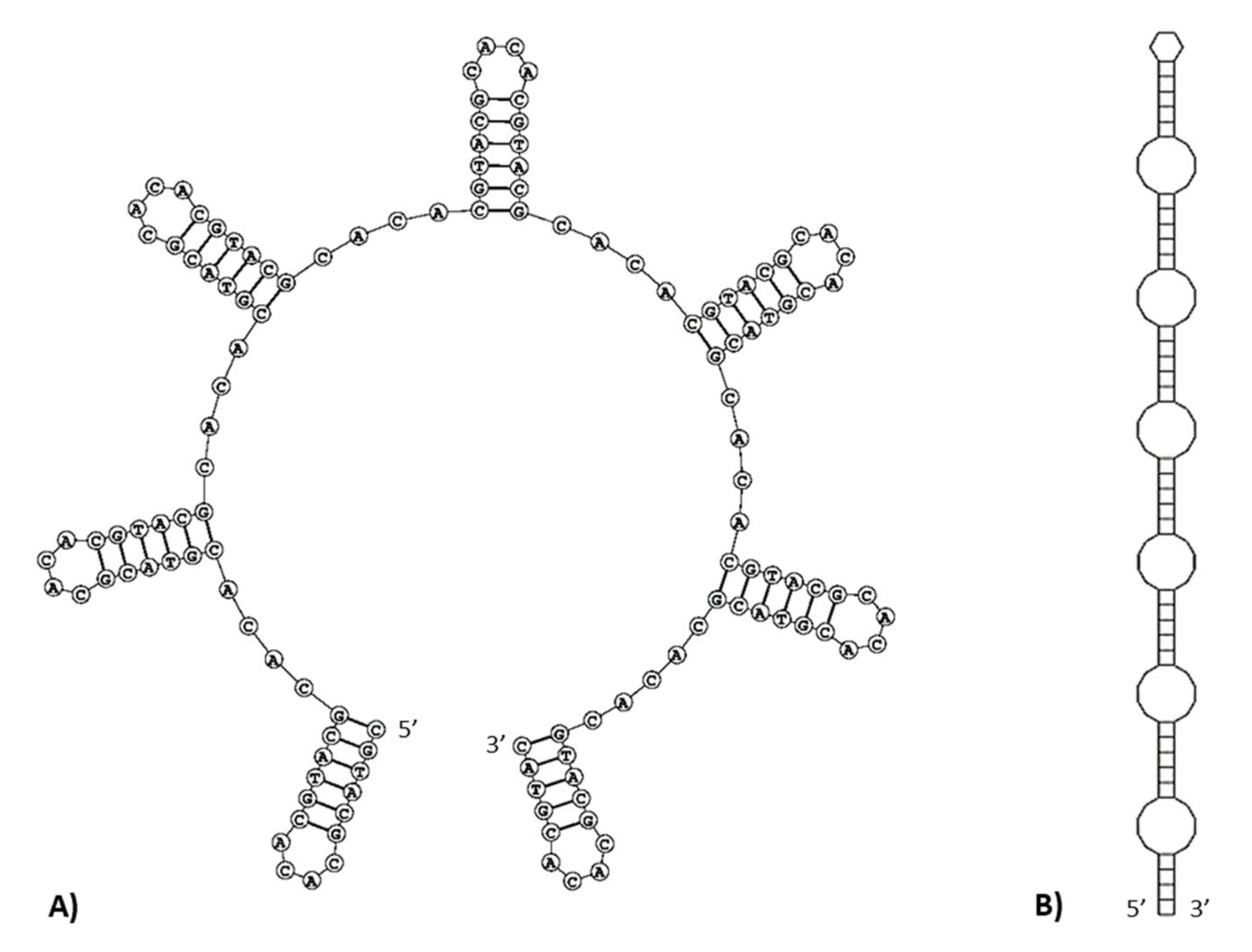
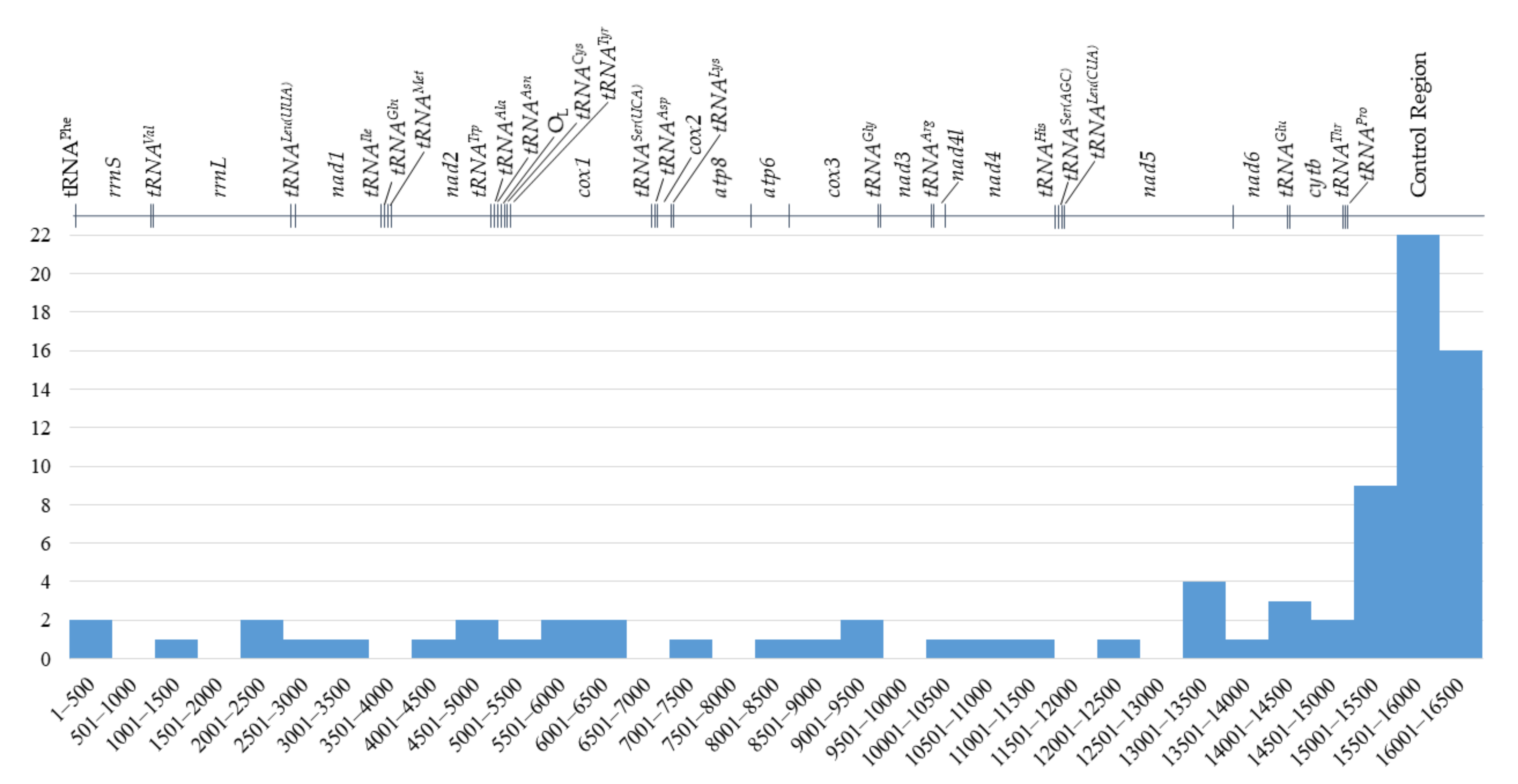
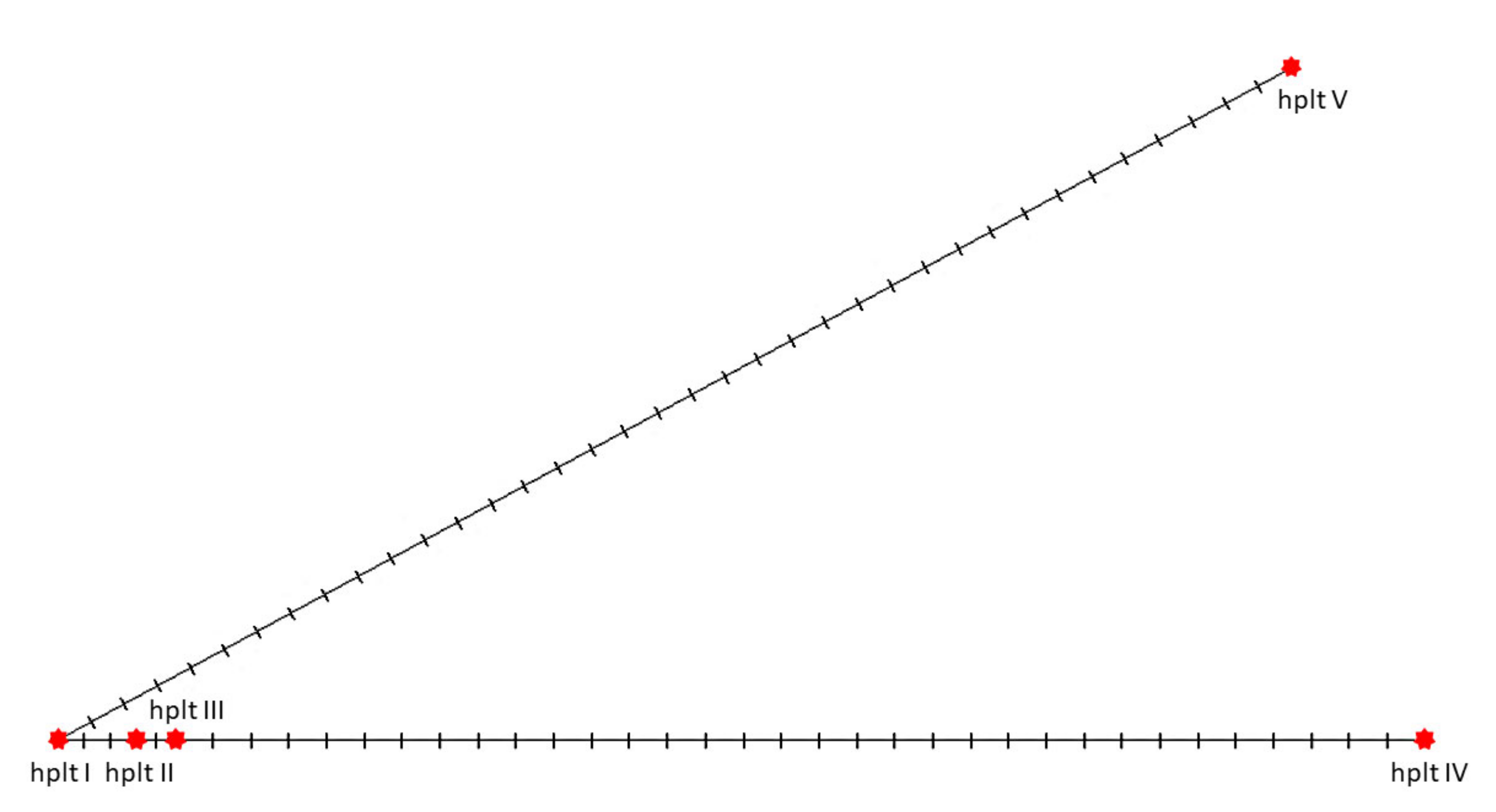
| Gene | Position | Size (bp) | Amino Acids Count 1 | Codon | Anticodon | Intergenic Nucleotide (bp) 2 | Strand 3 | ||
|---|---|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||||
| tRNAPhe | 1 | 69 | 69 | GAA | 0 | H | |||
| rrnS | 70 | 1028 | 959 | 0 | H | ||||
| tRNAVal | 1029 | 1096 | 68 | TAC | −2 | H | |||
| rrnL | 1095 | 2665 | 1571 | 0 | H | ||||
| tRNALeu(UUA) | 2666 | 2740 | 75 | TAA | 2 | H | |||
| nad1 | 2743 | 3698 | 956 | 318 | ATG | TA 5 | 0 | H | |
| tRNAIle | 3699 | 3767 | 69 | GAT | −3 | H | |||
| tRNAGln | 3765 | 3838 | 74 | TTG | 1 | L | |||
| tRNAMet | 3840 | 3908 | 69 | CAT | 0 | H | |||
| nad2 | 3909 | 4950 | 1042 | 347 | ATT 4 | T 5 | 0 | H | |
| tRNATrp | 4951 | 5017 | 67 | TCA | 10 | H | |||
| tRNAAla | 5028 | 5096 | 69 | TGC | 1 | L | |||
| tRNAAsn | 5098 | 5170 | 73 | GTT | 0 | L | |||
| OL | 5171 | 5205 | 35 | −3 | L | ||||
| tRNACys | 5203 | 5269 | 67 | GCA | 1 | L | |||
| tRNATyr | 5271 | 5338 | 68 | GTA | 1 | L | |||
| cox1 | 5340 | 6884 | 1545 | 514 | ATG | TAA | −3 | H | |
| tRNASer(UCA) | 6882 | 6950 | 69 | TGA | 5 | L | |||
| tRNAAsp | 6956 | 7022 | 67 | GTC | 0 | H | |||
| cox2 | 7023 | 7706 | 684 | 227 | ATG | TAA | 3 | H | |
| tRNALys | 7710 | 7776 | 67 | TTT | 1 | H | |||
| atp8 | 7778 | 7981 | 204 | 67 | ATG | TAA | −43 | H | |
| atp6 | 7939 | 8619 | 681 | 226 | ATG | TAA | −1 | H | |
| cox3 | 8619 | 9402 | 784 | 261 | ATG | T 5 | 0 | H | |
| tRNAGly | 9403 | 9471 | 69 | TCC | 0 | H | |||
| nad3 | 9472 | 9818 | 347 | 115 | ATA 4 | TA 5 | 0 | H | |
| tRNAArg | 9819 | 9886 | 68 | TCG | 0 | H | |||
| nad4l | 9887 | 10,183 | 297 | 98 | ATG | TAA | −7 | H | |
| nad4 | 10,177 | 11,554 | 1378 | 459 | ATG | T 5 | 0 | H | |
| tRNAHis | 11,555 | 11,623 | 69 | GTG | 0 | H | |||
| tRNASer(AGC) | 11,624 | 11,685 | 62 | GCT | 0 | H | |||
| tRNALeu(CUA) | 11,686 | 11,755 | 70 | TAG | 0 | H | |||
| nad5 | 11,756 | 13,576 | 1821 | 606 | ATT 4 | TAA | −17 | H | |
| nad6 | 13,560 | 14,090 | 531 | 176 | ATG | TAA | 3 | L | |
| tRNAGlu | 14,094 | 14,162 | 69 | TTC | 4 | L | |||
| Cytb | 14,167 | 15,306 | 1140 | 379 | ATG | AGA | 0 | H | |
| tRNAThr | 15,307 | 15,374 | 68 | TGT | −1 | H | |||
| tRNAPro | 15,374 | 15,439 | 66 | TGG | 0 | L | |||
| CR | 15,440 | 16,504 | 1065 | 0 | H | ||||
| Amino Acid | Codon | % | Amino Acid | Codon | % | Amino Acid | Codon | % | Amino Acid | Codon | % |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F | TTT | 2.65 | S | TCT | 1.42 | A | GCA | 2.39 | D | GAT | 0.58 |
| TTC | 3.44 | TCC | 1.79 | GCG | 0.18 | GAC | 1.18 | ||||
| L | TTA | 3.31 | TCA | 2.63 | Y | TAT | 1.39 | E | GAA | 2.08 | |
| TTG | 0.58 | TCG | 0.13 | TAC | 2.08 | GAG | 0.47 | ||||
| CTT | 1.89 | AGT | 0.47 | * | TAA | 0.32 | C | TGT | 0.29 | ||
| CTC | 1.92 | AGC | 1.05 | TAG | 0.00 | TGC | 0.39 | ||||
| CTA | 7.20 | P | CCT | 1.16 | AGA | 0.03 | W | TGA | 2.29 | ||
| CTG | 1.10 | CCC | 1.76 | AGG | 0.00 | TGG | 0.47 | ||||
| I | ATT | 3.99 | CCA | 2.05 | H | CAT | 0.60 | R | CGT | 0.16 | |
| ATC | 4.65 | CCG | 0.13 | CAC | 1.92 | CGC | 0.42 | ||||
| M | ATA | 5.28 | T | ACT | 2.08 | Q | CAA | 2.10 | CGA | 1.00 | |
| ATG | 1.18 | ACC | 2.15 | CAG | 0.21 | CGG | 0.11 | ||||
| V | GTT | 0.84 | ACA | 3.73 | N | AAT | 1.52 | G | GGT | 0.79 | |
| GTC | 1.10 | ACG | 0.42 | AAC | 2.44 | GGC | 1.26 | ||||
| GTA | 2.42 | A | GCT | 1.47 | K | AAA | 2.44 | GGA | 2.68 | ||
| GTG | 0.39 | GCC | 2.68 | AAG | 0.21 | GGG | 0.92 |
| Gene | ENC 1 | CBI 2 | G+C3s 3 |
|---|---|---|---|
| nad1 | 43.567 | 0.464 | 0.379 |
| nad2 | 31.270 | 0.597 | 0.313 |
| cox1 | 46.359 | 0.321 | 0.338 |
| cox2 | 45.559 | 0.417 | 0.326 |
| atp8 | 41.945 | 0.647 | 0.175 |
| atp6 | 39.575 | 0.454 | 0.382 |
| cox3 | 39.337 | 0.482 | 0.383 |
| nad3 | 44.332 | 0.561 | 0.288 |
| nad4l | 39.698 | 0.585 | 0.400 |
| nad4 | 43.570 | 0.454 | 0.393 |
| nad5 | 42.770 | 0.429 | 0.409 |
| nad6 | 39.644 | 0.565 | 0.243 |
| Cytb | 43.446 | 0.477 | 0.454 |
| overall of PCGs | 44.341 | 0.367 | 0.404 |
| Feature | tRNAPhe | tRNAVal | tRNALeu(UUA) | tRNAIle | tRNAGln | tRNAMet | tRNATrp | tRNAAla | tRNAAsn | tRNACys | tRNATyr | tRNASer(UCA) | tRNAAsp | tRNALys | tRNAGly | tRNAArg | tRNAHis | tRNASer(AGC) | tRNALeu(CUA) | tRNAGlu | tRNAThr | tRNAPro | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unmatched Base Pairs | U-G pairing 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 4 | 1 | 2 | 2 | 4 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 3 | 0 | 5 |
| Mismatching | A-A | A-G, U-U, U-U | A-C | U-U | A-A, A-C | U-U | |||||||||||||||||
| Stem (Arm) Size (bp) | Acceptor | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| TΨC | 5 | 4 | 5 | 5 | 3 | 5 | 5 | 5 | 5 | 4 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 5 | 5 | 5 | |
| Anticodon | 5 | 5 | 4 | 5 | 4 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 5 | 5 | 5 | 5 | |
| DHU | 4 | 4 | 4 | 3 | 4 | 4 | 4 | 4 | 3 | 4 | 3 | 3 | 4 | 5 | 4 | 4 | 4 | 0 | 4 | 4 | 3 | 4 | |
| Species | Family | Size (bp) | GC% | AT-Skew | GC-Skew | GenBank Accession No. | Percent Identity | Reference 1 |
|---|---|---|---|---|---|---|---|---|
| Mustela lutreola | Mustelidae | 16,504 | 39.9 | 0.093 | −0.308 | MW148603 | 100.00% | this study |
| Mustela putorius | 16,523 | 39.8 | 0.091 | −0.308 | HM106318 | 99.04% | d.s. | |
| Mustela eversmanni | 16,463 | 40.0 | 0.091 | −0.305 | NC_028013 | 98.72% | d.s. | |
| Mustela nigripes | 16,556 | 39.9 | 0.095 | −0.310 | NC_024942 | 98.31% | [48] | |
| Mustela sibirica | 16,558 | 39.8 | 0.092 | −0.303 | MN206976 | 96.99% | [49] | |
| Mustela itatsi | 16,027 | 39.5 | 0.091 | −0.304 | NC_034330 | 95.24% | [50] | |
| Mustela altaica | 16,521 | 39.7 | 0.086 | −0.301 | NC_021751 | 91.96% | [51] | |
| Mustela nivalis | 16,502 | 40.0 | 0.087 | −0.296 | MF459691 | 91.50% | [52] | |
| Mustela ermine | 16,500 | 40.1 | 0.112 | −0.324 | MW257230 | 91.32% | d.s. | |
| Mustela kathiah | 16,552 | 38.9 | 0.088 | −0.301 | HM106320 | 89.29% | d.s. | |
| Mustela frenata | 16,543 | 39.2 | 0.096 | −0.314 | HM106321 | 88.59% | d.s. | |
| Neogale vison | 16,552 | 38.6 | 0.093 | −0.312 | NC_020641 | 88.50% | [53] | |
| Lutra lutra | 16,536 | 42.0 | 0.111 | −0.310 | NC_011358 | 86.07% | d.s. | |
| Gulo gulo | 16,541 | 41.1 | 0.093 | −0.304 | NC_009685 | 86.02% | [54] | |
| Martes foina | 16,530 | 41.9 | 0.105 | −0.310 | NC_020643 | 86.00% | d.s. | |
| Meles meles | 16.442 | 38.9 | 0.091 | −0.316 | NC_011125 | 85.46% | [54] | |
| Procyon lotor | Procyonidae | 16,623 | 39.6 | 0.095 | −0.288 | NC_009126 | 82.33% | d.s. |
| Ailurus flugens | Ailuridae | 16,374 | 37.5 | 0.054 | −0.292 | NC_009691 | 82.30% | [55] |
| Canis lupus | Canidae | 16,727 | 39.7 | 0.048 | −0.287 | NC_002008 | 81.13% | [56] |
| Vulpes vulpes | 16,723 | 40.7 | 0.058 | −0.276 | KP342452 | 79.47% | [57] | |
| Ursus arctos | Ursidae | 17,020 | 41.3 | 0.053 | −0.239 | NC_003427 | 80.81% | [58] |
| Ailuropoda melanoleuca | 16,805 | 38.8 | 0.038 | −0.227 | NC_009492 | 80.73% | [59] |
| Species | M.er. | M.s. | M.ng. | M.k. | M.f. | M.p. | M.a. | M.nv. | M.ev. | M.i. | M.l. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M.er. | - | ||||||||||
| M.s. | 0.5967 | - | |||||||||
| M.ng. | 0.5989 | 0.6459 | - | ||||||||
| M.k. | 0.7195 | 0.6988 | 0.7102 | - | |||||||
| M.f. | 0.6944 | 0.6602 | 0.7204 | 0.6936 | - | ||||||
| M.p. | 0.6266 | 0.6346 | 0.5102 | 0.7240 | 0.3992 | - | |||||
| M.a. | 0.3230 | 0.7213 | 0.6900 | 0.7188 | 0.6077 | 0.7205 | - | ||||
| M.nv. | 0.7311 | 0.7194 | 0.7433 | 0.7200 | 0.7295 | 0.7406 | 0.7319 | - | |||
| M.ev. | 0.6372 | 0.6250 | 0.4969 | 0.7311 | 0.6843 | 0.4787 | 0.7364 | 0.7363 | - | ||
| M.i. | 0.6622 | 0.6429 | 0.6100 | 0.7184 | 0.6930 | 0.5848 | 0.7205 | 0.7341 | 0.5674 | - | |
| M.l. | 0.6346 | 0.6238 | 0.4975 | 0.7321 | 0.6845 | 0.4790 | 0.7361 | 0.7349 | 0.0241 | 0.5660 | - |
| Position (bp) 1 | C 2 | Hom. 3 | p-Value | Region |
|---|---|---|---|---|
| 15,348–15,514 | 0.861 | 0.957 | 0.0009 | ETAS domain (5′-end) |
| 15,677–15,825 | 0.865 | 0.951 | 0.0011 | CD domain (E-box, C-box, D-box) |
| 15,830–16,038 | 0.899 | 0.967 | 0 | CD domain (B-box, 3′-end) + CSB domain (5′-end, CSB1) |
| 16,421–16,476 | 0.862 | 0.962 | 0.0423 | CSB domain (LSP, HSP, 3′-end) |
| Sample Name | Sex | Age 1 | Date of Death | GenBank Accession No. |
|---|---|---|---|---|
| M.l.-M540-2016 | ♂ | 62 | June 2013 | MT304869 |
| M.l.-M598-2016 | ♂ | 61 | May 2014 | MW197423 |
| M.l.-M810-2016 | ♂ | 6 | November 2012 | MW148603 |
| M.l.-F490-2016 | ♀ | 67 | November 2012 | MW197424 |
| M.l.-F516-2016 | ♀ | 73 | May 2014 | MW197425 |
| M.l.-F835-2016 | ♀ | 20 | February 2014 | MW197426 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skorupski, J. Characterisation of the Complete Mitochondrial Genome of Critically Endangered Mustela lutreola (Carnivora: Mustelidae) and Its Phylogenetic and Conservation Implications. Genes 2022, 13, 125. https://doi.org/10.3390/genes13010125
Skorupski J. Characterisation of the Complete Mitochondrial Genome of Critically Endangered Mustela lutreola (Carnivora: Mustelidae) and Its Phylogenetic and Conservation Implications. Genes. 2022; 13(1):125. https://doi.org/10.3390/genes13010125
Chicago/Turabian StyleSkorupski, Jakub. 2022. "Characterisation of the Complete Mitochondrial Genome of Critically Endangered Mustela lutreola (Carnivora: Mustelidae) and Its Phylogenetic and Conservation Implications" Genes 13, no. 1: 125. https://doi.org/10.3390/genes13010125
APA StyleSkorupski, J. (2022). Characterisation of the Complete Mitochondrial Genome of Critically Endangered Mustela lutreola (Carnivora: Mustelidae) and Its Phylogenetic and Conservation Implications. Genes, 13(1), 125. https://doi.org/10.3390/genes13010125






