C3435T Polymorphism of the ABCB1 Gene in Polish Patients with Inflammatory Bowel Disease: A Case–Control and Meta-Analysis Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Controls
2.2. Genotyping
2.3. Phenotypic Assessment
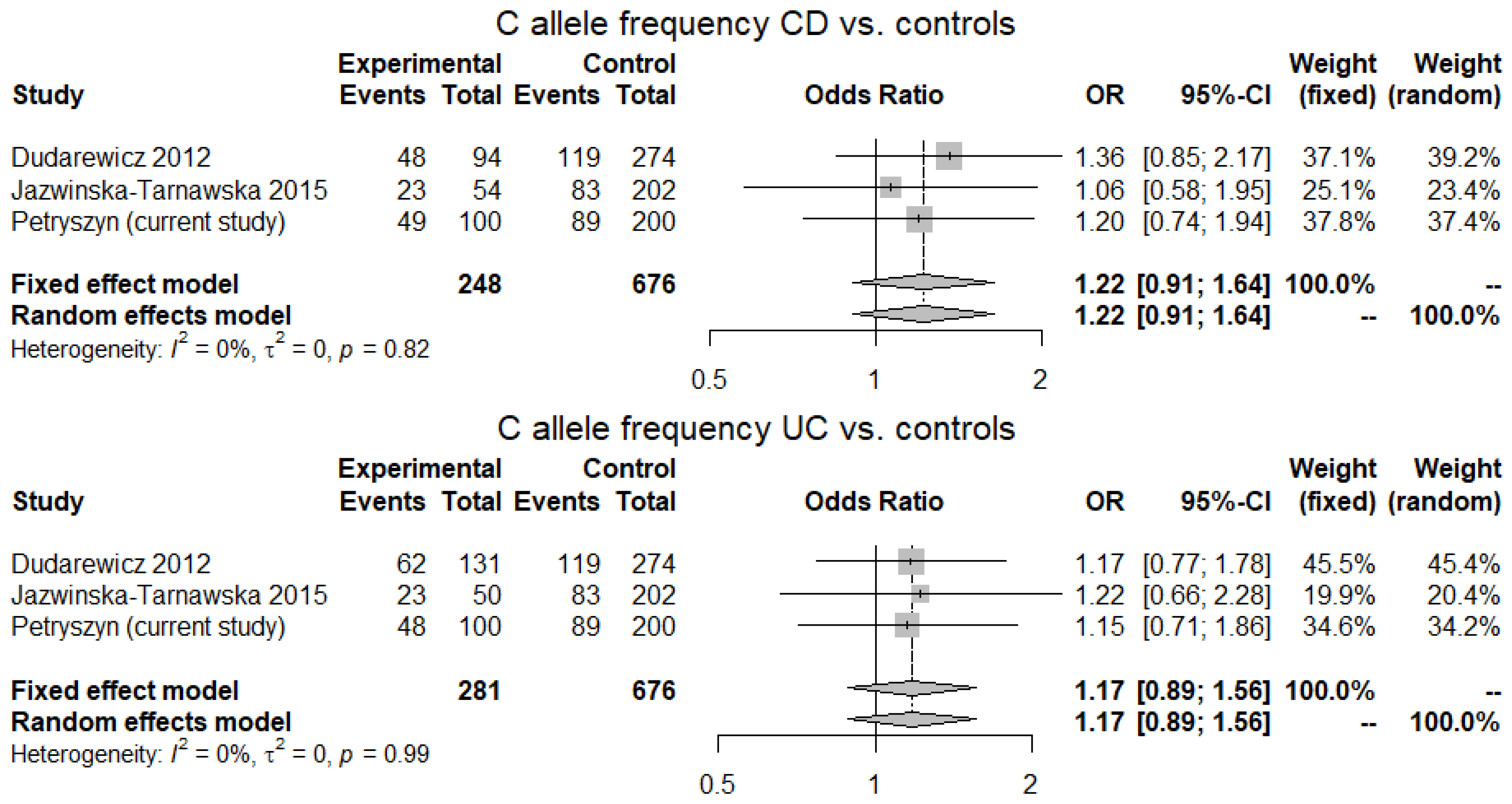
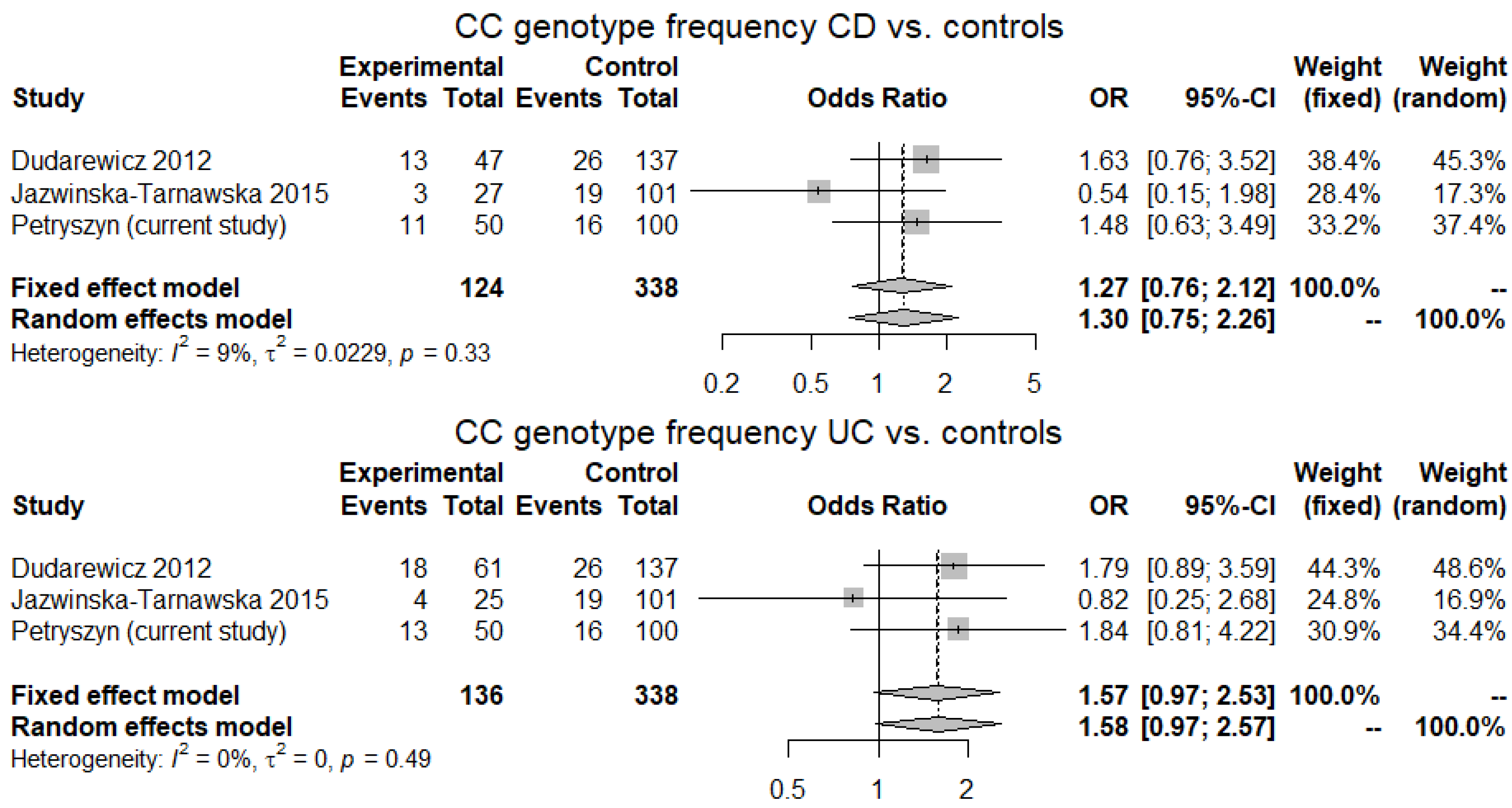
2.4. Meta-Analysis
2.5. Statistical Analysis
3. Results
3.1. Demographics and Clinical Characteristics
3.2. Effect of ABCB1 C3435T Polymorphism on Disease Susceptibility
3.3. Genotype–Phenotype Analysis
3.4. Meta-Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, M.; Gönczi, L.; Lakatos, P.L.; Burisch, J. The burden of inflammatory bowel disease in Europe in 2020. J. Crohn’s Colitis 2021, jjab029. [Google Scholar] [CrossRef]
- Guan, Q. A Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J. Immunol. Res. 2019, 2019, 7247238. [Google Scholar] [CrossRef]
- Magro, F.; Langner, C.; Driessen, A.; Ensari, A.; Geboes, K.; Mantzaris, G.J.; Villanacci, V.; Becheanu, G.; Nunes, P.B.; Cathomas, G.; et al. European consensus on the histopathology of inflammatory bowel disease. J. Crohn’s Colitis 2013, 7, 827–851. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Shivananda, S.; Lennard-Jones, J.; Logan, R.; Fear, N.; Price, A.; Carpenter, L.; Van Blankenstein, M. Incidence of inflammatory bowel disease across Europe: Is there a difference between north and south? Results of the European collaborative study on inflammatory bowel disease (EC-IBD). Gut 1996, 39, 690–697. [Google Scholar] [CrossRef]
- Kaplan, G.G.; Windsor, J.W. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 56–66. [Google Scholar] [CrossRef]
- Orholm, M.; Binder, V.; Sorensen, T.I.A.; Rasmussen, L.P.; Kyvik, K.O. Concordance of inflammatory bowel disease among Danish twins: Results of a nationwide study. Scand. J. Gastroenterol. 2000, 35, 1075–1081. [Google Scholar] [CrossRef]
- Halfvarson, J.; Bodin, L.; Tysk, C.; Lindberg, E.; Järnerot, G. Inflammatory bowel disease in a Swedish twin cohort: A long- term follow-up of concordance and clinical characteristics. Gastroenterology 2003, 124, 1767–1773. [Google Scholar] [CrossRef]
- Ogura, Y.; Bonen, D.; Inohara, N.; Nicolae, D.; Chen, F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.; et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cézard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’morain, C.A.; Gassull, M.; et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef]
- Bonen, D.K.; Ogura, Y.; Nicolae, D.L.; Inohara, N.; Saab, L.; Tanabe, T.; Chen, F.F.; Foster, S.J.; Duerr, R.H.; Brant, S.R.; et al. Crohn’s disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology 2003, 124, 140–146. [Google Scholar] [CrossRef]
- Hampe, J.; Grebe, J.; Nikolaus, S.; Solberg, C.; Croucher, P.J.P.; Mascheretti, S.; Jahnsen, J.; Moum, B.; Klump, B.; Krawczak, M.; et al. Association of NOD2 (CARD 15) genotype with clinical course of Crohn’s disease: A cohort study. Lancet 2002, 359, 1661–1665. [Google Scholar] [CrossRef]
- Schäffler, H.; Geiss, D.; Gittel, N.; Rohde, S.; Huth, A.; Glass, Ä.; Brandhorst, G.; Jaster, R.; Lamprecht, G. Mutations in the NOD2 gene are associated with a specific phenotype and lower anti-tumor necrosis factor trough levels in Crohn’s disease. J. Dig. Dis. 2018, 19, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Juanola, O.; Moratalla, A.; Gutiérrez, A.; Sempere, L.; Zapater, P.; Giménez, P.; Almenta, I.; Peiró, G.; González-Navajas, J.M.; Such, J.F.; et al. Anti-TNF-α loss of response is associated with a decreased percentage of FoxP3+ T cells and a variant NOD2 genotype in patients with Crohn’s disease. J. Gastroenterol. 2015, 50, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Uniken Venema, W.T.C.; Voskuil, M.D.; Dijkstra, G.; Weersma, R.K.; Festen, E.A.M. The genetic background of inflammatory bowel disease: From correlation to causality. J. Pathol. 2017, 241, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Mahid, S.S.; Minor, K.S.; Soto, R.E.; Hornung, C.A.; Galandiuk, S. Smoking and inflammatory bowel disease: A meta-analysis. Mayo Clin. Proc. 2006, 81, 1462–1471. [Google Scholar] [CrossRef]
- Higuchi, L.M.; Khalili, H.; Chan, A.T.; Richter, J.M.; Bousvaros, A.; Fuchs, C.S. A prospective study of cigarette smoking and the risk of inflammatory bowel disease in women. Am. J. Gastroenterol. 2012, 107, 1399–1406. [Google Scholar] [CrossRef]
- Porter, C.K.; Tribble, D.R.; Aliaga, P.A.; Halvorson, H.A.; Riddle, M.S. Infectious Gastroenteritis and Risk of Developing Inflammatory Bowel Disease. Gastroenterology 2008, 135, 781–786. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Higuchi, L.M.; Huang, E.S.; Khalili, H.; Richter, J.M.; Fuchs, C.S.; Chan, A.T. Aspirin, Nonsteroidal Anti-inflammatory Drug Use, and Risk for Crohn. Ann. Intern. Med. 2012, 156, 350–359. [Google Scholar] [CrossRef]
- Wawrzyniak, M.; Scharl, M. Genetics and epigenetics of inflammatory bowel disease. Swiss Med. Wkly. 2018, 148, w14671. [Google Scholar] [CrossRef]
- Ray, G.; Longworth, M.S. Epigenetics, DNA Organization, and Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2019, 25, 235–247. [Google Scholar] [CrossRef]
- Hornschuh, M.; Wirthgen, E.; Wolfien, M.; Singh, K.P.; Wolkenhauer, O.; Däbritz, J. The role of epigenetic modifications for the pathogenesis of Crohn’s disease. Clin. Epigenet. 2021, 13, 108. [Google Scholar] [CrossRef]
- Borst, P.; Evers, R.; Kool, M.; Wijnholds, J. A family of drug transporters: The multidrug resistance-associated proteins. J. Natl. Cancer Inst. 2000, 92, 1295–1302. [Google Scholar] [CrossRef]
- Marzolini, C.; Paus, E.; Buclin, T.; Kim, R.B. Polymorphisms in human MDR1 (P-glycoprotein): Recent advances and clinical relevance. Clin. Pharmacol. Ther. 2004, 75, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, I.; Kataoka, I.; Morishita, Y.; Hamada, H.; Tsuruo, T.; Itoyama, S.; Mori, S. Tissue Distribution of P-Glycoprotein Encoded by a Multidrug-Resistant Gene as Revealed by a Monoclonal Antibody, mrk 16. Cancer Res. 1988, 48, 1926–1929. [Google Scholar]
- Drozdzik, M.; Czekawy, I.; Oswald, S.; Drozdzik, A. Intestinal drug transporters in pathological states: An overview. Pharmacol. Rep. 2020, 72, 1173–1194. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Hrycyna, C.A.; Schoenlein, P.V.; Germann, U.A.; Pastan, I. Genetic analysis of the multidrug transporter. Annu. Rev. Genet. 1995, 29, 607–649. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M.; Eichelbaum, M.; Fromm, M.F. Genetic Polymorphisms of the Human MDR1 Drug Transporter. Annu. Rev. Pharmacol. Toxicol. 2003, 43, 285–307. [Google Scholar] [CrossRef]
- Hoffmeyer, S.; Burk, O.; Von Richter, O.; Arnold, H.P.; Brockmöller, J.; Johne, A.; Cascorbi, I.; Gerloff, T.; Roots, I.; Eichelbaum, M.; et al. Functional polymorphisms of the human multidrug-resistance gene: Multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 3473–3478. [Google Scholar] [CrossRef]
- Panwala, C.M.; Jones, J.C.; Viney, J.L. A novel model of inflammatory bowel disease: Mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J. Immunol. 1998, 161, 5733–5744. [Google Scholar]
- Petryszyn, P.W.; Wiela-Hojeńska, A. The importance of the polymorphisms of the ABCB1 gene in disease susceptibility, behavior and response to treatment in inflammatory bowel disease: A literature review. Adv. Clin. Exp. Med. 2018, 27, 1459–1463. [Google Scholar] [CrossRef] [PubMed]
- Onnie, C.M.; Fisher, S.A.; Pattni, R.; Sanderson, J.; Forbes, A.; Lewis, C.M.; Mathew, C.G. Associations of allelic variants of the multidrug resistance gene (ABCB1 or MDR1) and inflammatory bowel disease and their effects on disease behavior: A case-control and meta-analysis study. Inflamm. Bowel Dis. 2006, 12, 263–271. [Google Scholar] [CrossRef]
- Annese, V.; Valvano, M.; Palmieri, O.; Latiano, A.; Bossa, F.; Andriulli, A. Multidrug resistance 1 gene in inflammatory bowel disease: A meta-analysis. J. Gastroenterol. 2006, 12, 3636–3644. [Google Scholar] [CrossRef]
- Farrell, R.J.; Murphy, A.; Long, A.; Donnelly, S.; Cherikuri, A.; O’Toole, D.; Mahmud, N.; Keeling, P.W.N.; Weir, D.G.; Kelleher, D. High multidrug resistance (P-glycoprotein 170) expression in inflammatory bowel disease patients who fail medical therapy. Gastroenterology 2000, 118, 279–288. [Google Scholar] [CrossRef]
- Gomollón, F.; Dignass, A.; Annese, V.; Tilg, H.; Van Assche, G.; Lindsay, J.O.; Peyrin-Biroulet, L.; Cullen, G.J.; Daperno, M.; Kucharzik, T.; et al. 3rd European evidence-based consensus on the diagnosis and management of Crohn’s disease 2016: Part 1: Diagnosis and medical management. J. Crohn’s Colitis 2017, 11, 3–25. [Google Scholar] [CrossRef]
- Magro, F.; Gionchetti, P.; Eliakim, R.; Ardizzone, S.; Armuzzi, A.; Barreiro-de Acosta, M.; Burisch, J.; Gecse, K.B.; Hart, A.L.; Hindryckx, P.; et al. Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: Definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders. J. Crohn’s Colitis 2017, 11, 649–670. [Google Scholar] [CrossRef]
- Siegmund, W.; Ludwig, K.; Giessmann, T.; Dazert, P.; Schroeder, E.; Sperker, B.; Warzok, R.; Kroemer, H.K.; Cascorbi, I. The effects of the human MDR1 genotype on the expression of duodenal P-glycoprotein and disposition of the probe drug talinolol. Clin. Pharmacol. Ther. 2002, 72, 572–583. [Google Scholar] [CrossRef]
- Satsangi, J.; Silverberg, M.S.; Vermeire, S.; Colombel, J.F. The Montreal classification of inflammatory bowel disease: Controversies, consensus, and implications. Gut 2006, 55, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Jaźwińska-Tarnawska, E.; Jęśkowiak, I.; Waszczuk, E.; Mulak, A.; Głowacka, K.; Hurkacz, M.; Paradowski, L.; Zaleska, Z.; Wiela-Hojeńska, A. Genetic polymorphism of ABCB1 gene (C3435T) in patients with inflammatory bowel diseases. is there any gender dependency? Pharmacol. Rep. 2015, 67, 294–298. [Google Scholar] [CrossRef]
- Dudarewicz, M.; Barańska, M.; Rychlik-Sych, M.; Trzciński, R.; Dziki, A.; Skreţkowicz, J. C3435T polymorphism of the ABCB1/MDR1 gene encoding P-glycoprotein in patients with inflammatory bowel disease in a Polish population. Pharmacol. Rep. 2012, 64, 343–350. [Google Scholar] [CrossRef]
- Williams, C.N.; Kocher, K.; Lander, E.S.; Daly, M.J.; Rioux, J.D. Using a genome-wide scan and meta-analysis to identify a novel IBD locus and confirm previously identified IBD loci. Inflamm. Bowel Dis. 2002, 8, 375–381. [Google Scholar] [CrossRef]
- Barrett, J.C.; Hansoul, S.; Nicolae, D.L.; Cho, J.H.; Duerr, R.H.; Rioux, J.D.; Brant, S.R.; Silverberg, M.S.; Taylor, K.D.; Barmada, M.M.; et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008, 40, 955–962. [Google Scholar] [CrossRef]
- Ye, B.D.; McGovern, D.P. Genetic variation in IBD: Progress, clues to pathogenesis and possible clinical utility. Expert Rev. Clin. Immunol. 2016, 12, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.; Armuzzi, A.; Bunce, M.; MulcahyHawes, K.; Marshall, S.E.; Orchard, T.R.; Crawshaw, J.; Large, O.; De Silva, A.; Cook, J.T.; et al. The molecular classification of the clinical manifestations of Crohn’s disease. Gastroenterology 2002, 122, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M.; Schaeffeler, E.; Marx, C.; Fromm, M.F.; Kaskas, B.; Metzler, J.; Stange, E.; Herfarth, H.; Schoelmerich, J.; Gregor, M.; et al. Association between the C3435T MDR1 gene polymorphism and susceptibility for ulcerative colitis. Gastroenterology 2003, 124, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Ho, G.T.; Nimmo, E.R.; Tenesa, A.; Fennell, J.; Drummond, H.; Mowat, C.; Arnott, I.D.; Satsangi, J. Allelic variations of the multidrug resistance gene determine susceptibility and disease behavior in ulcerative colitis. Gastroenterology 2005, 128, 288–296. [Google Scholar] [CrossRef]
- Brinar, M.; Cukovic-Cavka, S.; Bozina, N.; Ravic, K.G.; Markos, P.; Ladic, A.; Cota, M.; Krznaric, Z.; Vucelic, B. MDR1 polymorphisms are associated with inflammatory bowel disease in a cohort of Croatian IBD patients. BMC Gastroenterol. 2013, 13, 1. [Google Scholar] [CrossRef]
- Farnood, A.; Naderi, N.; Moghaddam, S.J.M.; Noorinayer, B.; Firouzi, F.; Aghazadeh, R.; Daryani, N.E.; Zali, M.R. The frequency of C3435T MDR1 gene polymorphism in Iranian patients with ulcerative colitis. Int. J. Colorectal Dis. 2007, 22, 999–1003. [Google Scholar] [CrossRef]
- Croucher, P.; Mascheretti, S.; Foelsch, U.; Hampe, J.; Schreiber, S. Lack of Association Between the C3435T MDR1 Gene Polymorphism and Inflammatory Bowel Disease in Two Independent Northern European Populations Dear. Gastroenterology 2003, 125, 1919–1920. [Google Scholar] [CrossRef]
- Brant, S.R.; Panhuysen, C.I.M.; Nicolae, D.; Reddy, D.M.; Bonen, D.K.; Karaliukas, R.; Zhang, L.; Swanson, E.; Datta, L.W.; Moran, T.; et al. MDR1 Ala893 Polymorphism Is Associated with Inflammatory Bowel Disease. Am. J. Hum. Genet. 2003, 73, 1282–1292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Potočnik, U.; Ferkolj, I.; Glavač, D.; Dean, M. Polymorphisms in multidrug resistance 1 (MDR1) gene are associated with refractory Crohn disease and ulcerative colitis. Genes Immun. 2004, 5, 530–539. [Google Scholar] [CrossRef]
- Palmieri, O.; Latiano, A.; Valvano, R.; D’Incà, R.; Vecchi, M.; Sturniolo, G.C.; Saibeni, S.; Bossa, F.; Latiano, T.; Devoto, M.; et al. Multidrug resistance 1 gene polymorphisms are not associated with inflammatory bowel disease and response to therapy in Italian patients. Aliment. Pharmacol. Ther. 2005, 22, 1129–1138. [Google Scholar] [CrossRef]
- Ieiri, I.; Takane, H.; Otsubo, K. The MDR1 (ABCB1) gene polymorphism and its clinical implications. Clin. Pharmacokinet. 2004, 43, 553–576. [Google Scholar] [CrossRef]
- Glas, J.; Török, H.P.; Schiemann, U.; Folwaczny, C. MDR1 Gene Polymorphism in Ulcerative Colitis. Gastroenterology 2004, 126, 367. [Google Scholar] [CrossRef]
- Urcelay, E.; Mendoza, J.L.; Martín, M.C.; Mas, A.; Martínez, A.; Taxonera, C.; Fernandez-Arquero, M.; Díaz-Rubio, M.; De La Concha, E.G. MDR1 Gene: Susceptibility in Spanish Crohn’s disease and ulcerative colitis patients. Inflamm. Bowel Dis. 2006, 12, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.B.; Leake, B.F.; Choo, E.F.; Dresser, G.K.; Kubba, S.V.; Schwarz, U.I.; Taylor, A.; Xie, H.G.; McKinsey, J.; Zhou, S.; et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin. Pharmacol. Ther. 2001, 70, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Illmer, T.; Schuler, U.S.; Thiede, C.; Schwarz, U.I.; Kim, R.B.; Gotthard, S.; Freund, D.; Schäkel, U.; Ehninger, G.; Schaich, M. MDR1 gene polymorphisms affect therapy outcome in acute myeloid leukemia patients. Cancer Res. 2002, 62, 4955–4962. [Google Scholar]
- Nakamura, T.; Sakaeda, T.; Horinouchi, M.; Tamura, T.; Aoyama, N.; Shirakawa, T.; Matsuo, M.; Kasuga, M.; Okumura, K. Effect of the mutation (C3435t) at exon 26 of the MDR1 gene on expression level of MDR1 messenger ribonucleic acid in duodenal enterocytes of healthy Japanese subjects. Clin. Pharmacol. Ther. 2002, 71, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Ardizzone, S.; Maconi, G.; Bianchi, V.; Russo, A.; Colombo, E.; Cassinotti, A.; Penati, C.; Tenchini, M.L.; Porro, G.B. Multidrug resistance 1 gene polymorphism and susceptibility to inflammatory bowel disease. Inflamm. Bowel Dis. 2007, 13, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.F.; Chen, B.L.; Zhang, Q.S.; Zhu, Z.H.; Hu, B.; He, Y.; Gao, X.; Wang, Y.M.; Hu, P.J.; Chen, M.H.; et al. Contribution of MDR1 gene polymorphisms on IBD predisposition and response to glucocorticoids in IBD in a Chinese population. J. Dig. Dis. 2015, 16, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.L.; Urcelay, E.; Lana, R.; Martín, M.C.; López, N.; Guijarro, L.G.; Mayol, J.A.; Taxonera, C.; De La Concha, E.G.; Peña, A.S.; et al. MDR1 polymorphisms and response to azathioprine therapy in patients with Crohn’s disease. Inflamm. Bowel Dis. 2007, 13, 585–590. [Google Scholar] [CrossRef] [PubMed]
| Studies | Allele | Genotype | ||||
|---|---|---|---|---|---|---|
| C | T | CC | CT | TT | ||
| Jazwinska-Tarnawska 2015 [39] | CD = 27 | 23 | 31 | 3 | 17 | 7 |
| UC = 25 | 23 | 27 | 4 | 15 | 6 | |
| Controls = 101 | 83 | 119 | 19 | 45 | 37 | |
| Dudarewicz 2012 [40] | CD = 47 | 48 | 46 | 13 | 22 | 12 |
| UC = 61 | 62 | 60 | 18 | 26 | 17 | |
| Controls = 137 | 119 | 155 | 26 | 67 | 44 | |
| Characteristics | CD | UC | Controls | |
|---|---|---|---|---|
| Sex (M/F) (%) | 25/25 (50/50) | 32/18 (64/36) | 45/55 (45/55) | |
| Mean age (±SD) | 35.8 (±13.7) | 38.5 (±14.9) | 37.2 (±12.5) | |
| Mean age at diagnosis (±SD) | 27.6 (±13.8) | 29.3 (±13.1) | ||
| Disease location (CD) | L1 (ileal) | 12 | ||
| L2 (colonic) | 12 | |||
| L3 (ileocolonic) | 26 | |||
| Disease behaviour (CD) | B1 (inflammatory) | 10 | ||
| B2 (stricturing) | 35 | |||
| B3 (penetrating) | 15 | |||
| Disease extent (UC) | E1 (ulcerative proctitis) | 2 | ||
| E2 (left-sided colitis) | 16 | |||
| E3 (pancolitis) | 32 | |||
| Severe disease (UC) | 13 | |||
| 5-ASA | 47 | 48 | ||
| Immunosuppresant therapy | 31 | 22 | ||
| Corticosteroid use | 31 | 35 | ||
| Anti-TNF | 12 | 5 | ||
| IBD surgery | 17 | 7 | ||
| Subjects | Allele | Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C | OR (95% CI) | T | OR (95% CI) | CC | OR (95% CI) | CT | OR (95% CI) | TT | OR (95% CI) | |
| CD = 50 | 49 | 1.2 (0.72–1.99) | 51 | 0.84 (0.5–1.39) | 11 | 1.48 (0.56–3.76) | 27 | 0.92 (0.44–1.94) | 12 | 0.81 (0.34–1.88) |
| UC = 50 | 48 | 1.15 (0.69–1.92) | 52 | 0.87 (0.52–1.45) | 13 | 1.84 (0.73–4.55) | 22 | 0.62 (0.29–1.29) | 15 | 1.1 (0.48–2.46) |
| Controls = 100 | 89 | 111 | 16 | 56 | 28 | |||||
| Clinical Characteristics | Allele | Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C | OR (95% CI) | T | OR (95% CI) | CC | OR (95% CI) | CT | OR (95% CI) | TT | OR (95% CI) | |
| Age at diagnosis ≤ 25 years = 33 | 33 | 1.25 (0.69–2.26) | 33 | 0.8(0.44–1.46) | 7 | 1.41 (0.44–4.12) | 19 | 1.07 (0.45–2.58) | 7 | 0.69 (0.23–1.89) |
| Years since diagnosis ≥ 8 years = 23 | 21 | 1.05 (0.52–2.09) | 25 | 0.95 (0.48–1.92) | 5 | 1.45 (0.37–4.9) | 11 | 0.72 (0.26–1.98) | 7 | 1.12 (0.35–3.28) |
| Ileal disease (L1) = 12 | 12 | 1.25 (0.49–3.2) | 12 | 0.8 (0.31–2.06) | 4 | 2.6 (0.51–11.2) | 4 | 0.4 (0.08–1.59) | 4 | p = 0.7396, 1.28 (95% CI: 0.26–5.26) |
| Colonic disease (L2) = 12 | 16 | 2.48 (0.95–7.03) a | 8 | 0.4 (0.14–1.05) a | 4 | 2.6 (0.51–11.2) | 8 | 1.57 (0.39–7.58) | 0 | 0 (0–1.01) b |
| Ileocolonic disease (L3) = 26 | 21 | 0.85 (0.43–1.64) | 31 | 1.18 (0.61–2.33) | 3 | 0.69 (0.12–2.71) | 15 | 1.07 (0.41–2.86) | 8 | 1.14 (0.38–3.16) |
| Inflammatory disease (B1) = 10 | 14 | 2.9 (1–9.58) b | 6 | 0.35 (0.1–1) b | 4 | 3.45 (0.64–16.54) | 6 | 1.18 (0.26–6.03) | 0 | 0 (0–1.25) |
| Stricturing disease (B2) = 35 | 31 | 0.99 (0.55–1.78) | 39 | 1.01 (0.56–1.82) | 6 | 1.09 (0.32–3.28) | 19 | 0.93 (0.4–2.19) | 10 | 1.03 (0.39–2.58) |
| Penetrating disease (B3) = 15 | 10 | 0.62 (0.25–1.48) | 20 | 1.6 (0.67–4.03) | 1 | 0.38 (0.01–2.84) | 8 | 0.9 (0.26–3.16) | 6 | 1.71 (0.46–5.96) |
| Immunosuppresant therapy = 31 | 28 | 1.03 (0.55–1.89) | 34 | 0.97 (0.53–1.8) | 5 | 1.01 (0.26–3.26) | 18 | 1.09 (0.45–2.7) | 8 | 0.9 (0.31–2.39) |
| Corticosteroid use = 31 | 31 | 1.25 (0.68–2.3) | 31 | 0.8 (0.44–1.48) | 6 | 1.26 (0.36–3.85) | 19 | 1.24 (0.51–3.13) | 6 | 0.62 (0.19–1.77) |
| Anti-TNF = 12 | 9 | 0.75 (0.28–1.93) | 15 | 1.33 (0.52–3.63) | 2 | 1.05 (0.1–5.66) | 5 | 0.56 (0.13–2.23) | 5 | 1.83 (0.42–7.34) |
| IBD surgery = 17 | 11 | 0.6 (0.25–1.36) | 23 | 1.67 (0.74–4.02) | 1 | 0.33 (0.01–2.44) | 9 | 0.88 (0.28–2.87) | 7 | 1.79 (0.52–5.82) |
| Clinical Characteristics | Allele | Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C | T | CC | CT | TT | ||||||
| Age at diagnosis ≤ 25 years = 25 | 18 | 0.7 (0.35–1.39) | 32 | 1.42 (0.72–2.88) | 4 | 1 (0.22–3.56) | 10 | 0.53 (0.19–1.39) | 11 | 2.01 (0.73–5.44) |
| Years since diagnosis ≥ 8 years = 26 | 23 | 0.99 (0.51–1.91) | 29 | 1.01 (0.52–1.97) | 5 | 1.25 (0.32–4.12) | 13 | 0.79 (0.3–2.05) | 8 | 1.14 (0.38–3.16) |
| Left-sided colitis (E2) = 16 | 15 | 1.1 (0.48–2.49) | 17 | 0.91 (0.4–2.07) | 6 | 3.11 (0.81–11.15) | 3 | 0.18 (0.03–0.73) a | 7 | 1.99 (0.57–6.68) |
| Pancolitis (E3) = 32 | 32 | 1.25 (0.68–2.28) | 32 | 0.8 (0.44–1.47) | 7 | 1.47 (95% CI: 0.46–4.3) | 18 | 1.01 (0.42–2.46) | 7 | 0.72 (0.24–1.97) |
| Severe disease = 13 | 17 | 2.35 (0.94–6.28) b | 9 | 0.43 (0.16–1.07) b | 5 | 3.24 (0.74–13.03) b | 7 | 0.92 (0.24–3.56) | 1 | 0.22 (0–1.59) |
| Immunosuppresant therapy = 22 | 20 | 1.04 (0.51–2.11) | 24 | 0.96 (0.47–1.97) | 5 | 1.54 (0.39–5.22) | 10 | 0.66 (0.23–1.83) | 7 | 1.2 (0.37–3.54) |
| Corticosteroid use = 35 | 35 | 1.25 (0.7–2.23) | 35 | 0.8 (0.45–1.44) | 11 | 2.39 (0.88–6.36) | 13 | 0.47 (0.19–1.09) | 11 | 1.18 (0.46–2.9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petryszyn, P.; Dudkowiak, R.; Gruca, A.; Jaźwińska-Tarnawska, E.; Ekk-Cierniakowski, P.; Poniewierka, E.; Wiela-Hojeńska, A.; Głowacka, K. C3435T Polymorphism of the ABCB1 Gene in Polish Patients with Inflammatory Bowel Disease: A Case–Control and Meta-Analysis Study. Genes 2021, 12, 1419. https://doi.org/10.3390/genes12091419
Petryszyn P, Dudkowiak R, Gruca A, Jaźwińska-Tarnawska E, Ekk-Cierniakowski P, Poniewierka E, Wiela-Hojeńska A, Głowacka K. C3435T Polymorphism of the ABCB1 Gene in Polish Patients with Inflammatory Bowel Disease: A Case–Control and Meta-Analysis Study. Genes. 2021; 12(9):1419. https://doi.org/10.3390/genes12091419
Chicago/Turabian StylePetryszyn, Paweł, Robert Dudkowiak, Agnieszka Gruca, Ewa Jaźwińska-Tarnawska, Paweł Ekk-Cierniakowski, Elżbieta Poniewierka, Anna Wiela-Hojeńska, and Krystyna Głowacka. 2021. "C3435T Polymorphism of the ABCB1 Gene in Polish Patients with Inflammatory Bowel Disease: A Case–Control and Meta-Analysis Study" Genes 12, no. 9: 1419. https://doi.org/10.3390/genes12091419
APA StylePetryszyn, P., Dudkowiak, R., Gruca, A., Jaźwińska-Tarnawska, E., Ekk-Cierniakowski, P., Poniewierka, E., Wiela-Hojeńska, A., & Głowacka, K. (2021). C3435T Polymorphism of the ABCB1 Gene in Polish Patients with Inflammatory Bowel Disease: A Case–Control and Meta-Analysis Study. Genes, 12(9), 1419. https://doi.org/10.3390/genes12091419






