Genetic Diversity of Microneme Protein 2 and Surface Antigen 1 of Eimeria tenella
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chicken Faeces Samples and Genomic DNA Extraction
2.2. Amplifications of etmic2 and etsag1
2.3. Analyses of Genetic Diversity and Natural Selection in Korean etmic2 and etsag1
3. Results
3.1. Amplifications of etmic2 and etsag1 in Korean E. tenella
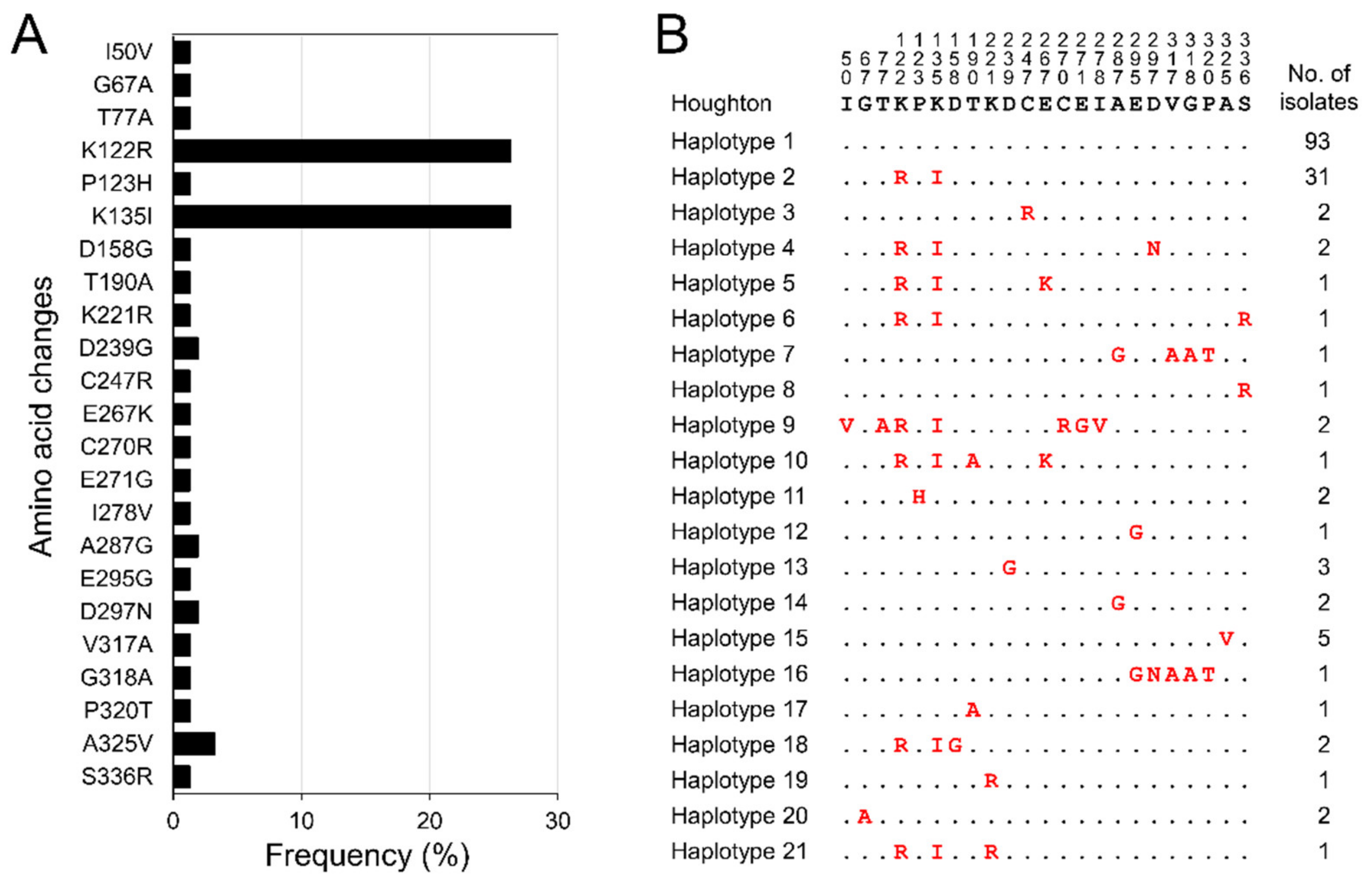
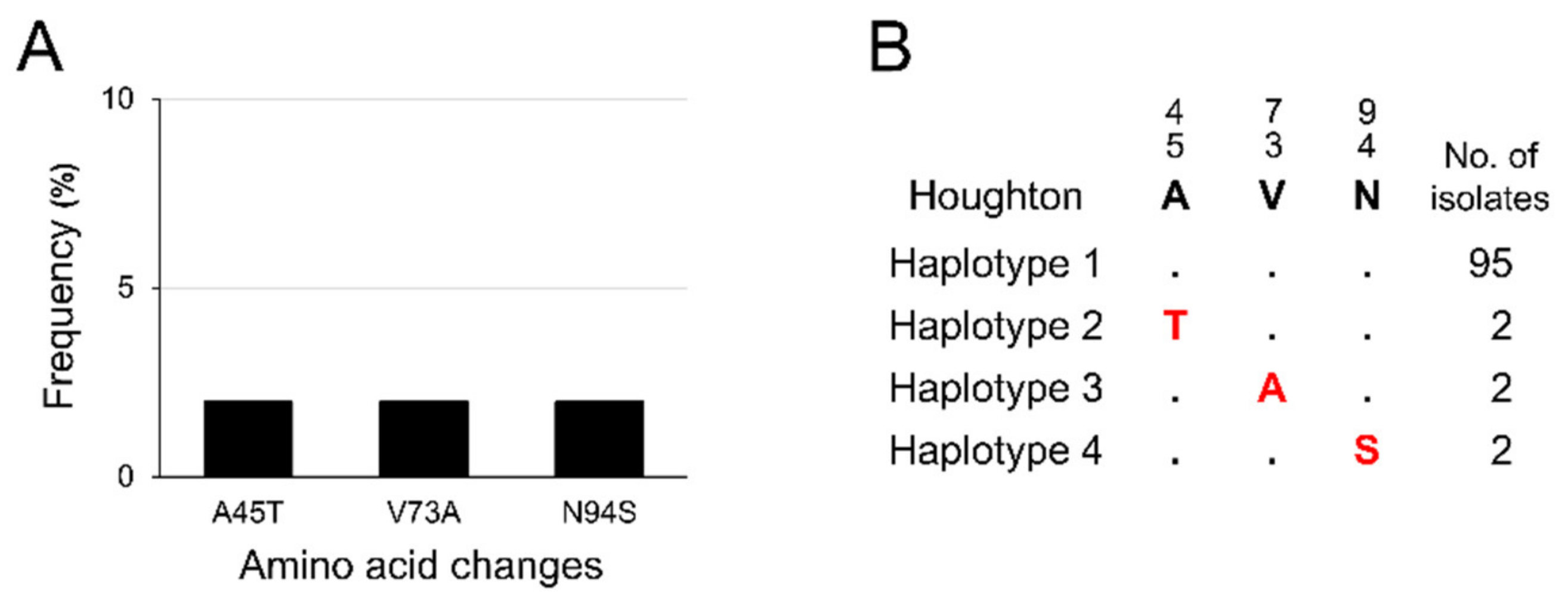
3.2. Polymorphism Patterns in Korean etmic2 and etsag1
3.3. Polymorphism Patterns of etmic2 and etsag1 in E. tenella with Different Geographical Origins
3.4. Nucleotide Diversity and Natural Selection of Korean etmic2 and etsag1
3.5. Association between Genetic Polymorphisms and Host Immune Pressure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blake, D.P.; Knox, J.; Dehaeck, B.; Huntington, B.; Rathinam, T.; Ravipati, V.; Ayoade, S.; Gilbert, W.; Adebambo, A.O.; Jatau, I.D.; et al. Re-calculating the cost of coccidiosis in chickens. Vet. Res. 2020, 51, 115. [Google Scholar] [CrossRef]
- Antonissen, G.; Eeckhaut, V.; Van Driessche, K.; Onrust, L.; Haesebrouck, F.; Ducatelle, R.; Moore, R.J.; Van Immerseel, F. Microbial shifts associated with necrotic enteritis. Avian Pathol. 2016, 45, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Chapman, H.D. Milestones in avian coccidiosis research: A review. Poult. Sci. 2014, 93, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Kim, W.H.; Jeong, J.; Yoo, J.; Kwon, Y.K.; Jung, B.Y.; Kwon, J.H.; Lillehoj, H.S.; Min, W. Prevalence and cross-immunity of Eimeria species on Korean chicken farms. J. Vet. Med. Sci. 2010, 72, 985–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirley, M.W.; Smith, A.L.; Tomley, F.M. The biology of avian Eimeria with an emphasis on their control by vaccination. Adv. Parasitol. 2005, 60, 285–330. [Google Scholar] [CrossRef]
- Clark, E.L.; Macdonald, S.E.; Thenmozhi, V.; Kundu, K.; Garg, R.; Kumar, S.; Ayoade, S.; Fornace, K.M.; Jatau, I.D.; Moftah, A.; et al. Cryptic Eimeria genotypes are common across the southern but not northern hemisphere. Int. J. Parasitol. 2016, 46, 537–544. [Google Scholar] [CrossRef] [Green Version]
- Noack, S.; Chapman, H.D.; Selzer, P.M. Anticoccidial drugs of the livestock industry. Parasitol. Res. 2019, 118, 2009–2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kant, V.; Singh, P.; Verma, P.K.; Bais, I.; Parmar, M.S.; Gopal, A.; Gupta, V. Anticoccidial Drugs Used in the Poultry: An Overview. Sci. Int. 2013, 1, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Abbas, R.Z.; Iqbal, Z.; Blake, D.; Khan, M.N.; Saleemi, M.K. Anticoccidial drug resistance in fowl coccidia: The state of play revisited. Worlds. Poult. Sci. J. 2011, 67, 337–350. [Google Scholar] [CrossRef]
- Soutter, F.; Werling, D.; Tomley, F.M.; Blake, D.P. Poultry Coccidiosis: Design and Interpretation of Vaccine Studies. Front. Vet. Sci. 2020, 7, 101. [Google Scholar] [CrossRef] [Green Version]
- Venkatas, J.; Adeleke, M.A. A review of Eimeria antigen identification for the development of novel anticoccidial vaccines. Parasitol. Res. 2019, 118, 1701–1710. [Google Scholar] [CrossRef] [PubMed]
- Sam-Yellowe, T.Y. Rhoptry organelles of the apicomplexa: Their role in host cell invasion and intracellular survival. Parasitol. Today 1996, 12, 308–316. [Google Scholar] [CrossRef]
- Ryan, R.; Shirley, M.; Tomley, F. Mapping and expression of microneme genes in Eimeria tenella. Int. J. Parasitol. 2000, 30, 1493–1499. [Google Scholar] [CrossRef]
- Yan, M.; Cui, X.; Zhao, Q.; Zhu, S.; Huang, B.; Wang, L.; Zhao, H.; Liu, G.; Li, Z.; Han, H.; et al. Molecular characterization and protective efficacy of the microneme 2 protein from Eimeria tenella. Parasite 2018, 25, 60. [Google Scholar] [CrossRef] [Green Version]
- Carruthers, V.B.; Tomley, F.M. Microneme proteins in apicomplexans. Subcell Biochem. 2008, 47, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Sathish, K.; Sriraman, R.; Subramanian, B.M.; Rao, N.H.; Balaji, K.; Narasu, M.L.; Srinivasan, V.A. Plant expressed EtMIC2 is an effective immunogen in conferring protection against chicken coccidiosis. Vaccine 2011, 29, 9201–9208. [Google Scholar] [CrossRef]
- Ding, X.; Lillehoj, H.S.; Dalloul, R.A.; Min, W.; Sato, T.; Yasuda, A.; Lillehoj, E.P. In ovo vaccination with the Eimeria tenella EtMIC2 gene induces protective immunity against coccidiosis. Vaccine 2005, 23, 3722–3740. [Google Scholar] [CrossRef]
- Shi, W.; Liu, Q.; Zhang, J.; Sun, J.; Jiang, X.; Geng, J.; Wang, F.; Xiao, Y.; Li, H.; Zhao, X. Co-expression of EtMic2 protein and chicken interleukin-18 for DNA vaccine against chicken coccidiosis. Res. Vet. Sci. 2014, 97, 64–70. [Google Scholar] [CrossRef]
- Sun, H.; Wang, L.; Wang, T.; Zhang, J.; Liu, Q.; Chen, P.; Chen, Z.; Wang, F.; Li, H.; Xiao, Y.; et al. Display of Eimeria tenella EtMic2 protein on the surface of Saccharomyces cerevisiae as a potential oral vaccine against chicken coccidiosis. Vaccine 2014, 32, 1869–1876. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, P.; Sun, H.; Liu, Q.; Wang, L.; Wang, T.; Shi, W.; Li, H.; Xiao, Y.; Wang, P.; et al. Pichia pastoris expressed EtMic2 protein as a potential vaccine against chicken coccidiosis. Vet. Parasitol. 2014, 205, 62–69. [Google Scholar] [CrossRef]
- Tabarés, E.; Ferguson, D.; Clark, J.; Soon, P.E.; Wan, K.L.; Tomley, F. Eimeria tenella sporozoites and merozoites differentially express glycosylphosphatidylinositol-anchored variant surface proteins. Mol. Biochem. Parasitol. 2004, 135, 123–132. [Google Scholar] [CrossRef]
- Jahn, D.; Matros, A.; Bakulina, A.Y.; Tiedemann, J.; Schubert, U.; Giersberg, M.; Haehnel, S.; Zoufal, K.; Mock, H.P.; Kipriyanov, S.M. Model structure of the immunodominant surface antigen of Eimeria tenella identified as a target for sporozoite-neutralizing monoclonal antibody. Parasitol. Res. 2009, 105, 655–668. [Google Scholar] [CrossRef]
- Haug, A.; Thebo, P.; Mattsson, J.G. A simplified protocol for molecular identification of Eimeria species in field samples. Vet. Parasitol. 2007, 146, 35–45. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X.; Li, W.H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Pastor-Fernández, I.; Kim, S.; Billington, K.; Bumstead, J.; Marugán-Hernández, V.; Küster, T.; Ferguson, D.J.P.; Vervelde, L.; Blake, D.P.; Tomley, F.M. Development of cross-protective Eimeria-vectored vaccines based on apical membrane antigens. Int. J. Parasitol. 2018, 48, 505–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kundu, K.; Garg, R.; Kumar, S.; Mandal, M.; Tomley, F.M.; Blake, D.P.; Banerjee, P.S. Humoral and cytokine response elicited during immunisation with recombinant Immune Mapped protein-1 (EtIMP-1) and oocysts of Eimeria tenella. Vet. Parasitol. 2017, 244, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, Y.; Yang, X.; Ke, Q.; Lei, W.; Mughal, M.N.; Fang, R.; Zhou, Y.; Shen, B.; Zhao, J. Genetic diversity and drug sensitivity studies on Eimeria tenella field isolates from Hubei Province of China. Parasit. Vectors 2017, 10, 137. [Google Scholar] [CrossRef] [Green Version]
- Blake, D.P.; Clark, E.L.; Macdonald, S.E.; Thenmozhi, V.; Kundu, K.; Garg, R.; Jatau, I.D.; Ayoade, S.; Kawahara, F.; Moftah, A.; et al. Population, genetic, and antigenic diversity of the apicomplexan Eimeria tenella and their relevance to vaccine development. Proc. Natl. Acad. Sci. USA 2015, 112, E5343–E5350. [Google Scholar] [CrossRef] [PubMed] [Green Version]


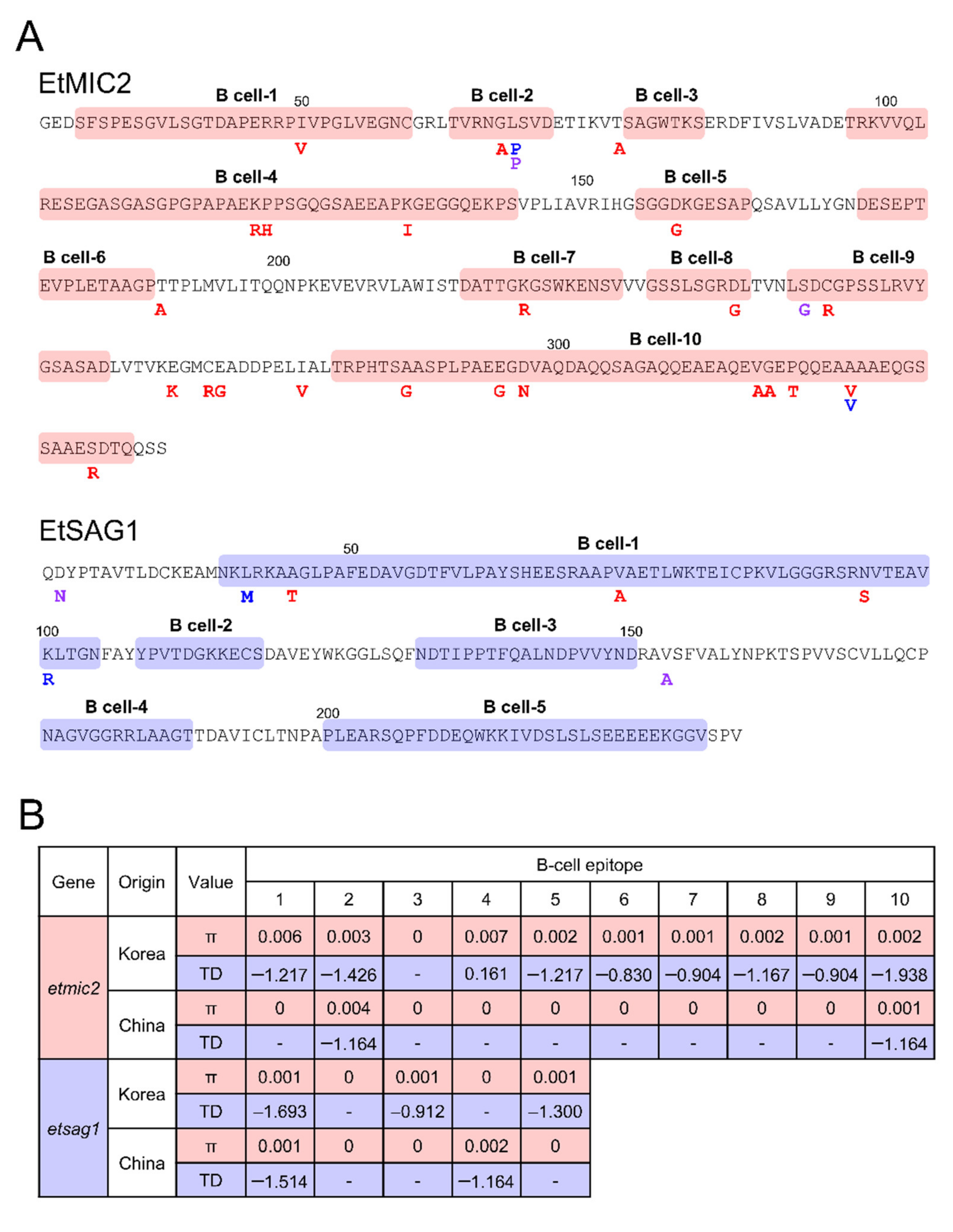
| Gene | Country | n | K | S | Eta | H | Hd ± SD | π ± SD | dN – dS | Tajima’s D | Fu & Li’s D | Fu & Li’s F |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| etmic2 | Korea | 156 | 1.76 | 33 | 33 | 30 | 0.674 ± 0.041 | 0.0019 ± 0.0002 | 0.000 | −2.0503 a | 1.3531 b | −0.0557 b |
| China | 21 | 0.19 | 2 | 2 | 2 | 0.095 ± 0.084 | 0.0002 ± 0.0002 | 0.000 | −1.5141 b | −2.0800 c | −2.2142 c | |
| etsag1 | Korea | 101 | 0.29 | 8 | 8 | 10 | 0.258 ± 0.058 | 0.0005 ± 0.0001 | −0.001 | −1.9600 a | 0.4505 b | −0.4427 b |
| China | 21 | 0.38 | 4 | 4 | 2 | 0.095 ± 0.084 | 0.0006 ± 0.0005 | −0.001 | −1.8733 a | −2.6730 a | −2.8255 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Võ, T.C.; Naw, H.; Flores, R.A.; Lê, H.G.; Kang, J.-M.; Yoo, W.G.; Kim, W.-H.; Min, W.; Na, B.-K. Genetic Diversity of Microneme Protein 2 and Surface Antigen 1 of Eimeria tenella. Genes 2021, 12, 1418. https://doi.org/10.3390/genes12091418
Võ TC, Naw H, Flores RA, Lê HG, Kang J-M, Yoo WG, Kim W-H, Min W, Na B-K. Genetic Diversity of Microneme Protein 2 and Surface Antigen 1 of Eimeria tenella. Genes. 2021; 12(9):1418. https://doi.org/10.3390/genes12091418
Chicago/Turabian StyleVõ, Tuấn Cường, Haung Naw, Rochelle A. Flores, Hương Giang Lê, Jung-Mi Kang, Won Gi Yoo, Woo-Hyun Kim, Wongi Min, and Byoung-Kuk Na. 2021. "Genetic Diversity of Microneme Protein 2 and Surface Antigen 1 of Eimeria tenella" Genes 12, no. 9: 1418. https://doi.org/10.3390/genes12091418
APA StyleVõ, T. C., Naw, H., Flores, R. A., Lê, H. G., Kang, J.-M., Yoo, W. G., Kim, W.-H., Min, W., & Na, B.-K. (2021). Genetic Diversity of Microneme Protein 2 and Surface Antigen 1 of Eimeria tenella. Genes, 12(9), 1418. https://doi.org/10.3390/genes12091418









