Pseudogene Transcripts in Head and Neck Cancer: Literature Review and In Silico Analysis
Abstract
1. Introduction
2. Pseudogenes
3. Pseudogene Transcripts
4. Pseudogene Transcripts in HNC
4.1. HNC in General
4.2. Oral Cancer
4.3. Laryngeal Cancer
| Pseudogene Transcript | Pseudogene–Gene Interaction | Tumor Effect | Clinical Outcome | Tumor Localization | Tumor HPV Status | Reference |
|---|---|---|---|---|---|---|
| PTENP1 | PTEN | Facilitates the aggressiveness of tumor | Poor prognosis | HN | Not specified | [25] |
| FKBP9P1 | PI3K/AKT | Facilitates the aggressiveness of tumor | Poor prognosis | HN | Not specified | [26] |
| LILRP1 | LILRB1 | Not specified | Poor prognosis | HN | Not specified | [24] |
| RP6-191P20.5 | VSIG | Not specified | Poor prognosis | HN | Not specified | [24] |
| RPL29P19 | PMEPA1 | Not specified | Poor prognosis | HN | Not specified | [24] |
| TAS2R2P | KLK5 | Not specified | Poor prognosis | HN | Not specified | [24] |
| ZBTB45P1 | HEATR1 | Not specified | Poor prognosis | HN | Not specified | [24] |
| PTTG3P | PTTG1 and PTTG2 | Facilitates the aggressiveness of tumor | Tumor development and progression | HN | Mixed | [27] |
| AC010677.5 | RPL23 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| TCEB2P2 | TCEB2 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| RPL37P2 | RPL37 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| PPIAP26 | PPIA | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| WTAPP1 | MMP1 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| UNGP3 | UNG | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| UBA52P8 | UBA52 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| RP11-490K7.4 | GTF2A2 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| UBA52P6 | UBA52 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| EIF4HP2 | EIF4H | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| AC114737.3 | FDPS | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| RP1-89D4.1 | RPS24 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| POLR2KP1 | POLR2K | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| CD8BP | CD8B | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| RP11-54C4.1 | RPLP1 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| UNGP1 | UNG | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| YWHAEP7 | YWHAE | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| NPM1P25 | NPM1 | Facilitates HPV16 infection | Tumor development | HN | Mixed | [28] |
| FTH1P3 | MMP1, PLAU, MMP3 and IL8 | Increased cell proliferation and migration | Tumor development and progression | Oral cavity | Not specified | [29] |
| FTH1P3 | miR-224-5p (FZD5) | Increased cell proliferation | Tumor development and progression | Oral cavity | Not specified | [30] |
| FTH1P3 | PI3K/Akt/GSK3β/Wnt/β-catenin | Increased cell proliferation and migration | Tumor development and progression | Oral cavity | Not specified | [31] |
| GTF2IRD2P1 | MMP1, PLAU, IL8 and MMP9 | Increased cell proliferation and migration | Tumor development and progression | Oral cavity | Not specified | [29] |
| PDIA3P | PLAU | Increased cell proliferation and migration | Tumor development and progression | Oral cavity | Not specified | [29] |
| PTENP1 | miR-21 (PTEN) | Increased cell proliferation | Tumor development | Oral cavity | Not specified | [32] |
| HMGA1P6 | HMGA2 | Facilitates the aggressiveness of tumor | Tumor development and progression | Larynx | Not specified | [33] |
| HMGA1P7 | HMGA2 | Facilitates the aggressiveness of tumor | Tumor development and progression | Larynx | Not specified | [33] |
| HLA-A*31012 | Not specified | Facilitates immune system escape | Tumor development | Larynx | Not specified | [34] |
| FTH1P3 | Not specified | Increased cell proliferation and migration | Tumor development and progression | Larynx | Not specified | [35] |
| DPY19L2P1 | Not specified | Not specified | Poor prognosis | Larynx | Not specified | [36] |
5. Materials and Methods
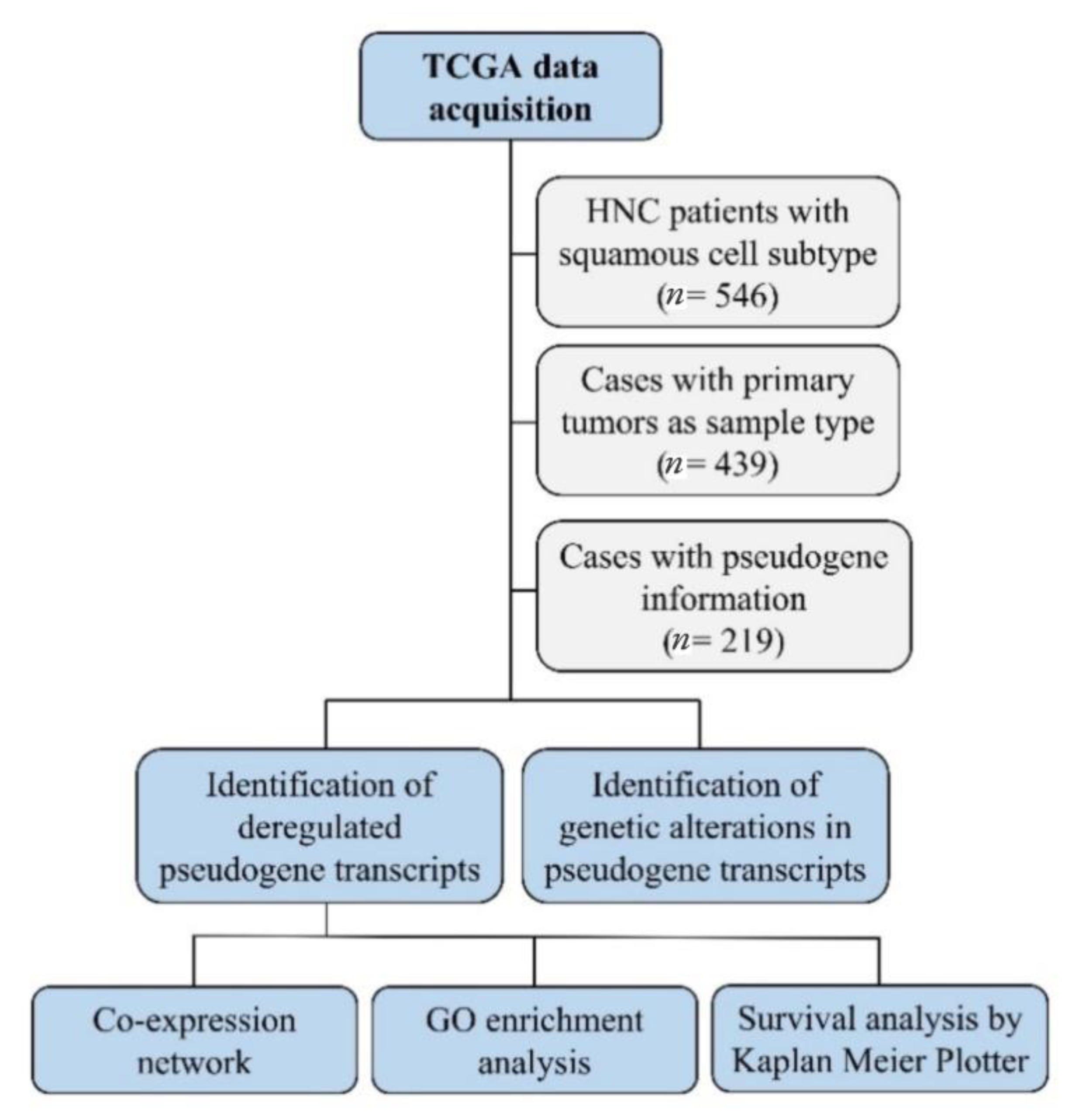
5.1. TCGA Data Analysis
5.2. Co-Expression Networks and GO Enrichment Analysis
5.3. Survival Analysis
6. Results and Discussion
6.1. TCGA Data Analysis
| Tumor Location and Pseudogene Transcript | Chromosome Location | Gene Family Function | Studies in Cancer | Reference |
|---|---|---|---|---|
| Head and neck (n = 219) | ||||
| SPATA31D5P | 9q21.32 | UV response and DNA repair | None | [59] |
| HERC2P3 | 15q11.1 | Cell growth and migration | Gastric | [44] |
| SPATA31C2 | 9q22.1 | UV response and DNA repair | None | [59] |
| MAGEB6P1 | Xp21.3 | Tumor-specific antigen | None | [60] |
| SLC25A51P1 | 6q12 | Mitochondrial NAD+ transporter | None | [61] |
| BAGE2 | 21p11.2 | Tumor-specific antigen | Lung, colon, and breast | [62] |
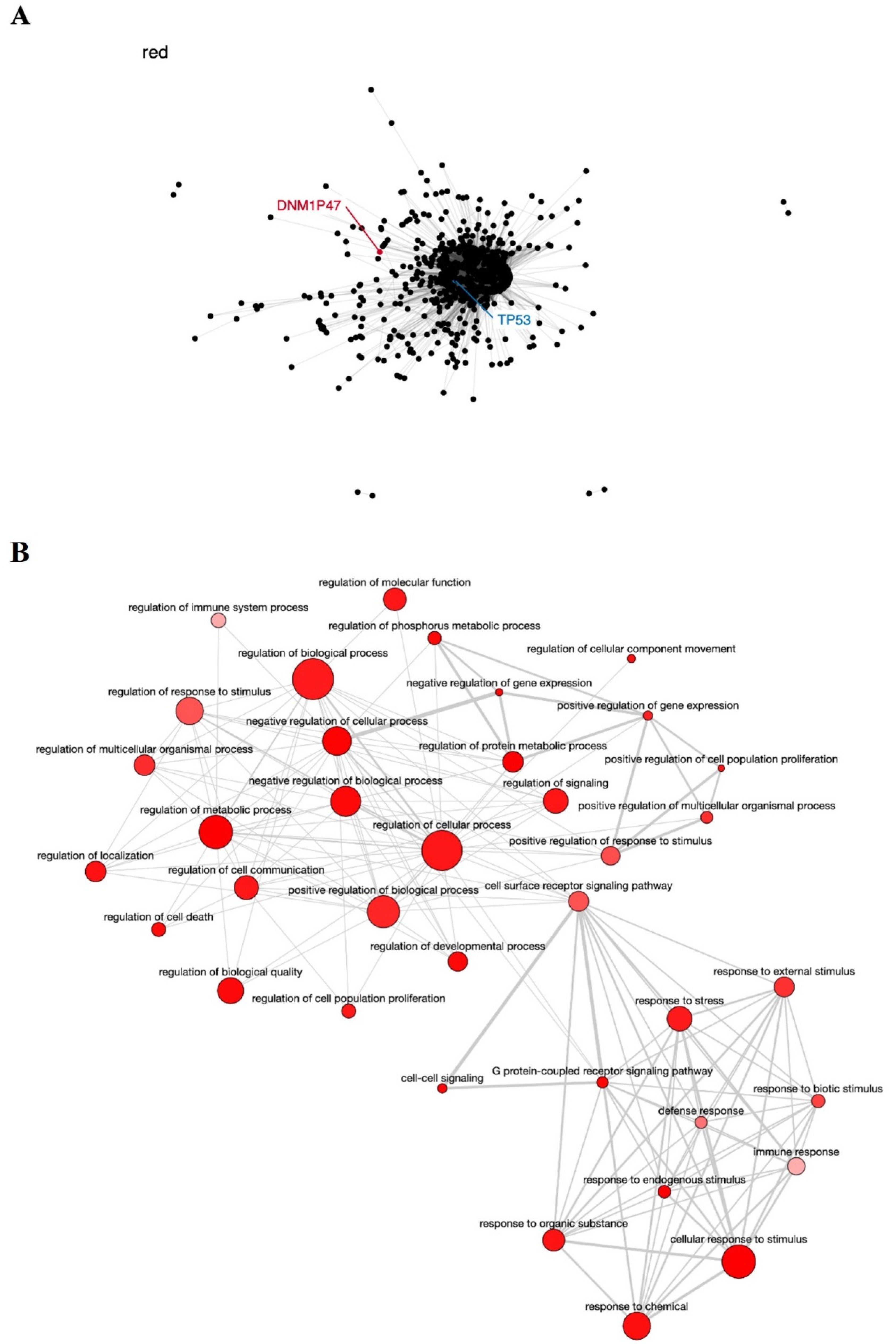
| DNM1P47 | 15q26.3 | Mitochondrial division | None | [63] |
| SPATA31C1 | 9q22.1 | UV response and DNA repair | None | [59] |
| ZNF733P | 7q11.21 | Transcription factor | None | [64] |
| OR2W5 | 1q44 | Cellular signaling | None | [65] |
| Oral cavity (n = 62) | ||||
| SPATA31D5P | 9q21.32 | UV response and DNA repair | None | [59] |
| NBPF25P | 1q21.1 | Neuronal modulation | None | [66] |
| HSP90AB2P | 4p15.33 | Cell proteostasis | None | [67] |
| NXF4 | Xq22.1 | RNA export from nucleus | None | [68] |
| FOLH1B | 11q14.3 | Metallopeptidase activity | Prostate | [46] |
| DNM1P47 | 15q26.3 | Mitochondrial division | None | [63] |
| BNIP3P1 | 14q12 | Autophagy and apoptosis | Breast cancer brain metastases | [47] |
| PKD1L2 | 16q23.2 | Transmembrane protein | Colorectal and breast | [48,49] |
| BAGE2 | 21p11.2 | Tumor-specific antigen | Lung, colon, and breast | [62] |
| ZNF658B | 9p12 | Transcription factor | None | [69] |
| Oropharynx (n = 51) | ||||
| POTEA | 8p11.1 | Apoptosis | Colorectal | [50] |
| MROH5 | 8q24.3 | Uncertain | None | |
| MSL3P1 | 2q37.1 | Transcription regulation | Renal and gastric | [51,70] |
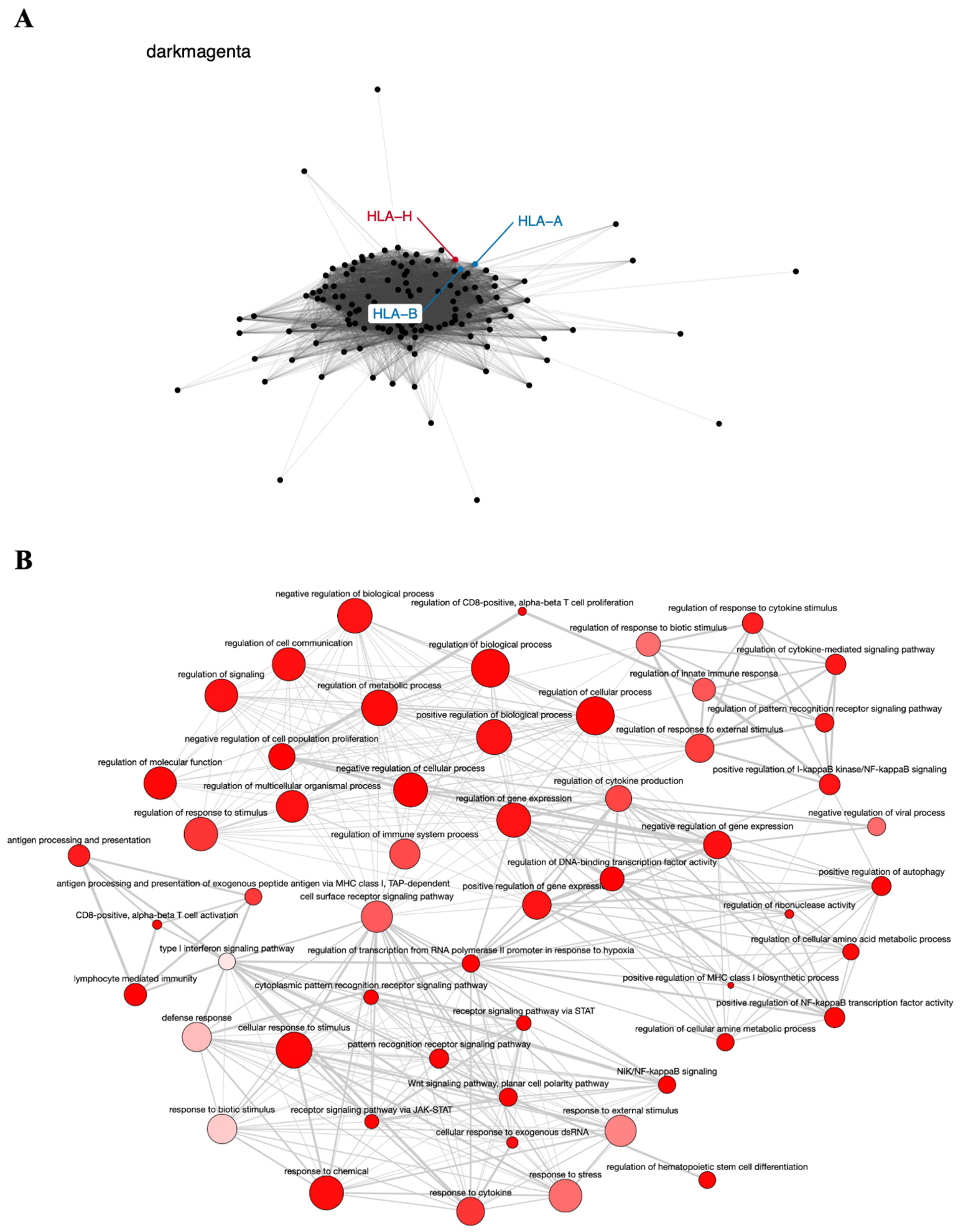
| HLA-H | 6p22.1 | Immune homeostasis | Cervical and lung | [52,53] |
| TUBB8P7 | 16q24.3 | Oocyte maturation | None | [71] |
| SLC7A5P2 | 16p12.2 | Amino acid transporter | None | [72] |
| DPY19L2P1 | 7p14.2 | Transmembrane protein | Larynx | [36] |
| TSSC2 | 11p15.4 | Tumor suppressor | None | [73] |
| SPATA31C2 | 9q22.1 | UV response and DNA repair | None | [59] |
| NXF4 | Xq22.1 | RNA export from nucleus | None | [68] |
| Hypopharynx (n = 8) | ||||
| DPY19L2P3 | 7p14.3 | Transmembrane protein | None | [36] |
| SPATA31D5P | 9q21.32 | UV response and DNA repair | None | [59] |
| GBA3 | 4p15.2 | Glucosylceramide hydrolysis | Liver | [54] |
| PLEKHM1P | 17q24.1 | Autophagy | None | [74] |
| DPY19L2P1 | 7p14.2 | Transmembrane protein | Larynx | [36] |
| MST1P2 | 1p36.13 | Cell invasion and apoptosis | Bladder and cervical | [55,56] |
| RP11-44F14.1 | 16q12.2 | Unknown | None | |
| ADAM21P1 | 14q24.2 | Cell adhesion and proliferation | None | [75] |
| MAGEB6P1 | Xp21.3 | Tumor-specific antigen | None | [60] |
| OR12D2 | 6p22.1 | Cellular signaling | None | [65] |
| Larynx (n = 98) | ||||
| HERC2P3 | 15q11.1 | Cell growth and migration | Gastric | [44] |
| SPATA31D5P | 9q21.32 | UV response and DNA repair | None | [59] |
| SPATA31C2 | 9q22.1 | UV response and DNA repair | None | [59] |
| SLC25A51P1 | 6q12 | Mitochondrial NAD+ transporter | None | [61] |
| MAGEB6P1 | Xp21.3 | Tumor-specific antigen | None | [60] |
| SPATA31C1 | 9q22.1 | UV response and DNA repair | None | [59] |
| BAGE2 | 21p11.2 | Tumor-specific antigen | Lung, colon, and breast | [62] |
| PNLIPRP2 | 10q25.3 | Lipase activity | Pancreas | [57] |
| ZNF733P | 7q11.21 | Transcription factor | None | [64] |
| DNM1P47 | 15q26.3 | Mitochondrial division | None | [63] |
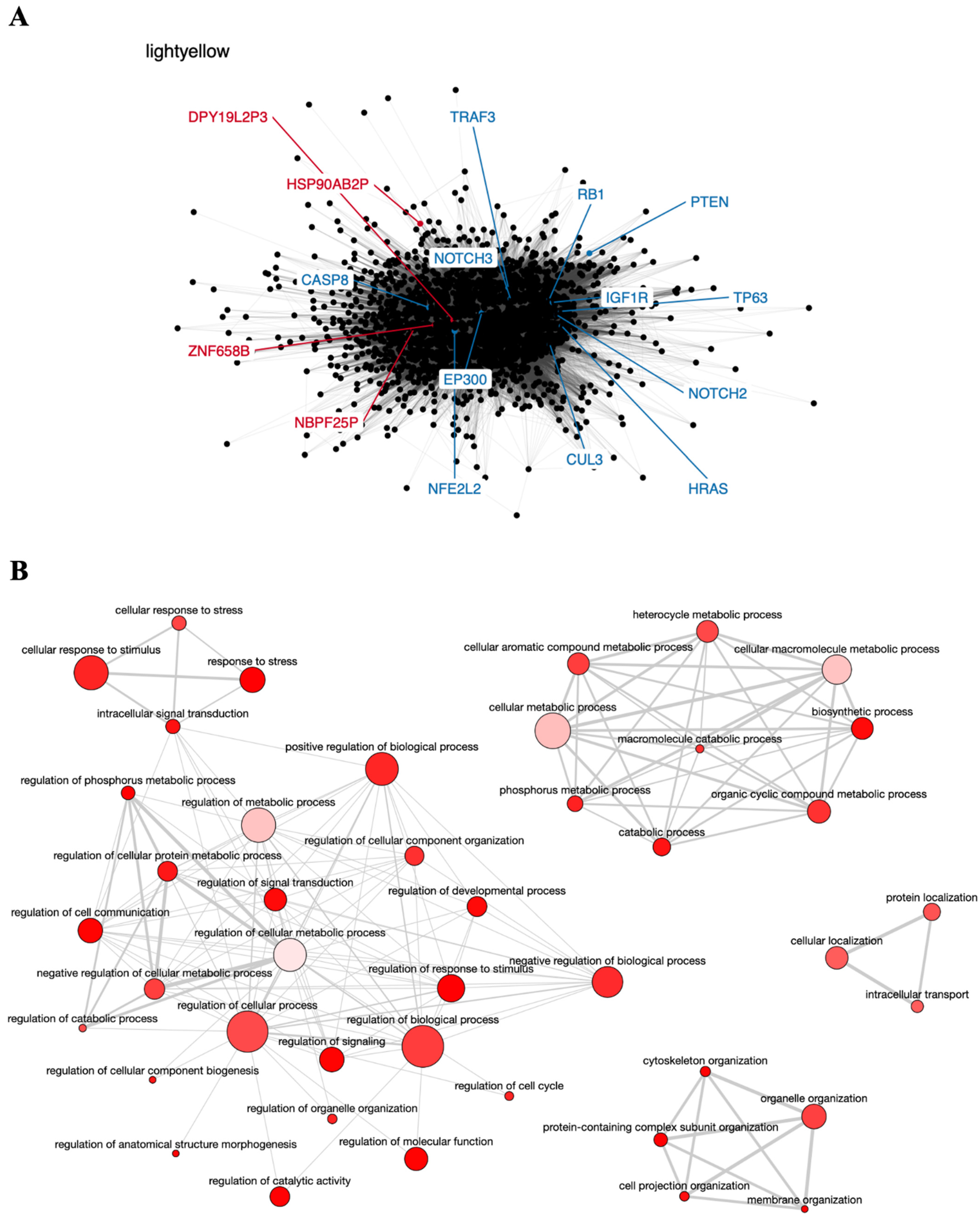
6.2. Co-Expression Networks and GO Enrichment Analysis
6.3. Survival Analysis
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Me, J.F.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Belcher, R.; Hayes, K.; Fedewa, S.; Chen, A.Y. Current treatment of head and neck squamous cell cancer. J. Surg. Oncol. 2014, 110, 551–574. [Google Scholar] [CrossRef]
- Mendenhall, W.; Werning, J.; Pfister, D. Treatment of head and neck cancers. In Cancer: Principles & Practice of Oncology, 9th ed.; DeVita, V.T., Lawrence, T.S., Rosenberg, S.A., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2011; pp. 729–780. [Google Scholar]
- Dayyani, F.; Etzel, C.J.; Liu, M.; Ho, C.-H.; Lippman, S.M.; Tsao, A.S. Meta-analysis of the impact of human papillomavirus (HPV) on cancer risk and overall survival in head and neck squamous cell carcinomas (HNSCC). Head Neck Oncol. 2010, 2, 15. [Google Scholar] [CrossRef]
- O’Sullivan, B.; Huang, S.H.; Su, J.; Garden, A.; Sturgis, E.M.; Dahlstrom, K.; Lee, N.; Riaz, N.; Pei, X.; Koyfman, S.A.; et al. Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): A multicentre cohort study. Lancet Oncol. 2016, 17, 440–451. [Google Scholar] [CrossRef]
- Siu, L.L.; Waldron, J.N.; Chen, B.E.; Winquist, E.; Wright, J.R.; Nabid, A.; Parulekar, W.R. Effect of Standard Radiotherapy With Cisplatin vs Accelerated Radiotherapy With Panitumumab in Locoregionally Advanced Squamous Cell Head and Neck Carcinoma: A Randomized Clinical Trial. JAMA Oncol. 2017, 3, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Bussmann, L.; Busch, C.-J.; Lörincz, B.B.; Rieckmann, T.; Block, A.R.; Knecht, R. Perspectives in chemosensitivity and chemoresistance assays and their implementation in head and neck cancer. Eur. Arch. Otorhinolaryngol. 2016, 273, 4073–4080. [Google Scholar] [CrossRef]
- Lacko, M.; Braakhuis, B.J.; Sturgis, E.M.; Boedeker, C.C.; Suárez, C.; Rinaldo, A.; Ferlito, A.; Takes, R.P. Genetic Susceptibility to Head and Neck Squamous Cell Carcinoma. Int. J. Radiat. Oncol. 2014, 89, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiar, S.M.; Ali, A.; Barh, D. Epigenetics in Head and Neck Cancer. Methods Mol. Biol. 2014, 1238, 751–769. [Google Scholar] [CrossRef]
- Jacq, C.; Miller, J.R.; Brownlee, G.G. A pseudogene structure in 5S DNA of Xenopus laevis. Cell 1977, 12, 109–120. [Google Scholar] [CrossRef]
- Tutar, Y. Pseudogenes. Comp. Funct. Genomics 2012, 2012, 424526. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Singh, D.; Yadava, A.; Srivastava, A.K. Molecular fossils “pseudogenes” as functional signature in biological system. Genes Genom. 2020, 42, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Balakirev, E.S.; Ayala, F.J. Pseudogenes: Are they “junk” or functional DNA? Annu. Rev. Genet. 2003, 37, 123–151. [Google Scholar] [CrossRef]
- Vinckenbosch, N.; Dupanloup, I.; Kaessmann, H. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. USA 2006, 103, 3220–3225. [Google Scholar] [CrossRef]
- Pan, Y.; Sun, C.; Huang, M.; Liu, Y.; Qi, F.; Liu, L.; Wen, J.; Liu, J.; Xie, K.; Ma, H.; et al. A genetic variant in pseudogene E2F3P1 contributes to prognosis of hepatocellular carcinoma. J. Biomed. Res. 2014, 28, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Lynn, H.; Sun, X.; Ayshiev, D.; Siegler, J.H.; Rizzo, A.N.; Karnes, J.H.; Gonzalez-Garay, M.; Wang, T.; Casanova, N.; Camp, S.M.; et al. Single nucleotide polymorphisms in the MYLKP1 pseudogene are associated with increased colon cancer risk in African Americans. PLoS ONE 2018, 13, e0200916. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Liu, H.; Du, M.; Zhang, G.; Lin, Y.; Ge, Y.; Wang, M.; Jin, G.; Zhao, Q.; Chu, H.; et al. A genetic variation in the CpG island of pseudogene GBAP1 promoter is associated with gastric cancer susceptibility. Cancer 2019, 125, 2465–2473. [Google Scholar] [CrossRef]
- Poliseno, L. Pseudogenes: Newly Discovered Players in Human Cancer. Sci. Signal. 2012, 5, re5. [Google Scholar] [CrossRef]
- Hu, X.; Yang, L.; Mo, Y.-Y. Role of Pseudogenes in Tumorigenesis. Cancers 2018, 10, 256. [Google Scholar] [CrossRef]
- Xiao-Jie, L.; Ai-Mei, G.; Li-Juan, J.; Jiang, X. Pseudogene in cancer: Real functions and promising signature. J. Med. Genet. 2014, 52, 17–24. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nat. Cell Biol. 2010, 465, 1033–1038. [Google Scholar] [CrossRef]
- Russo, F.; Fiscon, G.; Conte, F.; Rizzo, M.; Paci, P.; Pellegrini, M. Interplay Between Long Noncoding RNAs and MicroRNAs in Cancer. Methods Mol. Biol. 2018, 1819, 75–92. [Google Scholar] [CrossRef]
- Conte, F.; Fiscon, G.; Sibilio, P.; Licursi, V.; Paci, P. An Overview of the Computational Models Dealing with the Regulatory ceRNA Mechanism and ceRNA Deregulation in Cancer. Breast Cancer 2021, 2324, 149–164. [Google Scholar] [CrossRef]
- Xing, L.; Zhang, X.; Guo, M.; Zhang, X.; Liu, F. Application of Machine Learning in Developing a Novelty Five-Pseudogene Signature to Predict Prognosis of Head and Neck Squamous Cell Carcinoma: A New Aspect of “Junk Genes” in Biomedical Practice. DNA Cell Biol. 2020, 39, 709–723. [Google Scholar] [CrossRef]
- Liu, J.; Xing, Y.; Xu, L.; Chen, W.; Cao, W.; Zhang, C. Decreased expression of pseudogene PTENP1 promotes malignant behaviours and is associated with the poor survival of patients with HNSCC. Sci. Rep. 2017, 7, 41179. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-F.; Feng, L.; Shi, Q.; Ma, H.-Z.; He, S.-Z.; Hou, L.-Z.; Wang, R.; Fang, J.-G. Silencing novel long non-coding RNA FKBP9P1 represses malignant progression and inhibits PI3K/AKT signaling of head and neck squamous cell carcinoma in vitro. Chin. Med. J. 2020, 133, 2037–2043. [Google Scholar] [CrossRef] [PubMed]
- Grzechowiak, I.; Graś, J.; Szymańska, D.; Biernacka, M.; Guglas, K.; Poter, P.; Kolenda, T. The oncogenic roles of PTTG1 and PTTG2 Genes and Pseudogene PTTG3P in Head and Neck Squamous Cell Carcinomas. Diagnostics 2020, 10, 606. [Google Scholar] [CrossRef] [PubMed]
- Salyakina, D.; Tsinoremas, N.F. Non-coding RNAs profiling in head and neck cancers. Npj. Genom. Med. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Tian, L.; Ma, P.; Sun, Q.; Zhang, K.; Wang, G.; Liu, H.; Xu, B. Potential role of differentially expressed lncRNAs in the pathogenesis of oral squamous cell carcinoma. Arch. Oral Biol. 2015, 60, 1581–1587. [Google Scholar] [CrossRef]
- Zhang, C.-Z. Long non-coding RNA FTH1P3 facilitates oral squamous cell carcinoma progression by acting as a molecular sponge of miR-224-5p to modulate fizzled 5 expression. Gene 2017, 607, 47–55. [Google Scholar] [CrossRef]
- Liu, M.; Gao, X.; Liu, C.-L. Increased expression of lncRNA FTH1P3 promotes oral squamous cell carcinoma cells migration and invasion by enhancing PI3K/Akt/GSK3b/ Wnt/β-catenin signaling. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8306–8314. [Google Scholar]
- Gao, L.; Ren, W.; Zhang, L.; Li, S.; Kong, X.; Zhang, H.; Dong, J.; Cai, G.; Jin, C.; Zheng, D.; et al. PTENp1, a natural sponge of miR-21, mediates PTEN expression to inhibit the proliferation of oral squamous cell carcinoma. Mol. Carcinog. 2016, 56, 1322–1334. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A., Jr.; De Martino, M.; Esposito, F.; Fraggetta, F.; Neto, P.N.; Fernandes, P.V.; Santos, I.C.; Dias, F.L.; Nasciutti, L.E.; Da Costa, N.M.; et al. HMGA2, but not HMGA1, is overexpressed in human larynx carcinomas. Histopathology 2017, 72, 1102–1114. [Google Scholar] [CrossRef] [PubMed]
- Feenstra, M.; Bakema, J.; Verdaasdonk, M.; Rozemuller, E.; Tweel, J.V.D.; Slootweg, P.; de Weger, R.; Tilanus, M. Detection of a putativeHLA-A*31012 processed (intronless) pseudogene in a laryngeal squamous cell carcinoma. Genes Chromosom. Cancer 2000, 27, 26–34. [Google Scholar] [CrossRef]
- Yuan, H.; Jiang, H.; Wang, Y.; Dong, Y. Increased expression of lncRNA FTH1P3 predicts a poor prognosis and promotes aggressive phenotypes of laryngeal squamous cell carcinoma. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Zhang, G.; Fan, E.; Yue, G.; Zhong, Q.; Shuai, Y.; Wu, M.; Feng, G.; Chen, Q.; Gou, X. Five genes as a novel signature for predicting the prognosis of patients with laryngeal cancer. J. Cell. Biochem. 2019, 121, 3804–3813. [Google Scholar] [CrossRef]
- Eisen, M.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 1–43. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cai, H.; Xie, Y.; Jiang, H. Targeting long non-coding RNA HERC2P3 inhibits cell growth and migration in human gastric cancer cells. Int. J. Clin. Exp. Pathol. 2017, 10, 7632–7639. [Google Scholar]
- Brun, M.-E.; Ruault, M.; Ventura, M.; Roizès, G.; De Sario, A. Juxtacentromeric region of human chromosome 21: A boundary between centromeric heterochromatin and euchromatic chromosome arms. Gene 2003, 312, 41–50. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, H.; Zhou, H.; Gu, Y.; Bai, Y.; Yu, S.; An, R.; Qi, J. Identification of potential key genes and high-frequency mutant genes in prostate cancer by using RNA-Seq data. Oncol. Lett. 2018, 15, 4550–4556. [Google Scholar] [CrossRef]
- Schulten, H.-J.; Bangash, M.; Karim, S.; Dallol, A.; Hussein, D.; Merdad, A.; Althoubaity, F.; Al-Maghrabi, J.; Jamal, A.; Al-Ghamdi, F.; et al. Comprehensive molecular biomarker identification in breast cancer brain metastases. J. Transl. Med. 2017, 15, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Yao, Y.; Zhang, T.; Feng, F.; Zhou, C.; Xu, X.; Sun, C. A four-mRNA model to improve the prediction of breast cancer prognosis. Gene 2019, 721, 144100. [Google Scholar] [CrossRef]
- Park, C.; Kim, J.-I.; Hong, S.N.; Jung, H.M.; Kim, T.J.; Lee, S.; Kim, S.J.; Kim, H.C.; Kim, D.-H.; Cho, B.; et al. A copy number variation in PKD1L2 is associated with colorectal cancer predisposition in korean population. Int. J. Cancer 2016, 140, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Liu, W.; Mahdessian, H.; Bryant, P.; Ringdahl, J.; Timofeeva, M.; Farrington, S.M.; Dunlop, M.; Lindblom, A. Recurrent, low-frequency coding variants contributing to colorectal cancer in the Swedish population. PLoS ONE 2018, 13, e0193547. [Google Scholar] [CrossRef]
- Chen, B.; Wang, C.; Zhang, J.; Zhou, Y.; Hu, W.; Guo, T. New insights into long noncoding RNAs and pseudogenes in prognosis of renal cell carcinoma. Cancer Cell Int. 2018, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, A.; Samadder, S.; Das, P.; Mazumder, D.I.; Chatterjee, A.; Addya, S.; Mondal, R.; Roy, A.; Roychoudhury, S.; Panda, C.K. Deregulation of H19 is associated with cervical carcinoma. Genomics 2019, 112, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Wang, C.; Zhu, M.; Lu, Q.; Ma, Z.; Huang, M.; Dai, J.; Ma, H.; Jin, G.; Hu, Z.; et al. Fine-mapping the MHC region in Asian populations identified novel variants modifying susceptibility to lung cancer. Lung Cancer 2017, 112, 169–175. [Google Scholar] [CrossRef]
- Ying, J.F.; Zhang, Y.N.; Song, S.S.; Hu, Z.M.; He, X.L.; Pan, H.Y.; Zhang, C.W.; Wang, H.J.; Li, W.F.; Mou, X.Z. Decreased expression of GBA3 correlates with a poor prognosis in hepatocellular carcinoma patients. Neoplasma 2020, 67. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Li, Z.; Cao, L. LncRNA MST1P2/miR-133b axis affects the chemoresistance of bladder cancer to cisplatin-based therapy via Sirt1/p53 signaling. J. Biochem. Mol. Toxicol. 2020, 34, e22452. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhang, X.; Xu, Y.; Wang, J.; Li, Z.; Cui, X. Long noncoding RNA MST1P2 promotes cervical cancer progression by sponging with microRNA miR-133b. Bioengineered 2021, 12, 1851–1860. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Y.; Dai, G.; Wu, D.; Gao, Z.; Zhang, L.; Fan, Y. Screening and validating the core biomarkers in patients with pancreatic ductal adenocarcinoma. Math. Biosci. Eng. 2020, 17, 910–927. [Google Scholar] [CrossRef] [PubMed]
- Shaw, S.-W.; Chen, C.-P.; Cheng, P.-J.; Wang, T.-H.; Hou, J.-W.; Lin, C.-T.; Chang, S.-D.; Hwa, H.-L.; Lin, J.-L.; Chao, A.-S.; et al. Gene dosage change of TPTE and BAGE2 and breakpoint analysis in Robertsonian Down syndrome. J. Hum. Genet. 2007, 53, 136–143. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bekpen, C.; Xie, C.; Nebel, A.; Tautz, D. Involvement of SPATA31 copy number variable genes in human lifespan. Aging 2018, 10, 674–688. [Google Scholar] [CrossRef]
- Zamunér, F.T.; Karia, B.T.; de Oliveira, C.Z.; Santos, C.R.; Carvalho, A.L.; Vettore, A.L. A Comprehensive Expression Analysis of Cancer Testis Antigens in Head and Neck Squamous Cell Carcinoma Revels MAGEA3/6 as a Marker for Recurrence. Mol. Cancer Ther. 2015, 14, 828–834. [Google Scholar] [CrossRef]
- Luongo, T.S.; Eller, J.M.; Lu, M.-J.; Niere, M.; Raith, F.; Perry, C.; Bornstein, M.R.; Oliphint, P.; Wang, L.; McReynolds, M.R.; et al. SLC25A51 is a mammalian mitochondrial NAD. Nat. Cell Biol. 2020, 588, 174–179. [Google Scholar] [CrossRef]
- Yamada, R.; Takahashi, A.; Torigoe, T.; Morita, R.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; Kubo, T.; Watarai, K.; Kondo, T.; et al. Preferential expression of cancer/testis genes in cancer stem-like cells: Proposal of a novel sub-category, cancer/testis/stem gene. Tissue Antigens 2013, 81, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Imoto, Y.; Itoh, K.; Fujiki, Y. Molecular Basis of Mitochondrial and Peroxisomal Division Machineries. Int. J. Mol. Sci. 2020, 21, 5452. [Google Scholar] [CrossRef] [PubMed]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression Profile of Ectopic Olfactory Receptors Determined by Deep Sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar] [CrossRef]
- Wu, H.; Zhai, L.T.; Guo, X.X.; Rety, S.; Xi, X.G. The N-terminal of NBPF15 causes multiple types of aggregates and mediates phase transition. Biochem. J. 2020, 477, 445–458. [Google Scholar] [CrossRef]
- Gulla, A.; Kazlauskas, E.; Liang, H.; Strupas, K.; Petrauskas, V.; Matulis, D.; Eshleman, J.R. Heat Shock Protein 90 Inhibitor Effects on Pancreatic Cancer Cell Cultures. Pancreas 2021, 50, 625–632. [Google Scholar] [CrossRef]
- Tan, W.; Zolotukhin, A.S.; Tretyakova, I.; Bear, J.; Lindtner, S.; Smulevitch, S.V.; Felber, B.K. Identification and characterization of the mouse nuclear export factor (Nxf) family members. Nucleic Acids Res. 2005, 33, 3855–3865. [Google Scholar] [CrossRef]
- Francis, M.; Cheng, H.; Ma, P.; Grider, A. Genomic Characterization of the Zinc Transcriptional Regulatory Element Reveals Potential Functional Roles of ZNF658. Biol. Trace Element Res. 2019, 192, 83–90. [Google Scholar] [CrossRef]
- Tanabe, S.; Quader, S.; Ono, R.; Cabral, H.; Aoyagi, K.; Hirose, A.; Yokozaki, H.; Sasaki, H. Molecular Network Profiling in Intestinal—and Diffuse—Type Gastric Cancer. Cancers 2020, 12, 3833. [Google Scholar] [CrossRef]
- Zheng, W.; Hu, H.; Zhang, S.; Xu, X.; Gao, Y.; Gong, F.; Lu, G.; Lin, G. The comprehensive variant and phenotypic spectrum of TUBB8 in female infertility. J. Assist. Reprod. Genet. 2021, 1–12. [Google Scholar] [CrossRef]
- Nachef, M.; Ali, A.K.; Almutairi, S.M.; Lee, S.-H. Targeting SLC1A5 and SLC3A2/SLC7A5 as a Potential Strategy to Strengthen Anti-Tumor Immunity in the Tumor Microenvironment. Front. Immunol. 2021, 12, 624324. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.-J.; Lee, M.P.; Connors, T.D.; Johnson, L.A.; Burn, T.C.; Sub, K.; Landes, G.M.; Feinberg, A. A 2.5-Mb Transcript Map of a Tumor-Suppressing Subchromosomal Transferable Fragment from 11p15.5, and Isolation and Sequence Analysis of Three Novel Genes. Genomics 1997, 46, 9–17. [Google Scholar] [CrossRef]
- Gubas, A.; Karantanou, C.; Popovic, D.; Tascher, G.; Hoffmann, M.E.; Platzek, A.; Dawe, N.; Dikic, I.; Krause, D.S.; McEwan, D.G. The endolysosomal adaptor PLEKHM1 is a direct target for both mTOR and MAPK pathways. FEBS Lett. 2021, 595, 864–880. [Google Scholar] [CrossRef]
- Rocks, N.; Paulissen, G.; El Hour, M.; Quesada, F.; Crahay, C.; Gueders, M.; Foidart, J.; Noel, A.; Cataldo, D. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie 2008, 90, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N.; Morris, L.G.; Lee, W.; Chan, T.A. Unraveling the molecular genetics of head and neck cancer through genome-wide approaches. Genes Dis. 2014, 1, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Network CGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carron, J.; Della Coletta, R.; Lourenço, G.J. Pseudogene Transcripts in Head and Neck Cancer: Literature Review and In Silico Analysis. Genes 2021, 12, 1254. https://doi.org/10.3390/genes12081254
Carron J, Della Coletta R, Lourenço GJ. Pseudogene Transcripts in Head and Neck Cancer: Literature Review and In Silico Analysis. Genes. 2021; 12(8):1254. https://doi.org/10.3390/genes12081254
Chicago/Turabian StyleCarron, Juliana, Rafael Della Coletta, and Gustavo Jacob Lourenço. 2021. "Pseudogene Transcripts in Head and Neck Cancer: Literature Review and In Silico Analysis" Genes 12, no. 8: 1254. https://doi.org/10.3390/genes12081254
APA StyleCarron, J., Della Coletta, R., & Lourenço, G. J. (2021). Pseudogene Transcripts in Head and Neck Cancer: Literature Review and In Silico Analysis. Genes, 12(8), 1254. https://doi.org/10.3390/genes12081254






