Screening Potential Reference Genes in Tuta absoluta with Real-Time Quantitative PCR Analysis under Different Experimental Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Treatments
2.2.1. Developmental Stages
2.2.2. Adult Tissues
2.2.3. Insecticides
2.2.4. Temperatures
2.2.5. Plant Volatile Organic Compounds (VOCs)
2.3. Total RNA Extraction and cDNA Synthesis
2.4. Candidate Reference Gene Selection and Specific Primer Design
2.5. Standard Curve Construction and RT-qPCR
2.6. Statistical Analysis
3. Results
3.1. Acquisition of Candidate Reference Gene Sequences
3.2. Specificity and Amplification Efficiency of Specific RT-qPCR Primers
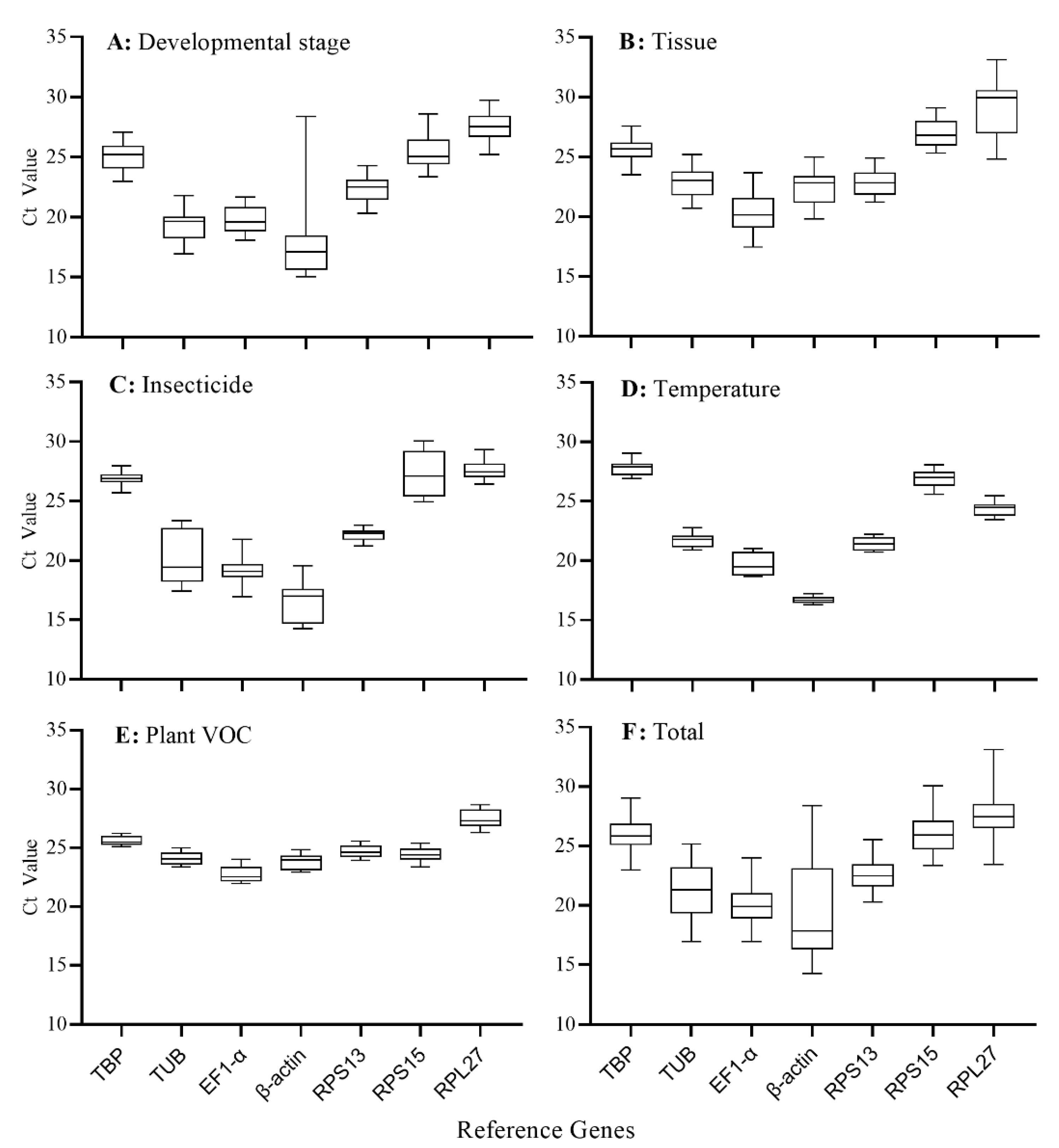
3.3. Expression Levels of the Candidate Reference Genes
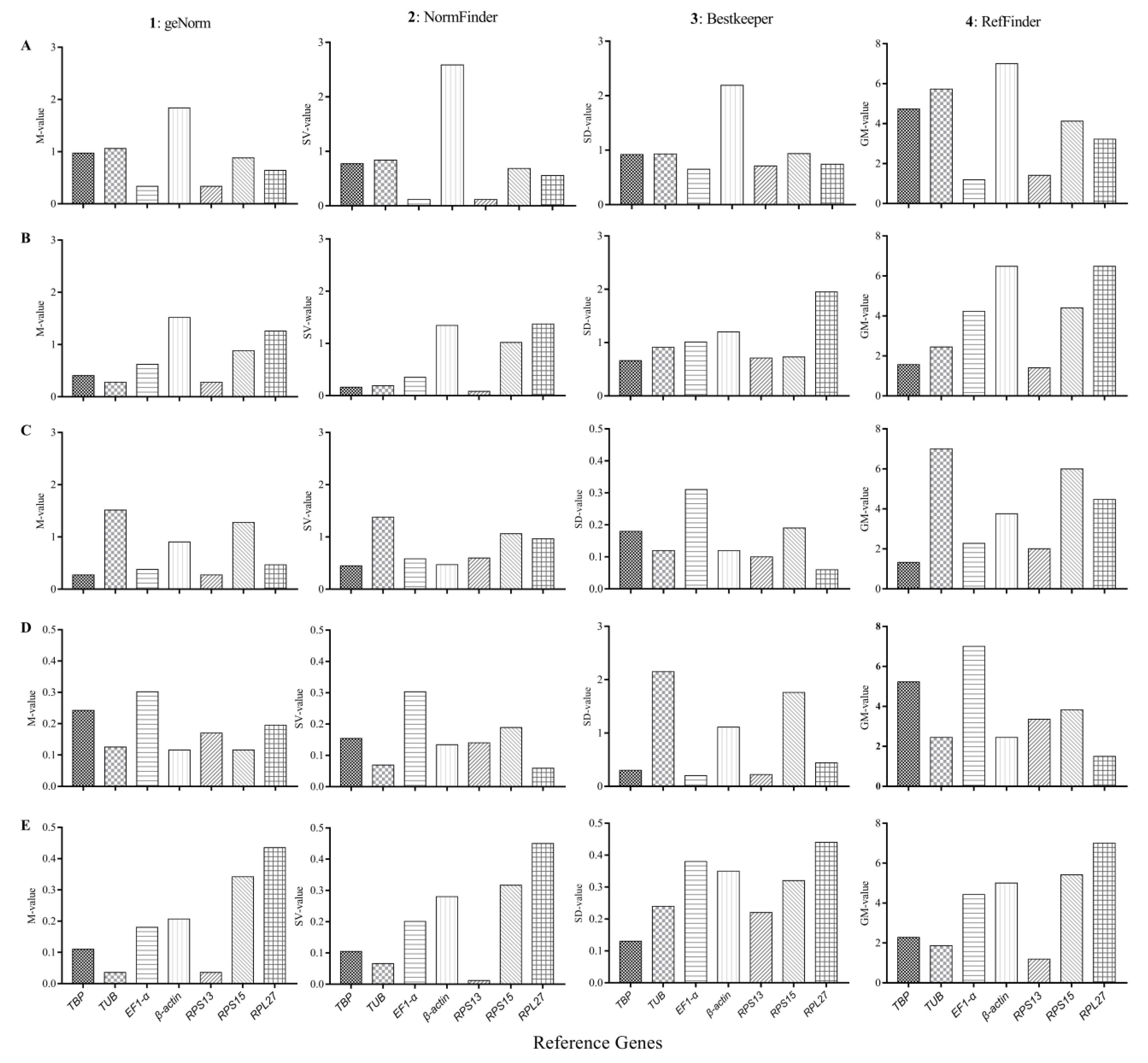
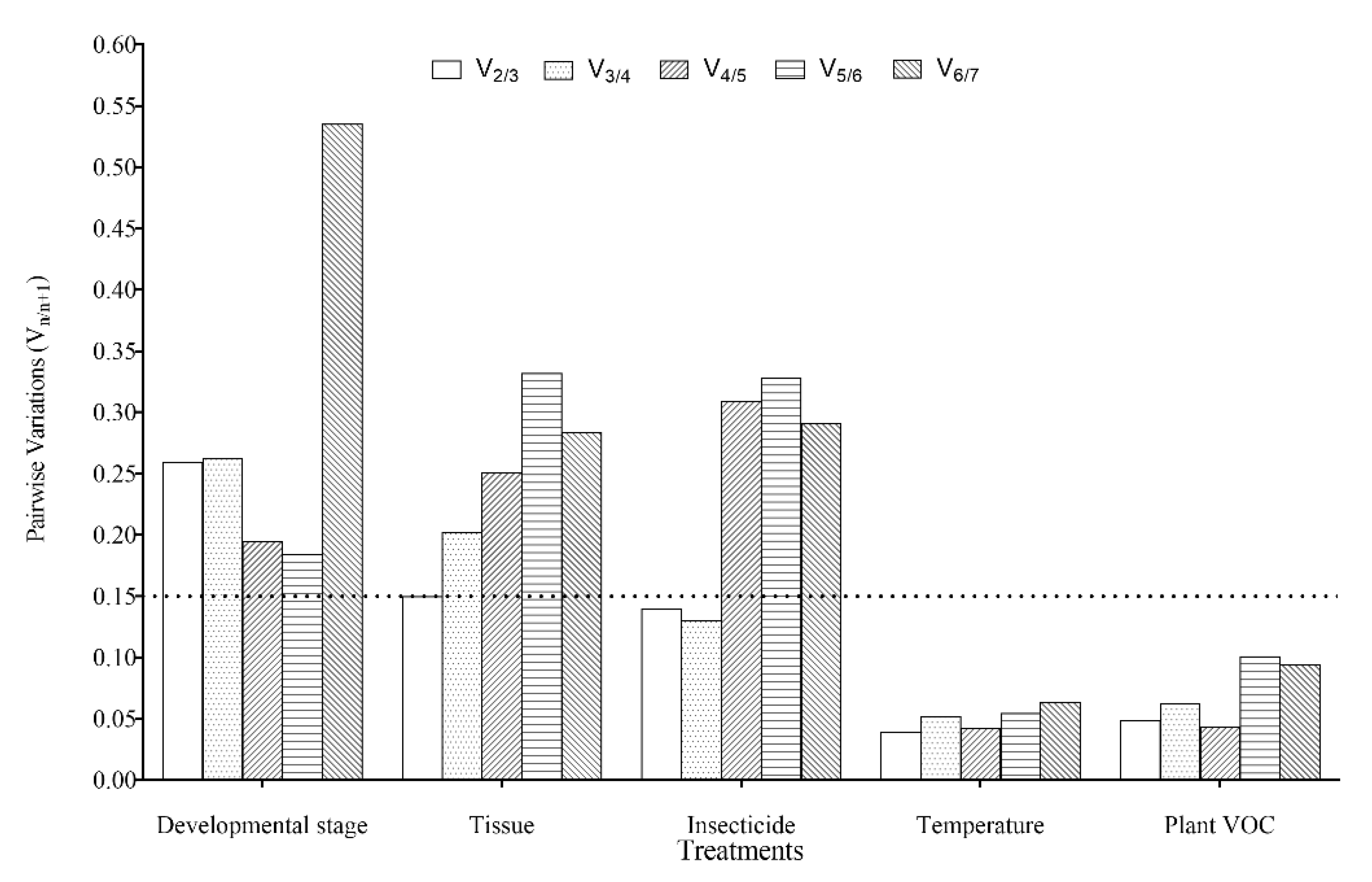
3.4. Expression Stability and Ranking of the Candidate Reference Genes
3.4.1. Developmental Stages
3.4.2. Adult Tissues
3.4.3. Insecticide-Induced Stress
3.4.4. Temperature-Induced Stress
3.4.5. Plant VOC-Induced Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Radonić, A.; Thulke, S.; Mackay, I.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalization; strategies and considerations. Genes. Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Lü, Y.N.; Zeng, M.; Jia, P.Y.; Lu, H.N.; Zhu, Y.B.; Li, S.; Cui, Y.Y.; Luan, Y.X. Selection of reference genes for normalization of gene expression in Thermobia domestica (Insecta: Zygentoma: Lepismatidae). Genes 2021, 12, 21. [Google Scholar] [CrossRef]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef] [Green Version]
- de Jonge, H.J.M.; Fehrmann, R.S.N.; de Bont, E.S.J.M.; Hofstra, R.M.W.; Gerbens, F.; Kamps, W.A.; de Vries, E.G.E.; van der Zee, A.G.J.; te Meerman, G.J.; ter Elst, A. Evidence based selection of housekeeping genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piazza, V.G.; Bartke, A.; Miquet, J.G.; Sotelo, A.I. Analysis of different approaches for the selection of reference genes in RT-qPCR experiments: A case study in skeletal muscle of growing mice. Int. J. Mol. Sci. 2017, 18, 1060. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Meth. 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Lee, P.D.; Sladek, R.; Greenwood, C.M.; Hudson, T.J. Control genes and variability: Absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002, 12, 292–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 467–470. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, L.; Mauriat, M.; Pelloux, J.; Bellini, C.; Wuytswinkel, O.V. Towards a systematic validation of references in real-time RT-PCR. Plant Cell 2008, 20, 1734–1735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, H.; Wu, X.Q.; Lyu, Y.P.; Zhou, H.Z.; Xie, X.Y.; Zhang, X.J.; Yang, H.T. Selection of reliable reference genes for gene expression studies in Botrytis cinerea. J. Microbiol. Meth. 2017, 142, 71. [Google Scholar] [CrossRef] [PubMed]
- Scharlaken, B.; de Graaf, D.C.; Goossens, K.; Brunain, M.; Peelman, L.J.; Jacobs, F.J. Reference gene selection for insect expression studies using quantitative real-time PCR: The head of the honeybee, Apis mellifera, after a bacterial challenge. J. Insect Sci. 2008, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- de Boer, M.E.; de Boer, T.E.; Mariën, J.; Timmermans, M.J.T.N.; Nota, B.; van Straalen, N.M.; Ellers, J.; Roelofs, D. Reference genes for qRT-PCR tested under various stress conditions in Folsomia candida and Orchesella cincta (Insecta, Collembola). BMC Mol. Biol. 2009, 10, 54. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.B.; Liu, Y.H.; Tang, P.A.; Zhou, A.W.; Wang, J.J. Validation of endogenous reference genes for insecticide-induced and developmental expression profiling of Liposcelis bostrychophila (Psocoptera: Liposcelididae). Mol. Biol. Rep. 2010, 37, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Arun, A.; Baumlé, V.; Amelot, G.; Nieberding, C.M. Selection and validation of reference genes for qRT-PCR expression analysis of candidate genes involved in olfactory communication in the butterfly Bicyclus anynana. PLoS ONE 2015, 3, e0120401. [Google Scholar] [CrossRef]
- Koramutla, M.K.; Aminedi, R.; Bhattacharya, R. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt.). Sci. Rep. 2016, 6, 25883. [Google Scholar] [CrossRef]
- Ponton, F.; Chapuis, M.P.; Pernice, M.; Sword, G.A.; Simpson, S.J. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 2011, 57, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.H.; Yuan, M.; Gao, X.W.; Kang, T.H.; Zhan, S.; Wan, H.; Li, J.H. Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS ONE 2013, 8, e68059. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.J.; Sun, L.L.; Zhang, Q.H.; Cao, C.W. Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene 2020, 749, 144712. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.; Wu, Q.; Liu, Y.; Zhou, X.; Zhou, X.G.; Zhang, Y. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792–802. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.L.; Jing, X.D.; Chen, W.; Bai, J.L.; Vasseur, L.; He, W.Y.; You, M.S. Selection of reference genes for expression analysis of plant-derived microRNAs in Plutella xylostella using qRT-PCR and ddPCR. PLoS ONE 2019, 14, e0220475. [Google Scholar] [CrossRef] [Green Version]
- Shu, B.; Zhang, J.J.; Cui, G.F.; Sun, R.R.; Sethuraman, V.; Yi, X.; Zhong, G.H. Evaluation of reference genes for real-time quantitative PCR analysis in larvae of Spodoptera litura exposed to azadirachtin stress conditions. Front. Physiol. 2018, 9, 372. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.X.; Wang, G.R.; Lu, Y.H.; Yang, Y.J.; Zheng, X.S.; Tian, J.C.; Lü, Z.X. Screening reference genes and evaluating of their expression stability for qRT-PCR normalization in Chilo suppressalis (Lepidoptera: Pyralididae). Chin. J. Rice Sci. 2019, 33, 75–84. [Google Scholar]
- Zhang, S.D.; An, S.H.; Li, Z.; Wu, F.M.; Yang, Q.P.; Liu, Y.C.; Cao, J.J.; Zhang, H.J.; Zhang, Q.W.; Liu, X.X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yuan, M.; Shakeel, M.; Zhang, Y.J.; Wang, S.L.; Wang, X.; Zhan, S.; Kang, T.H.; Li, J.H. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hubner) (Lepidoptera: Noctuidae). PLoS ONE 2014, 9, e84730. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lu, M.X.; Tang, X.T.; Du, Y.Z. Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: Noctuidae). PLoS ONE 2015, 10, e0115979. [Google Scholar] [CrossRef]
- Pan, H.P.; Yang, X.W.; Bidne, K.; Hellmich, R.L.; Siegfried, B.D.; Zhou, X.G. Selection of reference genes for RT-qPCR analysis in the monarch butterfly, Danaus plexippus (L.), a migrating bioindicator. PLoS ONE 2015, 10, e0129482. [Google Scholar] [CrossRef]
- Zhao, Z.J.; Wang, L.M.; Yue, D.M.; Ye, B.; Li, P.P.; Zhang, B.; Fan, Q. Evaluation of reference genes for normalization of RT-qPCR gene expression data for Trichoplusia ni cells during Antheraea pernyi (Lepidoptera: Saturniidae) multicapsid nucleo polyhedrovirus (AnpeNPV) infection. J. Insect Sci. 2019, 19, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Biondi, A.; Guedes, R.N.C.; Wan, F.H.; Desneux, N. Ecology, worldwide spread, and management of the invasive South American tomato pinworm, Tuta absoluta: Past, present, and future. Annu. Rev. Entomol. 2018, 63, 239–258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.F.; Liu, W.X.; Wan, F.H.; Xian, X.Q.; Zhang, Y.B.; Guo, J.Y. Bioecology, damage and management of the tomato leafminer Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae), a worldwide quarantine pest. J. Biosaf. 2018, 27, 155–163. [Google Scholar]
- Zhang, G.F.; Ma, D.Y.; Wang, Y.S.; Gao, Y.H.; Liu, W.X.; Zhang, R.; Fu, W.J.; Xian, X.Q.; Wang, J.; Meng, K.; et al. First report of the South American tomato leafminer, Tuta absoluta (Meyrick), in China. J. Integr. Agric. 2020, 19, 1912–1917. [Google Scholar] [CrossRef]
- Zhang, G.F.; Xian, X.Q.; Zhang, Y.B.; Zhang, R.; Ma, D.Y.; Liu, W.X.; Gao, Y.H.; Wang, J.; Yang, Z.L.; Li, Q.H.; et al. Warning of the dispersal of a newly invaded alien species, tomato leafminer Tuta absoluta (Meyrick), in China. Plant Prot. 2020, 46, 281–286. [Google Scholar]
- Desneux, N.; Wajnberg, E.; Wyckhuys, K.A.G.; Burgiog, G.; Arpaia, S.; Narvaéz-Vasquez, C.A.; González-Cabrera, J.; Ruescas, D.C.; Tabone, E.; Frandon, J.; et al. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest Sci. 2010, 83, 197–215. [Google Scholar] [CrossRef]
- Campos, M.R.; Biondi, A.; Adiga, A.; Guedes, R.N.C.; Desneux, N. From the Western Palaearctic region to beyond: Tuta absoluta ten years after invading Europe. J. Pest Sci. 2017, 90, 787–796. [Google Scholar] [CrossRef]
- Guedes, R.N.C.; Roditakis, E.; Campos, M.R.; Haddi, K.; Bielza, P.; Siqueira, H.A.A.; Tsagkarakou, A.; Vontas, J.; Nauen, R. Insecticide resistance in the tomato pinworm Tuta absoluta: Patterns, spread, mechanisms, management and outlook. J. Pest Sci. 2019, 92, 1329–1342. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Z.Q.; Hu, Y.; Tan, J.H.; Jia, J.; Xu, H.L.; Chen, X.H. Reference genes selection for quantitative gene expression studies in Pinus massoniana L. Trees 2016, 30, 685–696. [Google Scholar] [CrossRef]
- Nagy, N.A.; Németh, Z.; Juhász, E.; Póliska, S.; Rácz, R.; Kosztolányi, A.; ZoltánBarta, Z. Evaluation of potential reference genes for real-time qPCR analysis in a biparental beetle, Lethrus apterus (Coleoptera: Geotrupidae). Peer J. 2017, 5, e4047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, B.; Liang, P.; Gao, X.W. Cloning and evaluation of U6 snRNA gene as a reference gene for miRNA quantification in Plutella xylostella (Lepidoptera: Plutellidae). Acta Entomol. Sin. 2014, 57, 286–292. [Google Scholar]
- Ma, L.; Li, X.W.; Wang, S.M.; Wang, T.Z.; Lü, Y.B. Toxicity of six insecticides to tomato leaf miner moth in Yunnan. In Proceedings of the 2019 Annual Conference of Plant Protection Society of China, Guiyang, China, 23 October 2019. [Google Scholar]
- Zhang, G.F.; Zhang, Y.B.; Zhang, J.; Liu, W.X.; Wang, Y.S.; Wan, F.H.; Shu, C.L.; Liu, H.; Wang, F.L.; Zhao, L.; et al. Laboratory Toxicity and Field Control Efficacy of Biopesticide Bacillus thuringiensis G033A on the South American Tomato Leafminer Tuta absoluta (Meyrick), a New Invasive Alien Species in China. Chin. J. Biol. Control. 2020, 36, 175–183. [Google Scholar]
- Cuthbertson, A.G.S.; Mathers, J.J.; Blackburn, L.F.; Korycinska, A.; Luo, W.Q.; Jacobson, R.J.; Northing, P. Population development of Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae) under simulated UK glasshouse conditions. Insects 2013, 4, 185–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, J.C.; Picanço, M.C.; Bacci, L.; Guedes, R.N.C.; Santana, P.A.; Ferreira, D.O.; Chediak, M. Life table determination of thermal requirements of the tomato borer Tuta absoluta. J. Pest Sci. 2016, 89, 897–908. [Google Scholar] [CrossRef]
- Weid, D.; Rossier, D.; Lindup, M.; Tuberosa, J.; Widmer, A.; Col, J.D.; Kan, C.; Carleton, A.; Rodriguez, I. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat. Neurosci. 2015, 18, 1455. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Q. Identification of Odorant Receptors Responding to Myrcene and Ethyl Butyrate in Diaphorina citri. Master’s Thesis, Southwest University, Chongqing, China, 2018. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Provenzano, M.; Mocellin, S. Complementary techniques: Validation of gene expression data by quantitative real time PCR. Adv. Exp. Med. Biol. 2007, 593, 66. [Google Scholar] [CrossRef] [PubMed]
- Ladror, D.T.; Frey, B.L.; Scalf, M.; Levenstein, M.E.; Artymiuk, J.M.; Smith, L.M. Methylation of yeast ribosomal protein S2 is elevated during stationary phase growth conditions. Biochem. Biophys. Res. Commun. 2014, 445, 535–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef] [Green Version]
- Jank, T.; Belyi, Y.; Wirth, C.; Rospert, S.; Hu, Z.; Dengjel, J.; Tzivelekidis, T.; Andersen, G.R.; Hunte, C.; Schlosser, A.; et al. Protein glutaminylation is a yeast-specific posttranslational modification of elongation factor 1A. J. Biol. Chem. 2017, 292, 16014–16023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Jin, Y.; He, L.; Lu, W.C.; Li, M. Suitable reference gene selection for the different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. J. Insect Sci. 2010, 10, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.N. Cloning and Stability Analysis of Reference Gene from Phyllonorycter ringoniell. Master’s Thesis, Northwest A&F University, Xianyang, China, 2014. [Google Scholar]
- Li, H.B.; Dai, C.G.; Zhang, C.R.; He, Y.F.; Ran, H.Y.; Chen, S.H. Screening potential reference genes for quantitative real-time PCR analysis in the oriental armyworm, Mythimna separata. PLoS ONE 2018, 13, e0195096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Gene Name | Primer | Primer Sequences (5′-3′) | Tm (°C) | Amplicon Length (bp) |
|---|---|---|---|---|
| TUB | TUB-F TUB-R | GAGTGCATCTCAGTCCACGTT GAAGGAGTGGAAGATGAGGAAT | 55 | 417 |
| β-actin | β-actin-F β-actin-R | TCCTTGGAGATCCACATCTG CCCATCTACGAAGGTTACG | 53 | 590 |
| EF1-α | EF1-α-F EF1-α-R | GCCCTTCTCGCCTTCACCCTT GGGCACCGTTCCAATACCACC | 62 | 381 |
| RPL27 | RPL27-F RPL27-R | GTAAGATTATGAAACCTGGGAA ACCTAAGCTTTTGGAAGAACC | 51 | 395 |
| RPS13 | RPS13-F RPS13-R | CCCTACCTGGTTGAAACTGAC TGAAGCGGTGCTAGACTCATA | 51 | 376 |
| TBP | TBP-F TBP-R | TCCTCACCACTTGTCGGCT GTTGTCCGTGGCTCTCTGAT | 58 | 374 |
| RPS15 | RPS15-F RPS15-R | CGAGAAATACCCCAGAAATGT TTCTTCGCTTGTTGTTTGATT | 51 | 429 |
| Primer | Primer Sequences (5′-3′, F/R) | Amplicon Length (bp) | Tm (°C) | Efficiency (%) | R2 |
|---|---|---|---|---|---|
| RPL27 | CGTTACTCGGTAGACTTCA GTTCTTTCCACTCTTGTATCG | 126 | 60 | 92.7 | 0.992 |
| RPS13 | AGTTGCCCAAGTCAGATT TTCCAAGTGCTTCCTCAT | 133 | 60 | 102.0 | 0.999 |
| RPS15 | ATACGAAGTGAACAGTTAGC ACGGTCCATTCTTCCAAT | 92 | 60 | 103.8 | 0.990 |
| EF1-α | AGTCTCCTCATACATCAAGAAG CCTCCTTACGCTCAACAG | 143 | 60 | 100.7 | 0.998 |
| TBP | GTCTACTCCACACCTCAA CTGCCACTCGTTATCATTG | 77 | 60 | 110.2 | 0.993 |
| TUB | GGAGTCCAGATCGGTAAC GCTGAAGAAGGTGTTGAAC | 126 | 60 | 95.9 | 0.998 |
| β-actin | CAGGTCCTTACGGATGTC CTCTTCCAGCCTTCCTTC | 99 | 60 | 104.4 | 0.996 |
| Gene Name | Total Samples | Developmental Stage | Tissue | Insecticide | Temperature | Plant VOC | |||
|---|---|---|---|---|---|---|---|---|---|
| Treatment | Control | Treatment | Control | Treatment | Control | ||||
| TBP | 25.84 ± 1.28 b | 25.14 ± 1.15 b | 25.45 ± 0.98 b | 26.92 ± 0.51 a | 27.64 ± 0.52 a | 27.80 ± 0.65 a | 27.84 ± 0.64 a | 25.61 ± 0.39 b | 25.54 ± 0.35 b |
| TUB | 21.14 ± 2.22 d | 19.31 ± 1.27 d,e | 22.81 ± 1.16 c | 20.41 ± 2.29 b,c | 21.34 ± 0.44 d | 21.78 ± 0.60 d | 21.63 ± 0.74 d | 24.09 ± 0.51 c,d | 23.88 ± 0.47 d |
| EF1-α | 20.19 ± 1.50 d,e | 19.80 ± 1.04 d | 20.30 ± 1.47 d | 19.22 ± 1.04 c | 20.12 ± 0.83 e | 19.74 ± 0.89 e | 19.92 ± 0.85 e | 22.74 ± 0.69 e | 22.35 ± 0.37 e |
| β-actin | 19.36 ± 3.68 e | 18.11 ± 3.69 e | 22.42 ± 1.41 c | 16.33 ± 1.70 d | 16.50 ± 0.15 f | 16.71 ± 0.25 f | 16.58 ± 0.21 f | 23.83 ± 0.63 d | 23.69 ± 0.59 d |
| RPS13 | 22.61 ± 1.22 c | 22.30 ± 1.00 c | 22.83 ± 0.95 c | 22.16 ± 0.49 b | 21.28 ± 0.46 d | 21.44 ± 0.55 d | 21.46 ± 0.58 d | 24.72 ± 0.50 c | 24.51 ± 0.41 c |
| RPS15 | 26.08 ± 1.62 b | 25.35 ± 1.31 b | 26.94 ± 1.14 b | 27.39 ± 1.91 a | 26.57 ± 0.53 b | 26.93 ± 0.70 b | 26.69 ± 0.53 b | 24.44 ± 0.63 c,d | 24.08 ± 0.41 c,d |
| RPL27 | 27.47 ± 1.84 a | 27.49 ± 1.15 a | 28.88 ± 2.34 a | 27.63 ± 0.74 a | 24.17 ± 0.57 c | 24.37 ± 0.61 c | 24.38 ± 0.69 c | 27.45 ± 0.76 a | 26.99 ± 0.42 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, A.-P.; Wang, Y.-S.; Huang, C.; Lv, Z.-C.; Liu, W.-X.; Bi, S.-Y.; Wan, F.-H.; Wu, Q.; Zhang, G.-F. Screening Potential Reference Genes in Tuta absoluta with Real-Time Quantitative PCR Analysis under Different Experimental Conditions. Genes 2021, 12, 1253. https://doi.org/10.3390/genes12081253
Yang A-P, Wang Y-S, Huang C, Lv Z-C, Liu W-X, Bi S-Y, Wan F-H, Wu Q, Zhang G-F. Screening Potential Reference Genes in Tuta absoluta with Real-Time Quantitative PCR Analysis under Different Experimental Conditions. Genes. 2021; 12(8):1253. https://doi.org/10.3390/genes12081253
Chicago/Turabian StyleYang, An-Pei, Yu-Sheng Wang, Cong Huang, Zhi-Chuang Lv, Wan-Xue Liu, Si-Yan Bi, Fang-Hao Wan, Qiang Wu, and Gui-Fen Zhang. 2021. "Screening Potential Reference Genes in Tuta absoluta with Real-Time Quantitative PCR Analysis under Different Experimental Conditions" Genes 12, no. 8: 1253. https://doi.org/10.3390/genes12081253
APA StyleYang, A.-P., Wang, Y.-S., Huang, C., Lv, Z.-C., Liu, W.-X., Bi, S.-Y., Wan, F.-H., Wu, Q., & Zhang, G.-F. (2021). Screening Potential Reference Genes in Tuta absoluta with Real-Time Quantitative PCR Analysis under Different Experimental Conditions. Genes, 12(8), 1253. https://doi.org/10.3390/genes12081253






