Abstract
Cottonseed oil is rich in unsaturated fatty acids (UFAs) and serves as an edible oil in human nutrition. Reports suggest that acyl-coenzyme A: diacylglycerol acyltransferases (DGAT) and wax ester synthase/DGAT (WSD1) genes encode a key group of enzymes that catalyze the final step for triacylglycerol biosynthesis and enable an important rate-limiting process. However, their roles in oil biosynthesis and the fatty acid profile of cotton seed are poorly understood. Therefore, the aim of this study was to identify and characterize DGAT and WSD1 genes in cotton plants and examine their roles in oil biosynthesis, the fatty acid profile of cotton seeds, and abiotic stress responses. In this study, 36 GhDGAT and GhWSD1 genes were identified in upland cotton (G. hirsutum) and found to be clustered into four groups: GhDGAT1, GhDGAT2, GhDGAT3, and GhWSD1. Gene structure and domain analyses showed that the GhDGAT and GhWSD1 genes in each group are highly conserved. Gene synteny analysis indicated that segmental and tandem duplication events occurred frequently during cotton evolution. Expression analysis revealed that GhDGAT and GhWSD1 genes function widely in cotton development and stress responses; moreover, several environmental stress and hormone response-related cis-elements were detected in the GhDGAT and GhWSD1 promoter regions. The predicted target transcription factors and miRNAs imply an extensive role of GhDGAT and GhWSD1 genes in stress responses. Increases in GhDGAT3 gene expression with increases in cottonseed oil accumulation were observed. Transformation study results showed that there was an increase in C18:1 content and a decrease in C18:2 and C18:3 contents in seeds of Arabidopsis transgenic plants overexpressing GhDGAT3D compared with that of control plants. Overall, these findings contributed to the understanding of the functions of GhDGAT and GhWSD1 genes in upland cotton, providing basic information for further research.
1. Introduction
Cotton (Gossypium) is the fifth largest oil crop in the world and the most important natural fiber producing plant. Among cotton species, the upland cotton (Gossypium hirsutum, AD1) is the most cultivated, accounting for over 90% of cultivated cotton species due to its high fiber production and widespread environmental adaptation. G. hirsutum is believed to have resulted from the hybridization and genome doubling of two diploid ancestors, Gossypium arboreum (A2) and Gossypium raimondii (D5) [1]. Cottonseed oil is the primary byproduct of cotton processing, accounting for approximately 16% of the seed’s weight [2]. Moreover, cottonseed oil is an excellent edible oil for human consumption due to its high content of unsaturated fatty acids (UFA), consisting of 57.64% linoleic acid (C18:2), 22.92% palmitic acid (C16:0), 15.39% oleic acid (C18:1), and 2.22% stearic acid (C18:0) [3]. Additionally, high unsaturated fatty acid content has been linked to a reduced risk of cardiovascular diseases. Additionally, the large amount of cottonseed oil serves as a valuable resource for bioenergy [4].
Oil biosynthesis in plants includes two steps: de novo fatty acid biosynthesis in plastids and triacylglycerol (TAG) biosynthesis in the endoplasmic reticulum (ER) [5,6]. Lipid droplets (oil bodies) are the main type of seed oil stored in plants, with TAGs surrounded by steroleosins, oleosins, and caleosins. During TAG biosynthesis, glycerol-3-phosphate (G3P) serves as the substrate, while acyl-CoA serves as the acyl donor. In this pathway, glycerol-3-phosphate acyltransferase (GPAT) catalyzes the first acylation of G3P to yield lysophosphatidic acid (LPA), which is further acylated by lysophosphatidic acid acyltransferase (LPAAT) to produce phosphatidic acid (PA), followed by the conversion of PA to diacylglycerol (DAG) by phosphatidic acid phosphatase (PP). DGAT catalyzes the final acylation of the sn-3 position of DAG to form TAG, which is the committed step in acyl-CoA dependent TAG biosynthesis, known as the Kennedy pathway [7,8]. However, an acyl-CoA independent pathway also exists, in which phospholipid: diacylglycerol acyltransferase (PDAT) transfers an acyl moiety from phosphatidylcholine to a DAG sn-3 position to produce TAG [9,10]. Overall, both DGAT and PDAT play crucial roles in guiding carbon flux into TAG.
DGAT has been elaborately characterized to play a crucial role in DAG-to-TAG transformation. Three types of DGAT genes have been identified in plants: ER-localized DGAT1 and DGAT2, and soluble cytosolic DGAT3 [11,12]. DGAT1 is highly expressed in developing embryos of oilseed crops, and its expression level is correlated with oil accumulation during seed development [13,14]. Inactivation of DGAT1 results in decreased seed oil content in the Arabidopsis mutant AS11 [15]. ZmDGAT1 activation (by inserting a phenylalanine) increases the oil content of a maize (Zea mays) embryo [16]. However, compared with the role of DGAT1 in seed oil deposition, DGAT2 plays a minor role in regulating oil production [17]. There are no differences between AtDGAT2 mutants and the wild type (WT) in TAG accumulation [18]. However, AtDGAT2 in TAG biosynthesis was functionally confirmed via transient expression in Nicotiana benthamiana leaves [19]. Overexpression of Jatropha curcas JcDGAT1 or JcDGAT2 resulted in a 25% and 29.6% increase in oil production, respectively, compared with that of control plants [20]. The plant soluble cytosolic DGAT3 was first identified in peanuts (Arachis hypogaea) [21]. A study showed that Arabidopsis DGAT3 participated in the collecting of C18:2 and C18:3 into TAG when TAG breakdown was blocked [22]. AtDGAT3 is a metalloprotein involved in TAG biosynthesis [23]. Seed-specific expression of Camelina sativa CaDGAT3-3 significantly enhanced oil and unsaturated fatty acid accumulation, with higher levels of eicosanoic acid in tobacco seeds [24]. The reports of a previous study showed that the expression of GhDGAT3 was higher than that of GhDGAT1 and GhDGAT2 in developing cottonseeds, indicating the key role of GhDGAT3 in oil biosynthesis [3].
Furthermore, another enzyme of interest in the cotton plant is the WSD1 enzyme. WSD1 has a high level of wax synthase activity and an approximately 10-fold lower level of diacylglycerol acyltransferase activity than that of wax synthase activity [25], which is necessary for protection against desiccation [26]. Moreover, the WSD1 enzyme has been reported to play a considerable role in environmental adaptation and stress management [26,27].
Genome-wide analysis of the DGAT gene family has been performed in maize [28], oil palms [29], and soybeans [14,30]. A previous study also compared the structure and motifs of DGATs in upland cotton with that of oil palms [29]; however, a comprehensive analysis is still lacking. Therefore, the aim of this study was to comprehensively examine and characterize the DGAT and WSD1 gene families and to identify their functions in upland cotton. Based on the updated genome data [31], we performed a characteristic analysis of GhDGAT and GhWSD1 family genes in upland cotton. GhDGAT3D was functionally confirmed by overexpression in Arabidopsis. Our study aimed to improve the understanding of the roles of GhDGAT in oil biosynthesis and the function of DGAT genes in other oil crops.
2. Materials and Methods
2.1. Identification of DGAT and WSD1 Family Members in Gossypium
Genome datasets of Gossypium hirsutum acc. TM-1 (AD1, CRI_v1) and its ancestors, G. arboreum (A2, CRI_v1.0) and G. raimondii (D5, JGI_v2.1), were downloaded from the CottonGen website [32]. To identify candidate DGAT and WSD genes in cotton, the genome datasets were aligned against Arabidopsis DGAT and WSD amino acid (aa) sequences of AtDGAT1 (AT2G19450), AtDGAT2 (AT3G51520), AtDGAT3 (AT1G48300), and AtWSD1 (AT5G37300) for BLASTp search. The DGAT and WSD1 genes were annotated according to their corresponding orthologs in Arabidopsis and chromosomal location in upland cotton, and genes in G. hirsutum were named based on their homoeologs in each subgenome, with ‘A’ and ‘D’ representing homoeologs in the At and Dt subgenomes, respectively [33]. Whether a gene was from an At subgenome or Dt subgenome was judged by their DNA sequence homology with an A genome diploid species (G. arboreum) or a D genome diploid species (G. raimondii). The theoretical molecular weight (MW) and isoelectric point (pI) were predicted using ExPASy software [34]. Subcellular localization of DGAT and WSD1 proteins was evaluated via the WoLF PSORT server (https://wolfpsort.hgc.jp/, accessed on 28 June 2021). The regions 2 kb upstream of the start codon of GhDGAT and GhWSD1 promoter were subjected to the PlantCARE database [35] for cis-element searching.
2.2. Phylogenetic, Gene Structure, Conserved Domain, and Motifs Analysis
The muscle program in MEGA-X software [36] was used to perform multiple amino acid sequence alignments, and then the unrooted phylogenetic tree was constructed using the maximum likelihood (ML) method by bootstrap tests with 1000 replicates. The phylogenetic tree was visualized using the iTOL tool [37]. The structure of GhDGAT and GhWSD1 genes was obtained from the genomic dataset and exhibited by using a gene structure display server (GSDS) [38]. The conserved domains of GhDGAT and GhWSD1 proteins were searched using SMART software [39] and displayed using TBtools [40]. Amino acid sequences of GhDGAT and GhWSD1 in the G. hirsutum protein dataset were submitted to the MEME program to identify the conserved protein motifs [41]. A functional search of the conserved motifs was performed using the InterProScan database. To determine the evolutionary relationships, the transmembrane (TM) structures of GhDGAT and GhWSD1 proteins were predicted using the TMHMM-2.0 website (http://www.cbs.dtu.dk/services/TMHMM/#opennewwindow, accessed on 16 June 2020).
2.3. Chromosomal Location and Gene Synteny Analysis
The physical location on the chromosome of GhDGAT and GhWSD1 in G. hirsutum was acquired from the genomic datasets and displayed with Mapchart 2.2 software [42]. Gene synteny analysis was carried out using MCScanX software [43], and Circos [44] was used for graphical depiction.
2.4. Expression Pattern Analysis
The transcript level was calculated based on publicly released data. Gene expression datasets for developing cottonseeds were acquired by Hu et al [45]. RNA-Seq datasets of different tissues and cottonseed development in G. hirsutum were obtained from the BioProject with accession number PRJNA490626 [46]. Transcript levels were estimated as fragments per kilobase million (FPKM) value using HISAT and StringTie software tools [47].
2.5. Transcription Factors and miRNAs Targeting GhDGAT and GhWSD1 Homologs
Transcription factors regulating GhDGAT and GhWSD1 were predicted using PlantRegMap [48], with G. raimondii as the target. The full-length cDNA sequences of GhDGAT and GhWSD1 homologs were submitted to the psRNATarget website [49] for a potential miRNAs search against the G. hirsutum miRNA database. The relationships between the predicted TFs and miRNAs with GhDGAT and GhWSD1 were displayed by Cytoscape software [50].
2.6. Cotton Seedlings Treatments and Sampling
Seedlings of upland cotton, Zhongmiansuo 24 (ZM24) variety, were used for gene expression analysis in response to drought, salt, and cold stresses. Seedlings in three-leaf-stage were cultivated in Hoagland liquid medium containing 17% PEG6000 or 200 mM NaCl for drought and salt stresses, respectively, and were exposed to 4 °C conditions for cold stress. The leaves of the cotton seedlings were collected at eight time points (0, 0.5, 1, 3, 6, 12, 24, and 48 h) and rapidly frozen in liquid nitrogen for total RNA extraction.
2.7. Recombination Vector Construction and Arabidopsis Transformation
Based on the expression module, GhDAGT3D was selected for functional analysis via genetic transformation. The full-length cDNA of GhDGAT3D was cloned from the embryo of ZM24 upland cotton variety at 20 DPA. The ClonExpress® MultiS One Step Cloning Kit (Vazyme, Nanjing, China) was used to clone GhDGAT3D into the pCAMBIA2300 vector behind a 35S promoter, which we named p35S::GhDGAT3D recombined vector in this study. The recombined vector was transformed into Agrobacterium strain GV3101 using the heat shock method. Arabidopsis thaliana Columbia (Col-0) was grown in a greenhouse at 22 °C under a 16 h/8 h light/dark cycle. Transformation was performed using the floral dip method [51]. Positive plants were selected using kanamycin in selection media and verified by polymerase chain reaction (PCR) methods. The transgenic plants in the T1 generation with 3:1 positive: negative proportion were regarded as single-copy transgenic plants.
2.8. Oil Content and Fatty Acid Composition of Transgenic Arabidopsis
Transgenic Arabidopsis seeds were collected from the T3 generation and used for oil content and fatty acid composition analysis. Briefly, approximately 0.1 g of Arabidopsis seeds were ground to powder, and the fatty acid profile was detected using the Agilent 8890 gas chromograph (Agilent, Santa Clara, CA, USA) with C19:0 as the internal standard [52]. The statistically significant differences were determined by Student’s t-test.
2.9. RNA Isolation and Real-time Quantitative PCR (RT-qPCR)
Total RNA of the ZM24 upland cotton variety and transgenic Arabidopsis plants was isolated using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China) following the manufacturer’s protocol. After genomic DNA digestion, approximately 1 μg of total RNA was used for reverse transcription using the PrimeScriptTM RT reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Specific primers were designed using Primer6 software based on the coding nucleotide sequences of GhDGAT, GhWSD1, and oil-related genes in Arabidopsis (Table S1). RT-qPCR analysis was performed as described by Zhao et al [25], and the housekeeping cotton genes GhUBQ7 and Arabidopsis AtSYL8 were used as internal references. All experiments were performed in triplicates.
3. Results
3.1. Identification of GhDGAT and GhWSD Family Genes in Gossypium
Arabidopsis DGAT and WSD amino acid (aa) sequences of AtDGAT1 (AT2G19450), AtDGAT2 (AT3G51520), AtDGAT3 (AT1G48300), and AtWSD1 (AT5G37300) were used as queries for the BLASTp search of the Gossypium protein database. The results showed that 19, 19, and 36 DGAT and WSD genes were detected in the A2, D5, and AD1 genomes, respectively (Table 1 and Table S2). Eight DGAT and WSD genes were also identified in the cacao plant (Theobroma cacao), Malvaceae family. The phylogenetic tree showed that the 86 DGAT and WSD1 genes were classified into four groups: DGAT1, DGAT2, DGAT3, and WSD1 (Figure 1). Moreover, DGAT and WSD genes in the same group in different cotton genomes tended to form one clade, reflecting the orthologous relationships. The CDS length of the 36 GhDGAT and GhWSD1 genes in G. hirsutum ranged from 243 bp (GhDGAT2-2D) to 1533 bp (GhWSD1-6D). The amino acid sequences of the genes were also characterized. The molecular weight (MW) ranged from 9.06 kDa (GhDGAT2-2D) to 57.77 kDa (GhWSD1-6D), and the isoelectric point (pI) ranged from 7.66 (GhWSD1-1A) to 9.81 (GhDGAT2-2D). The subcellular locations of the GhDGAT and GhWSD proteins were predicted by the WoLF PSORT software, and 13 GhDGAT and GhWSD1 proteins were predicted to be located in the cytoplasm (Table 1).

Table 1.
Characterization of GhDGAT and GhWSD1 genes in G. hirsutum.a The top three most possible subcellular localizations of GhDGAT and GhWSD1 genes are shown. Abbreviations: chlo: chloroplast; cysk: cytoskeleton; cyto: cytoplasm; E.R.: endoplasmic reticulum; extr: extracellular; mito: mitochondrion; nucl: nucleus; pero: peroxisome; plas: plastid; vacu: vacuole.
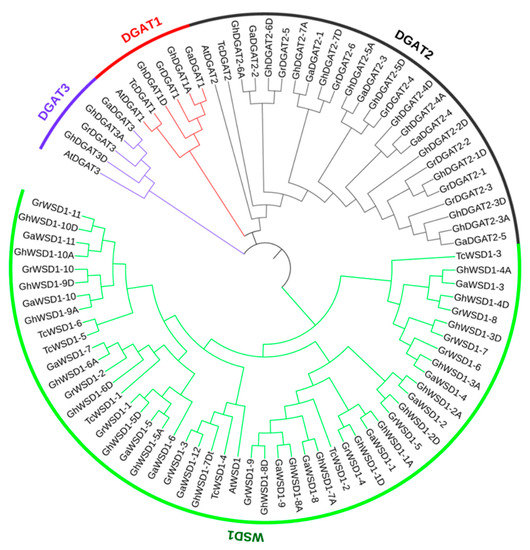
Figure 1.
Phylogenetic tree of the DGAT and WSD1 protein family. The phylogenetic tree was constructed using MEGA-X and displayed using iTOL software. At: Arabidopsis thaliana; Tc: Theobroma cacao; Gh: G. hirsutum; Ga: G. arboretum; Gr: G. raimondii.
3.2. Chromosomal Location and Gene Synteny of GhDGAT and GhWSD1 Genes in G. hirsutum
To reveal the homologous and homoeologous relationships of the GhDGAT and GhWSD1 genes, gene localization on chromosomes and gene duplication analyses were performed. We observed that most of the GhDGAT and GhWSD1 loci were highly parallel in the At and Dt subgenomes. The GhDGAT and GhWSD1 genes’ number and location on the At subgenomic chromosomes were similar to those on its homoeologous chromosomes in the Dt subgenome (Figure S1). The exception was GhDGAT2-1D and GhDGAT2-2D, which did not have homologs in the At subgenome, indicating that it might have been lost during evolution. However, the orthologs of GhDGAT2-1D and GhDGAT2-2D were present in G. raimondii (GrDGAT2-1 and GrDGAT2-2, respectively). Gene synteny analysis was performed to identify the duplicated genes of GhDGAT and GhWSD1 (Figure 2). The results showed that the cluster of GhDGAT2 might be duplicated genes, which belong to one synteny block. Moreover, GhWSD1-1A/D, GhWSD1-2A/D, GhWSD1-7, and GhWSD1-8A/D; GhWSD1-3A/D and GhWSD1-4A/D; GhWSD1-5A/D and GhWSD1-6A/D; and GhWSD1-5A/D and GhWSD1-6A/D might be duplicated genes (Figure 2). The results indicated that the GhDGAT and GhWSD1 genes were frequently duplicated during cotton evolution. According to the location of the duplicated genes, GhDGAT2-3A/D, GhDGAT2-4A/D, GhDGAT2-5A/D, GhDGAT2-6A/D, and GhDGAT2-7A/D; GhWSD1-1A/D and GhWSD1-2A/D; GhWSD1-1A/D and GhWSD1-2A/D; GhWSD1-1A/D and GhWSD1-2A/D; GhWSD1-3A/D and GhWSD1-4A/D; and GhWSD1-5A/D and GhWSD1-6A/D exhibited tandem duplication, whereas GhWSD1-9A/D and GhWSD1-10A/D might have resulted from segmental duplication.
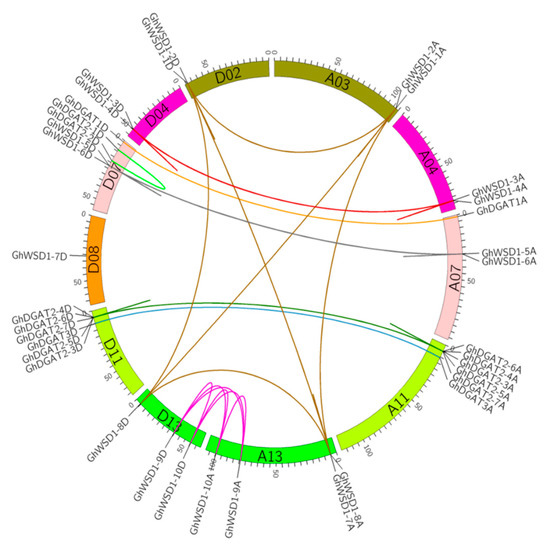
Figure 2.
The synteny relationship of GhDGAT and GhWSD1 genes. The relationship was visualized using Circos software. The homologous and homoeologous chromosomes in At and Dt subgenomes are displayed in the same color. The synteny relationship of GhDGAT and GhWSD1 genes are detectable in different colors. Light green lines: paralog genes of GhDGAT2-1D and GhDGAT2-2D; bottle green lines: duplicate genes of GhDGAT2; blue lines: paralog genes of GhDGAT3; dark brown lines: ortholog or paralog genes of GhWSD1-1, GhWSD1-2, GhWSD1-8, and GhWSD1-7A; red lines: ortholog or paralog genes of GhWSD1-3 and GhWSD1-4; orange lines: paralog genes of GhDGAT1; pink lines: ortholog or paralog genes of GhWSD1-9 and GhWSD1-10; grey lines: ortholog or paralog genes of GhWSD1-5 and GhWSD1-6.
3.3. The Conserved Structure and Motifs in GhDGAT and GhWSD1 Proteins
Upland cotton occupies the largest area of cultivated cotton globally and, therefore, more attention was paid to it during this study. A total of 36 GhDGAT and GhWSD1 proteins were used in the phylogenetic and gene structure analyses. The results showed that the phylogenetic relationships of GhDGAT and GhWSD1 proteins were in accordance with those of other cotton species (Figure 3A). The gene structure analysis showed that exon numbers ranged from 2 to 16. GhDGAT3D contains only one intron, most of the GhDGAT2 genes contain nine exons, whereas GhDGAT1 homoeologous genes contained 10 exons (Figure 3B). Consistently, GhDGAT and GhWSD1 genes with similar structures were grouped in the same clade.
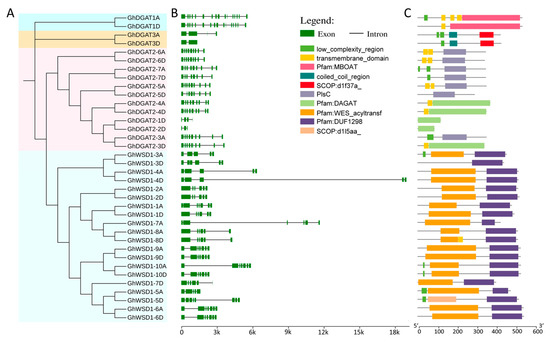
Figure 3.
Gene structure and conserved domains in GhDGAT and GhWSD1 members; (A) phylogenetic tree of GhDGAT and GhWSD1 proteins; (B) gene structure of exons and introns in GhDGAT and GhWSD1 genes; (C) the conserved domains in GhDGAT and GhWSD1 proteins.
Furthermore, the conserved domains were detected, GhDGAT1 homologs contained the MBOAT domain, GhDGAT3 homologs contained the SCOP:d1f317a domain; GhDGAT5, GhDGAT6, and GhDGAT7 homologs, and GhDGAT2-3A contained the PlsC domain. GhDGAT2-1D, GhDGAT2-2D, GhDGAT2-3D, and GhDGAT2-4 homologs contained the DAGAT domain. Characteristically, all the GhWSD1 proteins contained the DUF2198 domain, most of which have a WES_acyl transferase domain, except for GhWSD1-3D and GhWSD1-5D. However, only GhDGAT1, GhDGAT2-4, and GhDGAT2-6 homologs and GhDGAT2-3D and GhDGAT2-5A contained a transmembrane domain structure (Figure 3C). Additionally, the top 10 conserved motifs were identified in GhDGAT and GhWSD1 proteins; however, no conserved motifs were detected in GhDGAT1 and GhDGAT3 homologs (Figure S2). The annotation of motifs was in accord with the domains identified in GhDGAT and GhWSD1 proteins (Table S3, Figure 3C).
Transmembrane (TM) structures were predicted, and comparisons were made between GhDGAT1, GhDGAT2, GhDGAT3, and GhWSD1 proteins (Figure 4 and Figure S3). GhDGAT1 proteins contain nine TMs, whereas most of the GhDGAT2 proteins contain two TMs, except GhDGAT2-1D, GhDGAT2-2D, and GhDGAT2-5D. DGAT3 does not have TMs, consistent with its soluble nature. Consistently, only GhDGAT1 proteins had a membrane-bound acyltransferase (MBOAT) domain, whereas GhDGAT2, GhDGAT3, and GhWSD1 did not have an MBOAT domain (Figure 4 and Figure S3). These results support the hypothesis that DGAT1 and DGAT2 belong to different gene families and evolved separately during eukaryote evolution, as demonstrated by the phylogenetic tree (Figure 1).
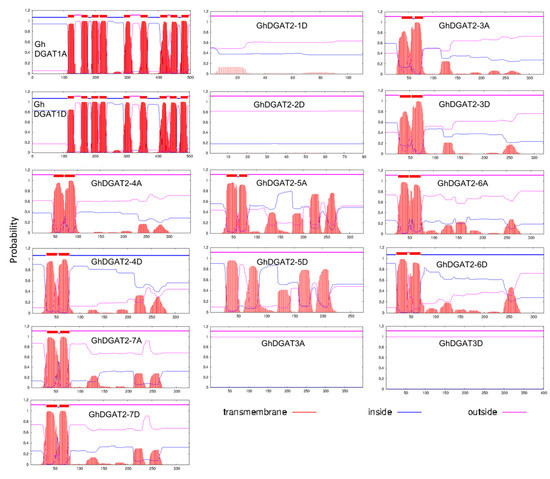
Figure 4.
Predicted transmembrane domain for GhDGAT proteins. Regions of GhDGAT amino acid sequences predicted to be located inside or outside the membrane are shown in blue and pink, respectively.
3.4. Cis-Elements in the GhDGAT and GhWSD1 Promoters
The 2 kb sequence upstream of the start codon (ATG) of GhDGAT and GhWSD1 genes was used to investigate the cis-elements in the promoter regions in the PlantCARE database. A total of 93 cis-elements were predicted in the promoter regions of the 36 GhDGAT and GhWSD1 genes. Several cis-elements were implicated in light response. Moreover, the roles of cis-elements in environmental stresses and hormone responses are highlighted in Figure 5. Among the predicted hormone response elements, ERE, ABRE, and CGTCA-motif were the most abundant, indicating that GhDGAT and GhWSD1 genes may primarily respond to ethylene, abscisic acid, and MeJA (Figure 5B). Ten environmental stress-related elements were identified, with most of them involving drought stress (MYC), stress response (STRE), and anaerobic induction (ARE) (Figure 5C).
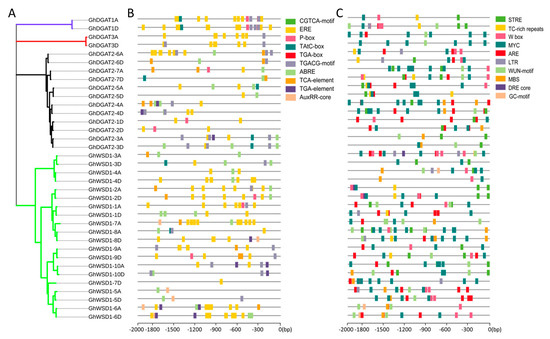
Figure 5.
cis-elements in GhDGAT and GhWSD1 promoter regions; (A) phylogenetic tree of GhDGAT and GhWSD1 proteins; (B) predicted cis-elements involved in plant hormones. ABRE: cis-acting element involved in abscisic acid responsiveness; AuxRR-core: cis-acting regulatory element involved in auxin responsiveness; AuxRE: part of an auxin-responsive element; CGTCA-motif: cis-acting regulatory element involved in MeJA-responsiveness; GARE-motif: gibberellin-responsive element; TGACG-motif: cis-acting regulatory element involved in MeJA-responsiveness; TGA-element: auxin-responsive element; TGA-box: part of auxin-responsive element; ERE: cis-acting ethylene responsive element; P-box: gibberellin-responsive element; (C) predicted cis-elements involved in environmental stress responses. GC-motif: enhancer-like element involved in anoxic specific inducibility; LTR: cis-acting element involved in low-temperature responsiveness; MBS: MYB binding site involved in drought-inducibility; STRE: stress response element; TC-rich repeats: cis-acting element involved in defense and stress responsiveness; WUN-motif: wound-responsive element; MYC: cis-acting element involved in drought stress; W box: cis-acting element involved in sugar metabolism and plant defense signaling; DRE core: dehydration-responsive element; ARE: cis-acting regulatory element essential for anaerobic induction.
3.5. Target Transcription Factors and miRNAs of GhDGAT and GhWSD Genes
Transcription factors (TFs) regulate the precise initiation of gene transcription. Therefore, we identified the target TFs of GhDGAT and GhWSD1 using the PlantRegMap server, and a total of 568 relationships were identified (Figure S4). GhWSD1-1D may be regulated by more TFs, such as the stress TFs ethylene response factor (ERF), DNA binding with one finger (Dof), or v-myb avian myeloblastosis viral oncogene homolog (MYB). Moreover, several GhDGAT and GhWSD1 genes were regulated by Dof, ERF, and NAC (NAM, ATAF, and CUC) TFs, indicating that these TFs regulate plant development and stress responses.
miRNAs have been widely studied in the regulation of gene expression, which plays an important role in abiotic stress responses. To explore the potential role of miRNAs in regulating GhDGAT and GhWSD1 genes, 26 putative miRNAs targeting 23 GhDGAT and GhWSD1 genes were predicted using the psRNATarget website, including 52 interaction relationships (Figure 6). We observed that GhWSD1-7Dt was the most targeted, interacting with seven miRNAs. ghr-miR71491 was the most regulated miRNA, and was involved in regulating four GhDGAT and GhWSD1 genes. Additionally, we observed that most GhDGAT and GhWSD1 homologs were regulated by the same miRNAs, suggesting similar functional roles.
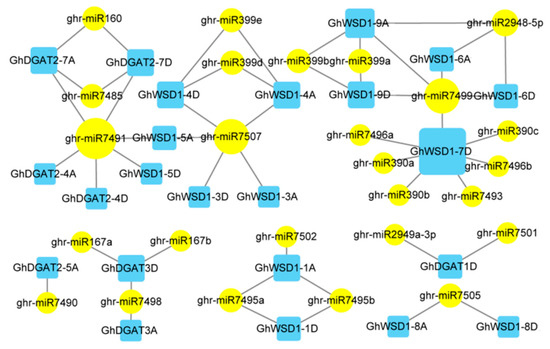
Figure 6.
miRNA targets GhDGAT and GhWSD1 genes. The predicted regulation miRNAs are marked with a round yellow background, the target GhDGAT and GhWSD1 genes are marked with blue rectangles. The regulation and targeting levels are shown with varying degrees.
3.6. Gene Expression Profile of Upland Cotton in Response to Abiotic Stresses
Furthermore, the expression profiles of the GhDGAT and GhWSD1 genes under abiotic stresses, including cold, drought, and salt stress were investigated at different time series (Figure 7). The GhDGAT or GhWSD1 genes that were not expressed in cotton leaves were unaffected. The results showed that there were differences in the expression profiles of the GhDGAT1, GhDGAT2, GhDGAT3, and GhWSD1-1 genes under the different abiotic stress conditions.
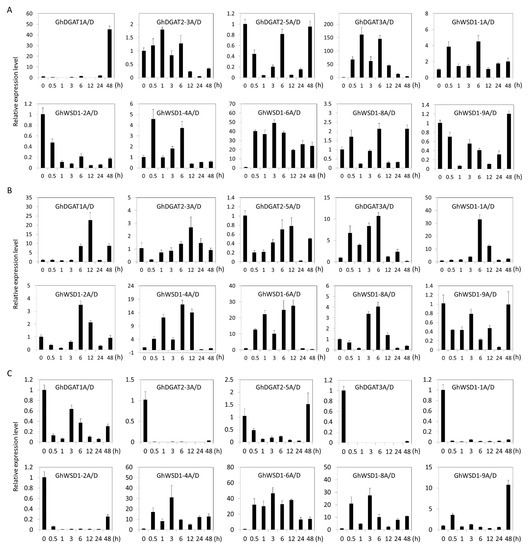
Figure 7.
Expression profiles of GhDGAT and GhWSD1 genes during abiotic stresses; (A) gene expression patterns under cold stress; (B) gene expression patterns under drought stress; (C) gene expression patterns under salt stress. Cold: 4 °C; drought: 17% PEG-6000; salt: 200 mM NaCl.
The expression of GhDGAT1 was upregulated at 48 h under cold stress. Additionally, the expression level of GhDGAT3A/D and GhWSD1-6A/D increased at several time points under cold stress (Figure 7A). However, most GhDGAT and GhWSD1 genes were upregulated under drought stress, except for GhDGAT2-5A/D and GhWSD1-9A/D, indicating that GhDGAT and GhWSD1 genes respond to drought stress (Figure 7B). The expression of GhDGAT1A/D, GhDGAT2-3A/D, GhDGAT2-5A/D, GhDGAT3A/D, GhWSD1-1A/D, and GhWSD1-2A/D was downregulated, and that of GhWSD1-4A/D, GhWSD1-6A/D, GhWSD1-8A/D, and GhWSD1-9A/D was upregulated under salt stress (Figure 7C). The expression profiles of GhDGAT and GhWSD1 genes under abiotic stress acted in cooperation with many environment response elements that were predicted in their promoter regions (Figure 5).
3.7. Expression Profiling of DGAT and WSD1 Genes in Cotton Development
Gene expression models are important for gene function analysis. The gene expression patterns of DGAT and WSD1 in developing cottonseeds of G. arboretum, G. raimondii, and G. hirsutum were investigated [45]. DGAT1 genes showed increased expression levels at later developmental stages of 30 and 40 days post anthesis (DPA). Compared with the higher expression levels of DGAT1 genes in G. arboretum and G. raimondii, there was an impaired expression of GhDGAT1 in G. hirsutum. Additionally, most DGAT2 genes exhibited low expression levels; however, GrDGAT2-3, GhDGAT2-3A/D, GaDGAT2-1, GhDGAT2-7D, and GrDGAT2-6 exhibited high expression levels (Figure S5A). Compared with DGAT1 and DGAT2, DGAT3 genes showed a more abundant expression in the diploid and tetraploid cotton species. For the WSD1 group, there was an increase in the expression profiles of GaWSD1-2, GhWSD1-1a, GrWSD1-4, GaWSD1-5, and GrWSD1-1, at 30 and 40 DPA, whereas only GrWSD1-2 was highly expressed at 10 and 20 DPA (Figure S5B). Similar expression patterns were observed in the same groups, and the different expression profiles in diploid and tetraploid cotton species indicated the evolution and differentiation of DGAT and WSD1 proteins.
Public expression datasets of upland cotton were used for gene expression analyses in different tissues and ovules at different fiber development stages [46]. The expression patterns of GhDGAT and GhWSD1 genes in developing ovules were consistent with those in developing cottonseed (Figure 8 and Figure S5). Several GhDGAT and GhWSD1 genes, including GhDGAT2-6, GhDGAT2-1, GhWSD1-3, GhWSD1-5, and GhWSD1-9 were barely expressed in upland cotton. GhDGAT1 and GhWSD1-10D genes were highly expressed in male reproductive organs (anthers and filaments). The GhDGAT2-4 gene was highly expressed in ovules during the early development stage (3 and 5 DO) and fiber rapid elongation stage (10 and 15 DF), indicating that it was involved in fiber elongation. GhDGAT2-7 was highly expressed in reproductive organs (torus, bract, and pistil) and fiber. However, GhWSD1-1 and GhWSD1-8 were preferentially expressed in reproductive organs. However, we highlight that GhDGAT3 genes were consistently and abundantly expressed in cotton development, and GhDGAT3D showed a higher expression level than that of GhDGAT3A in the developing cottonseed (Figure 8).
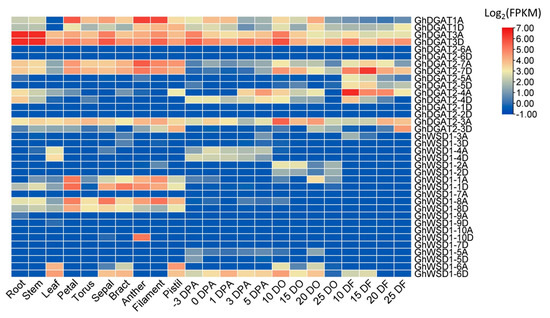
Figure 8.
Expression patterns of GhDGAT and GhWSD1 genes in upland cotton. DF: DPA fiber; DO: DPA ovule. Gene expression levels (FPKM) were averaged and normalized by a log2 scale.
3.8. Overexpression of GhDGAT3D Improves Oil Content in Arabidopsis Seeds
The recombination vector p35S::GhDGAT3D was transformed into Arabidopsis using the floral dip method. Several single transformation events were obtained, among which three single-copy transgenic lines overexpressing GhDGAT3D at the mRNA level were selected for further analysis (Figure S6). To confirm the role of GhDGAT3D in oil biosynthesis, the oil content and fatty acid composition of the homozygous T3 generation of Arabidopsis overexpressing GhDGAT3D was determined by gas chromatography. Total oil content was increased to 26.72%, 26.35%, and 28.25% in OE#1, OE#2, and OE#3 transgenic lines, respectively, compared with that of 21.37% in the control plant (Figure 9A). Additionally, there was an increase in C18:1 (26.82%, 28.97%, and 26.84% in OE#1, OE#2, and OE#3 transgenic lines, respectively, compared with that of 25.83% in the control) content and a decrease in C18:2 (24.30%, 24.33%, and 24.60%, compared with that of 25.42%) and C18:3 (33.50%, 33.03%, and 32.04%, compared with that of 35.25%) contents in Arabidopsis seeds (Figure 9B), indicating that GhDGAT3D was involved in regulating oil biosynthesis and fatty acid composition of cotton seeds.
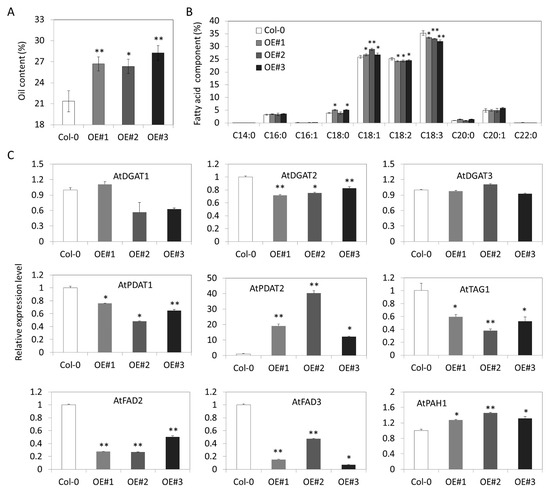
Figure 9.
Overexpression of GhDGAT3D increased oil content in Arabidopsis seeds; (A) total oil content in GhDGAT3D overexpressing Arabidopsis seeds; (B) fatty acid component in transgenic and the control Arabidopsis seed; (C) detection of oil biosynthesis related genes in transgenic Arabidopsis. * and ** indicate data were significantly different at p-value of 0.05 and 0.01 levels, respectively.
The expression levels of oil-related genes were investigated in transgenic Arabidopsis, including AtDGAT1, AtDGAT2, AtDGAT3, AtPDAT1, AtPDAT2, AtTAG1, AtFAD2, AtFAD3, and AtPAH2 (Figure 9C). Ectopic expression of GhDGAT3D did not affect the expression level of AtDGAT3, indicating that the alteration of the oil content and fatty acid composition of transgenic Arabidopsis resulted from the overexpression of GhDGAT3D. Additionally, there was a decrease in the expression levels of AtDGAT2, AtPDAT1, AtTAG1, AtFAD2, and AtFAD3, and an increase in the expression of AtPAH in GhDGAT3D overexpressing plants. The results indicated that the increase in C18:1 content and the decrease in C18:2 and C18:3 contents may result from the decrease in the expression levels of AtFAD2 and AtFAD3 in transgenic Arabidopsis. However, there was a significant increase in the expression of AtPDAT2 in GhDGAT3D overexpressing Arabidopsis, indicating a potential interaction relationship between DGAT3 and PDAT2 in plant seeds.
4. Discussion
4.1. Gene Duplication and Functional Diversification of GhDGAT and GhWSD1 Genes
The phylogenetic tree, transmembrane domain and expression analysis showed that the DGAT1, DGAT2, DGAT3 and WSD1 genes showed apparent differences (Figure 1, Figure 4, and Figure 8). These results indicate that they are divergent genes and may have a distinct origin, consistent with what is described in soybeans [53]. Gene duplication partakes a major role in the evolution of plant genomes. The results of the present study showed that GhDGAT and GhWSD1 genes were frequently duplicated during cotton evolution, with only one pair in each of the GhDGAT1 and GhDGAT3 genes, and over five pairs of GhDGAT2 homologs genes identified in upland cotton. It has been reported that there are two paralogs of DGAT2 genes in maize and five paralogs in soybean (Glycine max) [28,30]. Although, there is a close relationship between cacao and cotton, only one TcDGAT2 gene was found in cacao, whereas five paralogs were found in diploid cottons (G. arboretum and G. raimondii) (Figure 1), indicating that the duplication of GhDGAT2 genes occurred after the cotton genus separated from cacao. The multiple paralogs of GhDAGT2 in soybean and oilseeds, including cotton, confirmed the role of the genes in oil biosynthesis. The cluster of GhDGAT2 genes in the chromosomes of upland cotton showed that tandem duplication events occurred during cotton evolution. Gene duplication events also occurred in GhWSD1 genes, with only one AtWSD1 gene in Arabidopsis, six TcWSD1 genes in cacao, and over 10 WSD1 genes detected in diploid cottons (G. arboretum and G. raimondii). Additionally, tandem duplication events occurred in GhWSD1 genes, among which GhWSD1-1A/D, GhWSD1-2A/D, GhWSD1-3A/D and GhWSD1-4A/D, GhWSD1-5A/D, and GhWSD1-6A/D showed tandem duplication in upland cotton (Figure 2). Overall, the results showed that GhWSD1 genes were frequently duplicated before and after cotton division from cacao.
DGAT genes have been reported to be involved in TAG biosynthesis and abiotic stress responses [8,17,54]. Therefore, the expression profiles of GhDGAT and GhWSD1 genes were investigated in the present study. The results showed that several paralogs of GhDGAT2 genes were barely expressed in cotton, except GhDGAT2-7A/D, indicating that GhDGAT2 genes may have experienced functional diversification or shown gene redundancy during cotton evolution. GhWSD1 genes showed intricate expression patterns during cotton developmental stages and under abiotic stresses. Specifically, GhWSD1-1A/D responded to cold and drought stresses at several time points; however, the duplicate genes of GhWSD1-2A/D did not respond to any stress condition. The diverse expression patterns indicated that GhWSD1 genes also experienced functional diversification.
4.2. GhDGATs and GhWSD1s Response to Abiotic Stresses
DGAT1 appears to play a role in freezing and drought stress responses in Arabidopsis [54], Brassica napus [55] and Boechera stricta [56]. BsDGAT1 was higher expression in freeze-tolerant plants than freeze-susceptible plants; overexpression of AtDGAT1 increased freezing tolerance in Arabidopsis [56], whereas Arabidopsis DGAT1 (AtDGAT1) defective mutant lines were sensitive to freezing [54]. Additionally, overexpression of DGAT1 in B. napus is shown to reduce the negative effects of drought on seed oil content [55]. In the present study, there was an increase in the expression of GhDGAT1A/D homologs in Arabidopsis at 48 h under cold conditions. Additionally, there was an increase in the expression of GhDGAT1 at several time points under drought stress, indicating that GhDGAT1 was involved in cold and drought stress responses. The role of DGAT2 in abiotic stress remains unclear; however, the results of the present study showed that there was an increase in the expression of GhDGAT2-3A/D under cold and drought stresses, but it decreased invariably under salt stress (Figure 7). Additionally, there was a decreasing trend in the expression of GhDGAT2-5A/D in response to drought and salt stress conditions. The inconsistent expression patterns of GhDGAT2 in response to different environmental stresses indicated the complicated role of GhDGAT2 in environmental adaptation. The function of DGAT3 in abiotic stress is scarcely reported. GhDGAT3 genes were highly expressed in the root, stem, and other tissues. Meanwhile, there was an increase in the expression of GhDGAT3 genes under cold and drought stress, whereas the expression of the gene was reduced under salt stress conditions (Figure 7). Moreover, several MYC cis-elements were found in the GhDGAT3A/D promoter region, as MYC is a cis-acting element involved in drought stress (Figure 5C). These results indicate that GhDGAT3 may be involved in cold and drought stress responses in cotton.
Patwari et al. (2019) reported that WSD1 responds to drought stress [26]. WSD1 is upregulated in grape berries in response to drought [57]. An R2R3-type MYB94 transcription factor activates the Arabidopsis cuticular wax biosynthesis gene WSD1 and may be important in the plant response to drought stress [58]. Another AP2/ERF-type transcription factor, WRINKLED4, binding the WSD1 promoter specifically, controls cuticular wax biosynthesis [59]. In the present study, GhWSD1-1 genes were highly expressed under drought stress, confirming the function of GhWSD1 in the drought response (Figure 7B). Additionally, there was an increase in the expression profiles of GhWSD1-4A/D, GhWSD1-6A/D, GhWSD1-8A/D, and GhWSD1-9A/D under cold and salt stress at several time points, indicating the multiple roles of the GhWSD1 genes in the spread of upland cotton to different regions (Figure 7A,C).
4.3. Role of GhDGATs in Oil Biosynthesis Regulation
DGAT1 has been functionally confirmed in oil biosynthesis in Arabidopsis, soybean, and oilseed rape [17]. Expression analysis revealed that DGAT1 was abundantly expressed in developing embryos in several oilseed crops and its transcript level according to oil accumulation in developing seed [60]. In the present study, there was an increase in the expression of GhDGAT1 in cotton seeds at 10 and 20 DO (Figure 8), which corresponded with the rapid oil accumulation stage in cottonseed, indicating that GhDGAT1 was important in TAG biosynthesis. Moreover, there were high expression levels of GhDGAT1 in the petals, anthers, and filaments of upland cotton, indicating that GhDGAT1 might be involved in the reproductive development of upland cotton. It was reported that TAG production via DGAT1 and DGAT2 occurs in a distinct ER subdomain; moreover, it has been reported that tung tree DGAT1 and DGAT2 proteins are localized to different ER regions and differ in substrate preference [61]. The expression level of DGAT2 was found to be higher in unusual or polyunsaturated fatty acids accumulating in developing seeds. Cyperus esculentus CeDGAT2b has been shown to have a substrate preference for UFA for TAG synthesis [62]. Ectopic overexpression of CeDGAT2 has been shown in enhanced oil and C18:1 accumulation in tobacco leaves [63]. In the present study, only GhDGAT2-3A and GhDGAT2-7D exhibited high expression levels during cottonseed development, whereas the other 10 paralogs of GhDAGT2 genes were barely expressed. However, we found that GhDGAT2-7A and GhDGAT2-4D were abundantly expressed at 10 to 20 DPA during fiber development, indicating that the two genes may be involved in fiber elongation.
Few studies have focused on the role of DGAT3 in TAG biosynthesis. To date, only AtDGAT3 and CsDGAT3 are confirmed as metalloproteins involved in TAG biosynthesis in plants [23,24]. The DGAT3 protein has not been identified in mossy or algal species [28], indicating that it may have evolved during plant evolution. GhDGAT3 is regarded as a key candidate gene for the total triglyceride pool [64]. GmDGAT3 has the highest transcript levels when compared to other GmDGAT genes in developing soybean seeds, suggesting that GmDGAT3 is probably involved in TAG synthesis [53]. The expression level of GhDGAT3 is significantly higher than that of GhDGAT1 and GhDGAT2 during cottonseed development [3]. In the present study, we observed that GhDGAT3 was highly expressed in the ovule of upland cotton and during fiber development (Figure 8). Moreover, there was a significant increase in the oil content of GhDGAT3D overexpressing Arabidopsis transgenic plants compared with that of the control plants, indicating that GhDGAT3 was involved in oil biosynthesis. Additionally, there was a decrease in the C18:2 and C18:3 contents and an increase in the C18:1 content of the seeds of GhDGAT3D, overexpressing Arabidopsis transgenic plants (Figure 9B). These results are consistent with the transcript levels of AtFAD2 and AtFAD3 being weakened in transgenic Arabidopsis (Figure 9C). In the present study, the expression assay results showed that most GhWSD1 genes were barely or not expressed in developing cottonseed. Moreover, it has been reported that the WSD1 enzyme shows deficient levels of diacylglycerol acyltransferase activity [25]. However, few studies have confirmed the role of WSD1 in oil biosynthesis. Overall, the WSD1 enzyme may be more involved in environmental stress responses than in oil biosynthesis.
5. Conclusions
In summary, GhDGATs and GhWSD1s were identified and classified in upland cotton; additionally, their roles in stress responses, oil biosynthesis, and fatty acid composition were also elucidated. The findings of this study showed that WSD1 genes were mostly involved in stress responses, whereas DGAT genes were involved in both oil synthesis, fatty acid composition, and abiotic stress responses. Overall, the findings of this study contribute to the understanding of DGAT and WSD1 genes in fatty acid biosynthesis and abiotic stress responses in cotton.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12071045/s1, Table S1. Primers used in this study. Table S2. Identification and nomenclature of DGAT and WSD1 genes in G. arboreum and G. raimondii. Table S3. Annotation of the conserved motifs identified in GhDGAT and GhWSD1. Figure S1. Location of GhDGAT and GhWSD1 genes in the upland cotton genome. Figure S2. Conserved motifs identified in GhDGAT and GhWSD1 proteins; (A) phylogenetic tree of GhDGAT and GhWSD1 proteins; (B) top 10 conserved motifs; (C) logos of the ten conserved motifs in GhDGAT and GhWSD1 proteins. Figure S3. Predicted transmembrane domain of GhWSD1 proteins. Regions of GhWSD1 amino acid sequences predicted to be located inside or outside the membrane are shown in blue and pink, respectively. Figure S4. Predicted target TFs of GhDGAT and GhWSD1 genes. The predicted regulation miRNAs are depicted as round in yellow background, the target GhDGAT and GhWSD1 genes as rectangles in blue background. Figure S5. Expression patterns of GhDGAT and GhWSD1 genes in developing cottonseed; (A) transcript levels of GhDGAT genes in developing cottonseed; (B) expression profiles of GhWSD1 genes in cottonseed development. DPA: days post anthesis. Gene expression levels (FPKM) were averaged and normalized by log2 scale. Figure S6. Identification of the positive transgenic Arabidopsis in RNA level. AtSYL8: the internal reference; OE#1, OE#2, OE#3: transgenic Arabidopsis lines.
Author Contributions
Conceptualization, Y.-P.Z.; formal analysis, Y.-X.H. and F.-G.L.; investigation, Y.-P.Z., N.W., J.-L.S., W.-J.L. and C.C.; writing-original draft, Y.-P.Z.; writing-review and editing, Y.-X.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 31901578), the “Seven Crop Breeding” National Major Project (grant no. 2016YFD0101006); the Postdoctoral Starting Research Fund of Henan Province (grant no. 201903001), and the State Key Laboratory of Cotton Biology (grant no. CB2020A14 and CB2019B04).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Materials.
Acknowledgments
We really appreciate the help and support of all teachers and students in our research team.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Surinder, S.; Chapman, K.; Green, A. Bridging Traditional and Molecular Genetics in Modifying Cottonseed Oil; Springer: London, UK, 2009; pp. 353–382. [Google Scholar]
- Zhao, Y.; Wang, Y.; Huang, Y.; Cui, Y.; Hua, J. Gene network of oil accumulation reveals expression profiles in developing embryos and fatty acids composition in Upland cotton. J. Plant Physiol. 2018, 228, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Napier, J.A.; Clemente, T.E.; Cahoon, E.B. New frontiers in oilseed biotechnology: Meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr. Opin. Biotechnol. 2011, 22, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weselake, R.J.; Murphy, D.J. (Eds.) Plant Lipids Biol. Util; Blackwell Publishing: Oxford, UK, 2005; pp. 162–225. [Google Scholar]
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934–940. [Google Scholar]
- Maraschin, F.D.S.; Kulcheski, F.R.; Segatto, A.L.A.; Trenz, T.S.; Barrientos-Diaz, O.; Margis-Pinheiro, M.; Margis, R.; Turchetto-Zolet, A.C. Enzymes of glycerol-3-phosphate pathway in triacylglycerol synthesis in plants: Function, biotechnological application and evolution. Prog. Lipid Res. 2019, 73, 46–64. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Stahl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H. Phospholipid: Diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef] [Green Version]
- Stahl, U.; Carlsson, A.S.; Lenman, M.; Dahlqvist, A.; Huang, B.; Banas, W.; Banas, A.; Stymne, S. Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis. Plant Physiol. 2004, 135, 1324–1335. [Google Scholar] [CrossRef] [Green Version]
- Lung, S.C.; Weselake, R.J. Diacylglycerol acyltransferase: A key mediator of plant triacylglycerol synthesis. Lipids 2006, 41, 1073–1088. [Google Scholar] [CrossRef]
- Liu, Q.; Siloto, R.M.P.; Lehner, R.; Stone, S.J.; Weselake, R.J. Acyl-CoA:diacylglycerol acyltransferase: Molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 2012, 51, 350–377. [Google Scholar] [CrossRef]
- Li, R.; Yu, K.; Hildebrand, D.F. DGAT1, DGAT2 and PDAT expression in seeds and other tissues of epoxy and hydroxy fatty acid accumulating plants. Lipids 2010, 45, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Hatanaka, T.; Yu, K.; Wu, Y.; Fukushige, H.; Hildebrand, D. Soybean oil biosynthesis: Role of diacylglycerol acyltransferases. Funct. Integr. Genom. 2013, 13, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Wei, Y.; Jako, C.; Kumar, A.; Selvaraj, G.; Taylor, D.C. The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 1999, 19, 645–653. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Allen, W.B.; Roesler, K.; Williams, M.E.; Zhang, S.; Li, J.; Glassman, K.; Ranch, J.; Nubel, D.; Solawetz, W.; et al. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat. Genet. 2008, 40, 367–372. [Google Scholar] [CrossRef]
- Xu, Y.; Caldo, K.M.P.; Pal-Nath, D.; Ozga, J.; Lemieux, M.J.; Weselake, R.J.; Chen, G. Properties and biotechnological applications of acyl-CoA: Diacylglycerol acyltransferase and phospholipid: Diacylglycerol acyltransferase from terrestrial plants and microalgae. Lipids 2018, 53, 663–688. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.L.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.R.; Shrestha, P.; Yin, F.; Petrie, J.R.; Singh, S.P. AtDGAT2 is a functional acyl-CoA: Diacylglycerol acyltransferase and displays different acyl-CoA substrate preferences than AtDGAT1. FEBS Lett. 2013, 587, 2371–2376. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.T.; He, H.; Xu, C.J.; Fu, Q.; Tao, Y.B.; Xu, R.; Xu, Z.F. Overexpression of type 1 and 2 diacylglycerol acyltransferase genes (JcDGAT1 and JcDGAT2) enhances oil production in the woody perennial biofuel plant Jatropha curcas. Plants 2021, 10, 699. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Enugutti, B.; Rajakumari, S.; Rajasekharan, R. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol. 2006, 141, 1533–1543. [Google Scholar] [CrossRef] [Green Version]
- Hernández, M.L.; Whitehead, L.; He, Z.; Gazda, V.; Gilday, A.; Kozhevnikova, E.; Vaistij, F.E.; Larson, T.R.; Graham, I.A. A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol. 2012, 160, 215–225. [Google Scholar] [CrossRef] [Green Version]
- Aymé, L.; Arragain, S.; Canonge, M.; Baud, S.; Touati, N.; Bimai, O.; Jagic, F.; Louis-Mondésir, C.; Briozzo, P.; Fontecave, M.; et al. Arabidopsis thaliana DGAT3 is a [2Fe-2S] protein involved in TAG biosynthesis. Sci. Rep. 2018, 8, 17254. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Gao, Y.; Zhang, F.; Liu, B.; Ji, C.; Xue, J.; Yuan, L.; Li, R. Functional characterization of an novel acyl-CoA:diacylglycerol acyltransferase 3-3 (CsDGAT3-3) gene from Camelina sativa. Plant Sci. 2021, 303, 110752. [Google Scholar] [CrossRef]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patwari, P.; Salewski, V.; Gutbrod, K.; Kreszies, T.; Dresen-Scholz, B.; Peisker, H.; Steiner, U.; Meyer, A.J.; Schreiber, L.; Dörmann, P. Surface wax esters contribute to drought tolerance in Arabidopsis. Plant J. 2019, 98, 727–744. [Google Scholar] [CrossRef]
- Abdullah, H.M.; Rodriguez, J.; Salacup, J.M.; Castañeda, I.S.; Schnell, D.J.; Pareek, A.; Dhankher, O.P. Increased cuticle waxes by overexpression of WSD1 improves osmotic stress tolerance in Arabidopsis thaliana and Camelina sativa. Int. J. Mol. Sci. 2021, 22, 5173. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Xu, X.; Gu, Y.; Zhao, Y.; Zhao, X.; He, L.; Zhao, C.; Li, Z.; Xu, J. Genome-wide characterization and expression profiling of diacylglycerol acyltransferase genes from maize. Genome 2018, 61, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Rosli, R.; Chan, P.L.; Chan, K.L.; Amiruddin, N.; Low, E.L.; Singh, R.; Harwood, J.L.; Murphy, D.J. In silico characterization and expression profiling of the diacylglycerol acyltransferase gene family (DGAT1, DGAT2, DGAT3 and WS/DGAT) from oil palm, Elaeis guineensis. Plant Sci. 2018, 275, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Bi, R.; Li, S.; Zhou, D.; Bai, Y.; Jing, G.; Zhang, K.; Zhang, W. Genome-wide analysis and functional characterization of Acyl-CoA: Diacylglycerol acyltransferase from soybean identify GmDGAT1A and 1B roles in oil synthesis in Arabidopsis seeds. J. Plant Physiol. 2019, 242, 153019. [Google Scholar] [CrossRef]
- Yang, Z.; Ge, X.; Yang, Z.; Qin, W.; Sun, G.; Wang, Z.; Li, Z.; Liu, J.; Wu, J.; Wang, Y.; et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Yu, J.; Jung, S.; Cheng, C.H.; Ficklin, S.P.; Lee, T.; Zheng, P.; Jones, D.; Percy, R.G.; Main, D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014, 42, D1229–D1236. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Guo, A.; Wang, Y.; Hua, J. Evolution of PEPC gene family in Gossypium reveals functional diversification and GhPEPC genes responding to abiotic stresses. Gene 2019, 698, 61–71. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.J.; Hovav, R.; Grover, C.E.; Faigenboim-Doron, A.; Kadmon, N.; Page, J.T.; Udall, J.A.; Wendel, J.F. Evolutionary conservation and divergence of gene coexpression networks in Gossypium (cotton) seeds. Genome Biol. Evol. 2016, 8, 3765–3783. [Google Scholar]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Guo, Y.; Yan, J.; Zhang, J.; Song, T.; Rocheford, T.; Li, J.S. Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor. Appl. Genet. 2010, 120, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Turchetto-Zolet, A.C.; Christoff, A.P.; Kulcheski, F.R.; Loss-Morais, G.; Margis, R.; Margis-Pinheiro, M. Diversity and evolution of plant diacylglycerol acyltransferase (DGATs) unveiled by phylogenetic, gene structure and expression analyses. Genet. Mol. Biol. 2016, 39, 524–538. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.J.; Yang, Y.C.; Zhou, Y.; Huang, L.P.; Xu, L.; Chen, Q.F.; Yu, L.J.; Xiao, S. Diacylglycerol Acyltransferase and Diacylglycerol Kinase modulate triacylglycerol and phosphatidic acid production in the plant response to freezing stress. Plant Physiol. 2018, 177, 1303–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weselake, R.J.; Shah, S.; Tang, M.; Quant, P.A.; Snyder, C.L.; Furukawa-Stoffer, T.L.; Zhu, W.; Taylor, D.C.; Zou, J.; Kumar, A.; et al. Metabolic control analysis is helpful for informed genetic manipulation of oilseed rape (Brassica napus) to increase seed oil content. J. Exp. Bot. 2008, 59, 3543–3549. [Google Scholar] [CrossRef] [Green Version]
- Arisz, S.A.; Heo, J.Y.; Koevoets, I.T.; Zhao, T.; van Egmond, P.; Meyer, A.J.; Zeng, W.; Niu, X.; Wang, B.; Mitchell-Olds, T.; et al. Diacylglycerol Acyltransferase1 contributes to freezing tolerance. Plant Physiol. 2018, 177, 1410–1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimopoulos, N.; Tindjau, R.; Wong, D.C.J.; Matzat, T.; Haslam, T.; Song, C.; Gambetta, G.A.; Kunst, L.; Castellarin, S.D. Drought stress modulates cuticular wax composition of the grape berry. J. Exp. Bot. 2020, 71, 3126–3141. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 2015, 56, 48–60. [Google Scholar] [CrossRef] [Green Version]
- Park, C.S.; Go, Y.S.; Suh, M.C. Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 2016, 88, 257–270. [Google Scholar] [CrossRef]
- Lu, C.L.; de Noyer, S.B.; Hobbs, D.H.; Kang, J.; Wen, Y.; Krachtus, D.; Hills, M.J. Expression pattern of diacylglycerol acyltransferase-1, an enzyme involved in triacylglycerol biosynthesis, in Arabidopsis thaliana. Plant Mol. Biol. 2003, 52, 31–41. [Google Scholar] [CrossRef]
- Shockey, J.M.; Gidda, S.K.; Chapital, D.C.; Kuan, J.C.; Dhanoa, P.K.; Bland, J.M.; Rothstein, S.J.; Mullen, R.T.; Dyer, J.M. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 2006, 18, 2294–2313. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Ji, H.; Yang, Z. Functional characterization of three novel genes encoding diacylglycerol acyltransferase (DGAT) from oil-rich tubers of Cyperus esculentus. Plant Cell Physiol. 2020, 61, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, Y.; Gao, H.; Chen, Y.; Wang, X.; Xue, J.; Jia, X.; Li, R. Ectopic overexpression of a type-II DGAT (CeDGAT2-2) derived from oil-rich tuber of Cyperus esculentus enhances accumulation of oil and oleic acid in tobacco leaves. Biotechnol. Biofuels 2021, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Hovav, R.; Faigenboim-Doron, A.; Kadmon, N.; Hu, G.; Zhang, X.; Gallagher, J.P.; Wendel, J.F. A transcriptome profile for developing seed of polyploid cotton. Plant Genome 2015, 8, 1. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).