Characterization of ETFDH and PHGDH Mutations in a Patient with Mild Glutaric Aciduria Type II and Serine Deficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Consideration
2.2. Biochemical Analysis
2.3. DNA Extraction and Whole Exome Sequencing
2.4. In Silico Analysis
3. Results
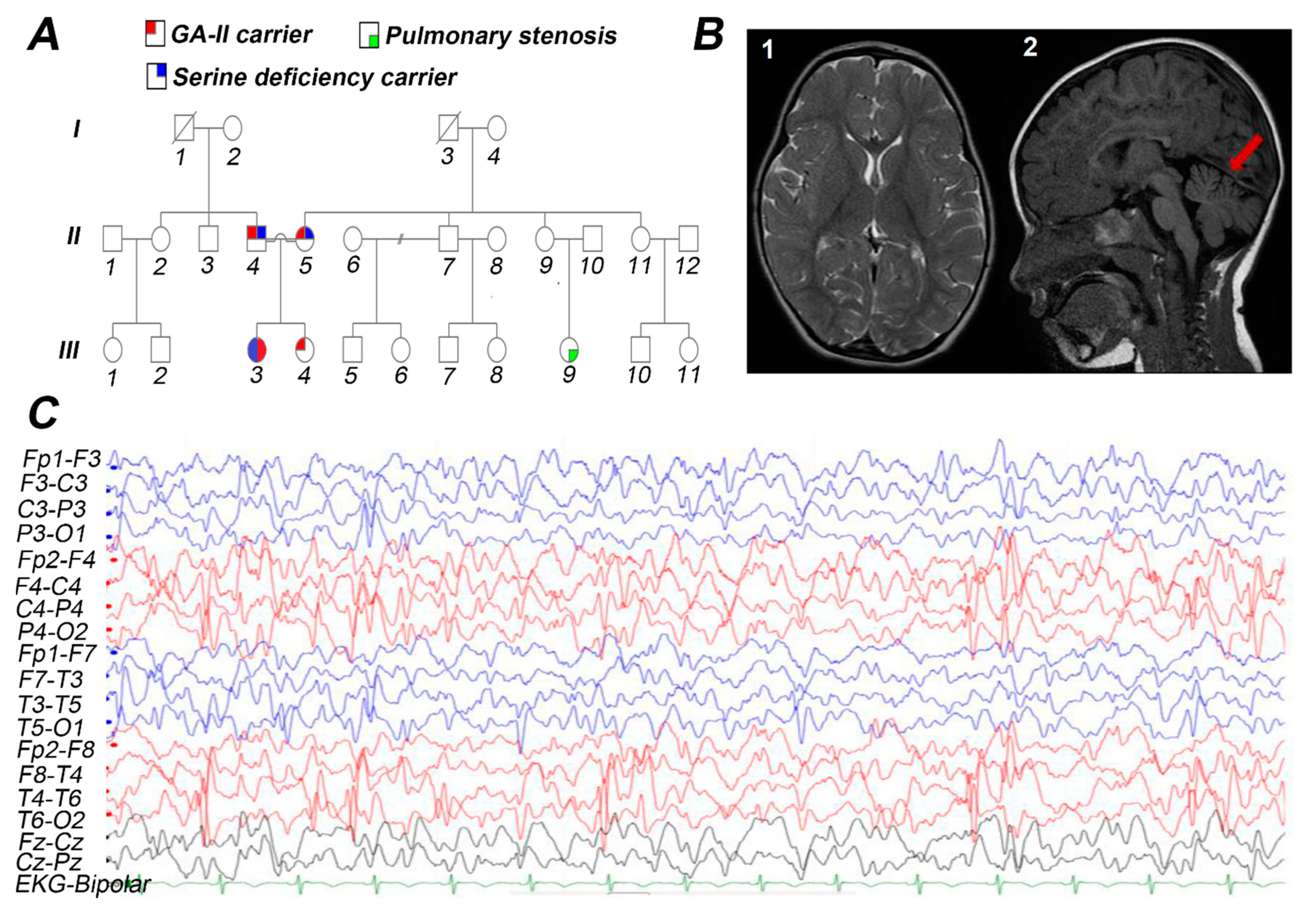
3.1. Case Presentation
3.2. Biochemical Studies
3.3. Molecular Findings
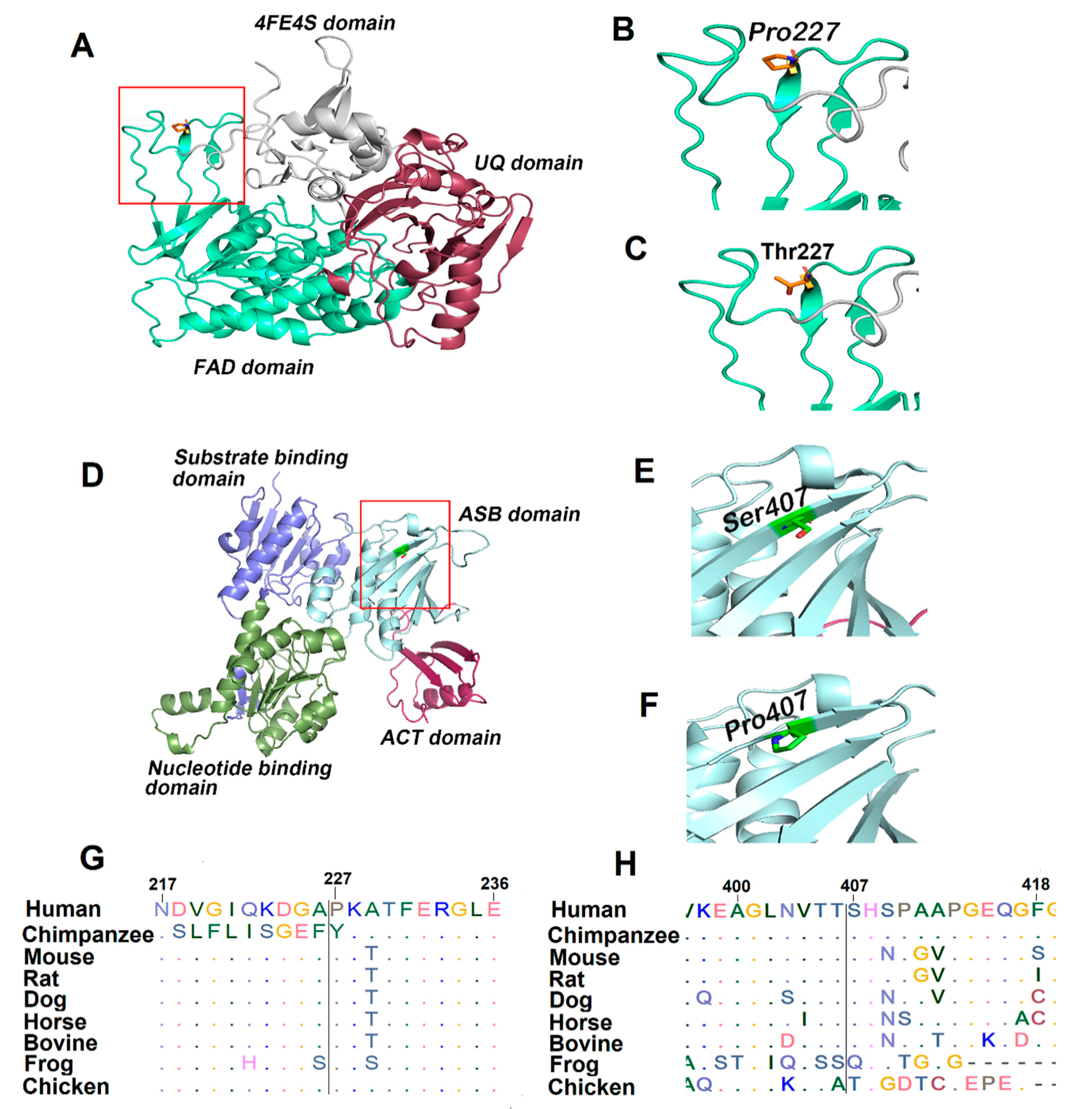
3.4. Bioinformatics Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Shioya, A.; Takuma, H.; Yamaguchi, S.; Ishii, A.; Hiroki, M.; Fukuda, T.; Sugie, H.; Shigematsu, Y.; Tamaoka, A. Amelioration of acylcarnitine profile using bezafibrate and riboflavin in a case of adult-onset glutaric acidemia type 2 with novel mutations of the electron transfer flavoprotein dehydrogenase (ETFDH) gene. J. Neurol. Sci. 2014, 346, 350–352. [Google Scholar] [CrossRef]
- Domizio, S.; Romanelli, A.; Brindisino, P.; Puglielli, C.; Conte, E.; Domizio, R.; Sgarrella, M.C.; Sabatino, G. Glutaric aciduria type II: A case report. Int. J. Immunopathol. Pharmacol. 2005, 18, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Beresford, M.W.; Pourfarzam, M.; Turnbull, D.M.; Davidson, J.E. So doctor, what exactly is wrong with my muscles? Glutaric aciduria type II presenting in a teenager. Neuromuscul. Disord. 2006, 16, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Ou, M.; Zhu, L.; Zhang, Y.; Zhou, J.; Chen, X.; Yang, L.; Li, T.; Su, X.; Hu, Q.; Wang, W. A novel electron transfer flavoprotein dehydrogenase (ETFDH) gene mutation identified in a newborn with glutaric acidemia type II: A case report of a Chinese family. BMC Med. Genet. 2020, 21, 98. [Google Scholar] [CrossRef]
- Angle, B.; Burton, B.K. Risk of sudden death and acute life-threatening events in patients with glutaric acidemia type II. Mol. Genet. Metab. 2008, 93, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Kobayashi, H.; Bo, R.; Takahashi, T.; Purevsuren, J.; Hasegawa, Y.; Taketani, T.; Fukuda, S.; Ohkubo, T.; Yokota, T.; et al. Clinical, biochemical and molecular investigation of adult-onset glutaric acidemia type II: Characteristics in comparison with pediatric cases. Brain Dev. 2016, 38, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Olsen, R.K.; Koňaříková, E.; Giancaspero, T.A.; Mosegaard, S.; Boczonadi, V.; Mataković, L.; Veauville-Merllié, A.; Terrile, C.; Schwarzmayr, T.; Haack, T.B. Riboflavin-responsive and-non-responsive mutations in FAD synthase cause multiple acyl-CoA dehydrogenase and combined respiratory-chain deficiency. Am. J. Hum. Genet. 2016, 98, 1130–1145. [Google Scholar] [CrossRef]
- Goodman, S.I.; Binard, R.J.; Woontner, M.R.; Frerman, F.E. Glutaric acidemia type II: Gene structure and mutations of the electron transfer flavoprotein:ubiquinone oxidoreductase (ETF:QO) gene. Mol. Genet. Metab. 2002, 77, 86–90. [Google Scholar] [CrossRef]
- Schiff, M.; Froissart, R.; Olsen, R.K.; Acquaviva, C.; Vianey-Saban, C. Electron transfer flavoprotein deficiency: Functional and molecular aspects. Mol. Genet. Metab. 2006, 88, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Frerman, F.E.; Kim, J.J. Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc. Natl. Acad. Sci. USA 2006, 103, 16212–16217. [Google Scholar] [CrossRef]
- Klomp, L.W.; De Koning, T.J.; Malingré, H.E.; Van Beurden, E.A.; Brink, M.; Opdam, F.L.; Duran, M.; Jaeken, J.; Pineda, M.; Van Maldergem, L. Molecular characterization of 3-phosphoglycerate dehydrogenase deficiency—a neurometabolic disorder associated with reduced L-serine biosynthesis. Am. J. Hum. Genet. 2000, 67, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Jaeken, J.; Detheux, M.; Van Maldergem, L.; Foulon, M.; Carchon, H.; Van Schaftingen, E. 3-Phosphoglycerate dehydrogenase deficiency: An inborn error of serine biosynthesis. Arch. Dis. Child. 1996, 74, 542–545. [Google Scholar] [CrossRef] [PubMed]
- De Koning, T.J.; Duran, M.; Dorland, L.; Gooskens, R.; Van Schaftingen, E.; Jaeken, J.; Blau, N.; Berger, R.; Poll-The, B.T. Beneficial effects of L-serine and glycine in the management of seizures in 3-phosphoglycerate dehydrogenase deficiency. Ann. Neurol. Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 1998, 44, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Hausler, M.G.; Jaeken, J.; Monch, E.; Ramaekers, V.T. Phenotypic heterogeneity and adverse effects of serine treatment in 3-phosphoglycerate dehydrogenase deficiency: Report on two siblings. Neuropediatrics 2001, 32, 191–195. [Google Scholar] [CrossRef]
- Tabatabaie, L.; Klomp, L.; Rubio-Gozalbo, M.; Spaapen, L.; Haagen, A.; Dorland, L.; De Koning, T. Expanding the clinical spectrum of 3-phosphoglycerate dehydrogenase deficiency. J. Inherit. Metab. Dis. Off. J. Soc. Study Inborn Errors Metab. 2011, 34, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, F.; Kariminejad, A.; Kahlert, A.K.; Morrison, P.J.; Gumus, E.; Mathews, K.D.; Darbro, B.W.; Amor, D.J.; Walsh, M.; Sznajer, Y.; et al. Expanding the genotypic and phenotypic spectrum of severe serine biosynthesis disorders. Hum. Mutat. 2020, 41, 1615–1628. [Google Scholar] [CrossRef]
- Glinton, K.E.; Benke, P.J.; Lines, M.A.; Geraghty, M.T.; Chakraborty, P.; Al-Dirbashi, O.Y.; Jiang, Y.; Kennedy, A.D.; Grotewiel, M.S.; Sutton, V.R.; et al. Disturbed phospholipid metabolism in serine biosynthesis defects revealed by metabolomic profiling. Mol. Genet. Metab. 2018, 123, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Byers, H.M.; Bennett, R.L.; Malouf, E.A.; Weiss, M.D.; Feng, J.; Scott, C.R.; Jayadev, S. Novel Report of Phosphoserine Phosphatase Deficiency in an Adult with Myeloneuropathy and Limb Contractures. Jimd Rep. 2016, 30, 103–108. [Google Scholar] [CrossRef]
- Pineda, M.; Vilaseca, M.; Artuch, R.; Santos, S.; González, M.G.; Sau, I.; Aracil, A.; Van Schaftingen, E.; Jaeken, J. 3-phosphoglycerate dehydrogenase deficiency in a patient with West syndrome. Dev. Med. Child Neurol. 2000, 42, 629–633. [Google Scholar] [CrossRef]
- Pind, S.; Slominski, E.; Mauthe, J.; Pearlman, K.; Swoboda, K.J.; Wilkins, J.A.; Sauder, P.; Natowicz, M.R. V490M, a common mutation in 3-phosphoglycerate dehydrogenase deficiency, causes enzyme deficiency by decreasing the yield of mature enzyme. J. Biol. Chem. 2002, 277, 7136–7143. [Google Scholar] [CrossRef]
- Dey, S.; Grant, G.A.; Sacchettini, J.C. Crystal structure of Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase: Extreme asymmetry in a tetramer of identical subunits. J. Biol. Chem. 2005, 280, 14892–14899. [Google Scholar] [CrossRef] [PubMed]
- Vreken, P.; Van Lint, A.; Bootsma, A.; Overmars, H.; Wanders, R.; Van Gennip, A. Quantitative plasma acylcarnitine analysis using electrospray tandem mass spectrometry for the diagnosis of organic acidaemias and fatty acid oxidation defects. J. Inherit. Metab. Dis. 1999, 22, 302–306. [Google Scholar] [CrossRef]
- Babu, S.V.; Shareef, M.M.; Shetty, A.P.; Shetty, K.T. HPLC method for amino acids profile in biological fluids and inborn metabolic disorders of aminoacidopathies. Indian J. Clin. Biochem. 2002, 17, 7–26. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, R.C.; Pollard, L.; Scott, A.I.; Huguenin, S.; Goodman, S.; Sun, Q. Laboratory analysis of organic acids, 2018 update: A technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2018, 20, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Bendl, J.; Musil, M.; Stourac, J.; Zendulka, J.; Damborsky, J.; Brezovsky, J. PredictSNP2: A Unified Platform for Accurately Evaluating SNP Effects by Exploiting the Different Characteristics of Variants in Distinct Genomic Regions. PLoS Comput. Biol. 2016, 12, e1004962. [Google Scholar] [CrossRef]
- Capra, J.A.; Singh, M. Predicting functionally important residues from sequence conservation. Bioinformatics 2007, 23, 1875–1882. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2020. Available online: http://www.pymol.org (accessed on 15 March 2021).
- Xue, Y.; Zhou, Y.; Zhang, K.; Li, L.; Kayoumu, A.; Chen, L.; Wang, Y.; Lu, Z. Compound heterozygous mutations in electron transfer flavoprotein dehydrogenase identified in a young Chinese woman with late-onset glutaric aciduria type II. Lipids Health Dis. 2017, 16, 185. [Google Scholar] [CrossRef] [PubMed]
- Nilipour, Y.; Fatehi, F.; Sanatinia, S.; Bradshaw, A.; Duff, J.; Lochmüller, H.; Horvath, R.; Nafissi, S. Multiple acyl-coenzyme A dehydrogenase deficiency shows a possible founder effect and is the most frequent cause of lipid storage myopathy in Iran. J. Neurol. Sci. 2020, 411, 116707. [Google Scholar] [CrossRef] [PubMed]
- Law, L.-K.; Tang, N.L.; Hui, J.; Fung, S.L.; Ruiter, J.; Wanders, R.J.; Fok, T.-F.; Lam, C.W. Novel mutations in ETFDH gene in Chinese patients with riboflavin-responsive multiple acyl-CoA dehydrogenase deficiency. Clin. Chim. Acta 2009, 404, 95–99. [Google Scholar] [CrossRef]
- Wang, Z.-Q.; Chen, X.-J.; Murong, S.-X.; Wang, N.; Wu, Z.-Y. Molecular analysis of 51 unrelated pedigrees with late-onset multiple acyl-CoA dehydrogenation deficiency (MADD) in southern China confirmed the most common ETFDH mutation and high carrier frequency of c. 250G>A. J. Mol. Med. 2011, 89, 569–576. [Google Scholar] [CrossRef]
- Grant, G.A. D-3-Phosphoglycerate Dehydrogenase. Front. Mol. Biosci. 2018, 5, 110. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Furuya, S.; Osuka, S.; Mitoma, J.; Shinoda, Y.; Watanabe, M.; Azuma, N.; Tanaka, H.; Hashikawa, T.; Itohara, S.; et al. Targeted disruption of the mouse 3-phosphoglycerate dehydrogenase gene causes severe neurodevelopmental defects and results in embryonic lethality. J. Biol. Chem. 2004, 279, 3573–3577. [Google Scholar] [CrossRef]
- Tabatabaie, L.; De Koning, T.; Geboers, A.; Van Den Berg, I.; Berger, R.; Klomp, L. Novel mutations in 3-phosphoglycerate dehydrogenase (PHGDH) are distributed throughout the protein and result in altered enzyme kinetics. Hum. Mutat. 2009, 30, 749–756. [Google Scholar] [CrossRef]
| Analyte | Result | Interpretation | Reference Range | Unit |
|---|---|---|---|---|
| Acetylcarnitine, C2 | 7.32 | N | 2.14–15.89 | nmol/mL |
| Propionylcarnitine, C3 | 1.16 | H | <0.55 | nmol/mL |
| Iso-/Butyrylcarnitine, C4 | 0.38 | N | <0.46 | nmol/mL |
| Isovaleyrl-/2-Methylbutyrylcarnitine, C5 | 0.28 | N | <0.38 | nmol/mL |
| Glutarylcarnitine, C5:DC | 0.07 | H | <0.06 | nmol/mL |
| Hexanoylcarnitine, C6 | 0.19 | H | <0.14 | nmol/mL |
| 3-OH-hexanoylcarnitine, C6-OH | 0.01 | N | <0.08 | nmol/mL |
| Octanoylcarnitine, C8 | 0.47 | H | <0.19 | nmol/mL |
| Octenoylcarnitine, C8:1 | 0.20 | N | <0.48 | nmol/mL |
| Decanoylcarnitine, C10 | 0.77 | H | <0.27 | nmol/mL |
| Decenoylcarnitine, C10:1 | 0.20 | N | <0.25 | nmol/mL |
| Dodecanoylcarnitine, C12 | 0.28 | H | <0.18 | nmol/mL |
| 3-OH-dodecanoylcarnitine, C12-OH | 0.03 | N | <0.06 | nmol/mL |
| Tetradecanoylcarnitine, C14 | 0.12 | H | <0.11 | nmol/mL |
| Tetradecenoylcarnitine, C14:1 | 0.20 | H | <0.16 | nmol/mL |
| 3-OH-tetradecanoylcarnitine, C14-OH | 0.01 | N | <0.04 | nmol/mL |
| Hexadecanoylcarnitine, C16 | 0.13 | N | <0.36 | nmol/mL |
| Hexadecenoylcarnitine, C16:1 | 0.04 | N | <0.15 | nmol/mL |
| 3-OH-hexadecanoylcarnitine, C16-OH | 0.01 | N | <0.78 | nmol/mL |
| Stearoylcarnitine, C18 | 0.05 | N | <0.10 | nmol/mL |
| Oleylcarnitine, C18:1 | 0.16 | N | <0.25 | nmol/mL |
| Amino Acid | Before Treatment | After Treatment | Reference Range (1–5 Years) | Unit |
|---|---|---|---|---|
| Plasma serine | 42 | 184 | 115–169 | µmol/L |
| Plasma Glycine | 96 | 145 | 175–283 | µmol/L |
| CSF serine | 6 | NP | 56–103 | µmol/L |
| CSF glycine | 8 | NP | 6.8–15 | µmol/L |
| Gene | Missense Substitutions | Zygosity | Polyphen | SIFT | PROVEAN | PANTHER | DANN | GWAVA | Funseq2 | FATHMM | Frequency | JSD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETFDH | c. 679C>A; p. Pro227Thr | Homozygous | 0.989 D | 0.01 D | −7.52 D | 0.95 D | 0.997 D | 0.45 Unknown | 4 D | 0.991 D | 0.000025 | 0.782 |
| PHGDH | c. 1219T>C; p. Ser407Pro | Homozygous | 0.736 D | 0.041 D | −1.79 N | 0.27 B | 0.994 D | 0.28 D | 4 D | 0.620 N | ND | 0.766 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, A.; Dhahouri, N.A.; Almesmari, F.S.A.; Fathalla, W.M.; Jasmi, F.A. Characterization of ETFDH and PHGDH Mutations in a Patient with Mild Glutaric Aciduria Type II and Serine Deficiency. Genes 2021, 12, 703. https://doi.org/10.3390/genes12050703
Ali A, Dhahouri NA, Almesmari FSA, Fathalla WM, Jasmi FA. Characterization of ETFDH and PHGDH Mutations in a Patient with Mild Glutaric Aciduria Type II and Serine Deficiency. Genes. 2021; 12(5):703. https://doi.org/10.3390/genes12050703
Chicago/Turabian StyleAli, Amanat, Nahid Al Dhahouri, Fatmah Saeed Ali Almesmari, Waseem Mahmoud Fathalla, and Fatma Al Jasmi. 2021. "Characterization of ETFDH and PHGDH Mutations in a Patient with Mild Glutaric Aciduria Type II and Serine Deficiency" Genes 12, no. 5: 703. https://doi.org/10.3390/genes12050703
APA StyleAli, A., Dhahouri, N. A., Almesmari, F. S. A., Fathalla, W. M., & Jasmi, F. A. (2021). Characterization of ETFDH and PHGDH Mutations in a Patient with Mild Glutaric Aciduria Type II and Serine Deficiency. Genes, 12(5), 703. https://doi.org/10.3390/genes12050703





