Characterization of Four Orphan Receptors (GPR3, GPR6, GPR12 and GPR12L) in Chickens and Ducks and Regulation of GPR12 Expression in Ovarian Granulosa Cells by Progesterone
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Enzymes, Primers and Antibodies
2.2. Animal Tissues
2.3. RNA Extraction, RT-PCR, and Quantitative Real-Time PCR Assays
2.4. Cloning the cDNAs of Chicken, Duck, Zebrafish, and Pig GPR3, GPR6, GPR12, and GPR12L
2.5. Sequence Alignment and Phylogenetic Analysis
2.6. Detection of the Basal Constitutive Activity of Human, Pig, Chicken, Duck, and Zebrafish GPR3, GPR6, GPR12, and GPR12L
2.7. Western Blot
2.8. qPCR Detection of GPR12 Expression in Chicken Granulosa Cell (GC) and Oocytes
2.9. Effect of E2, P4 and DHT5α on GPR12 Expression in Cultured 6–8 mm Follicle GCs
2.10. Promoter Analysis of Chicken GPR12 Gene
2.11. Detection of the Basal Constitutive Activity of GPR12 in Cultured GCs
2.12. Data Analysis
3. Results
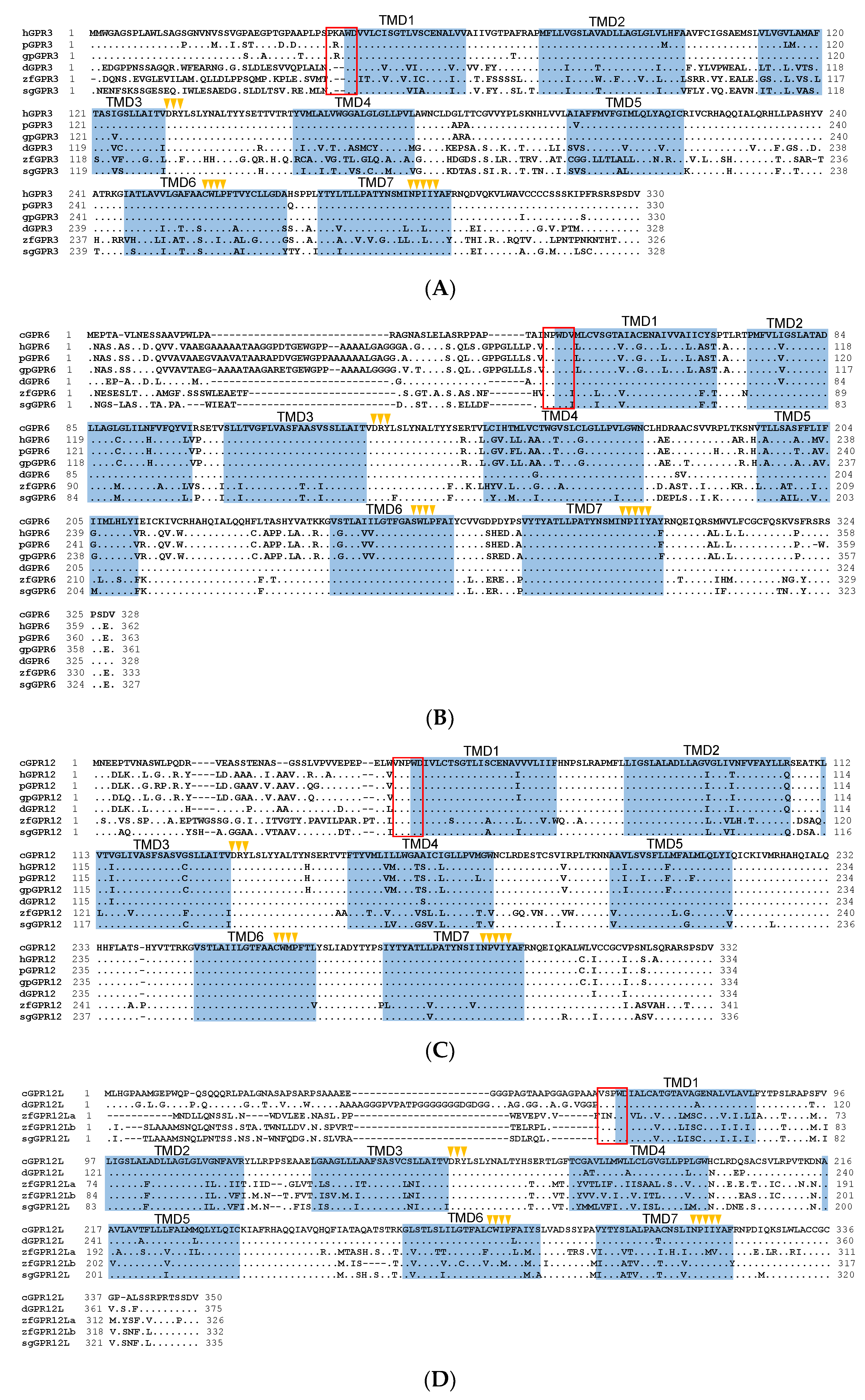
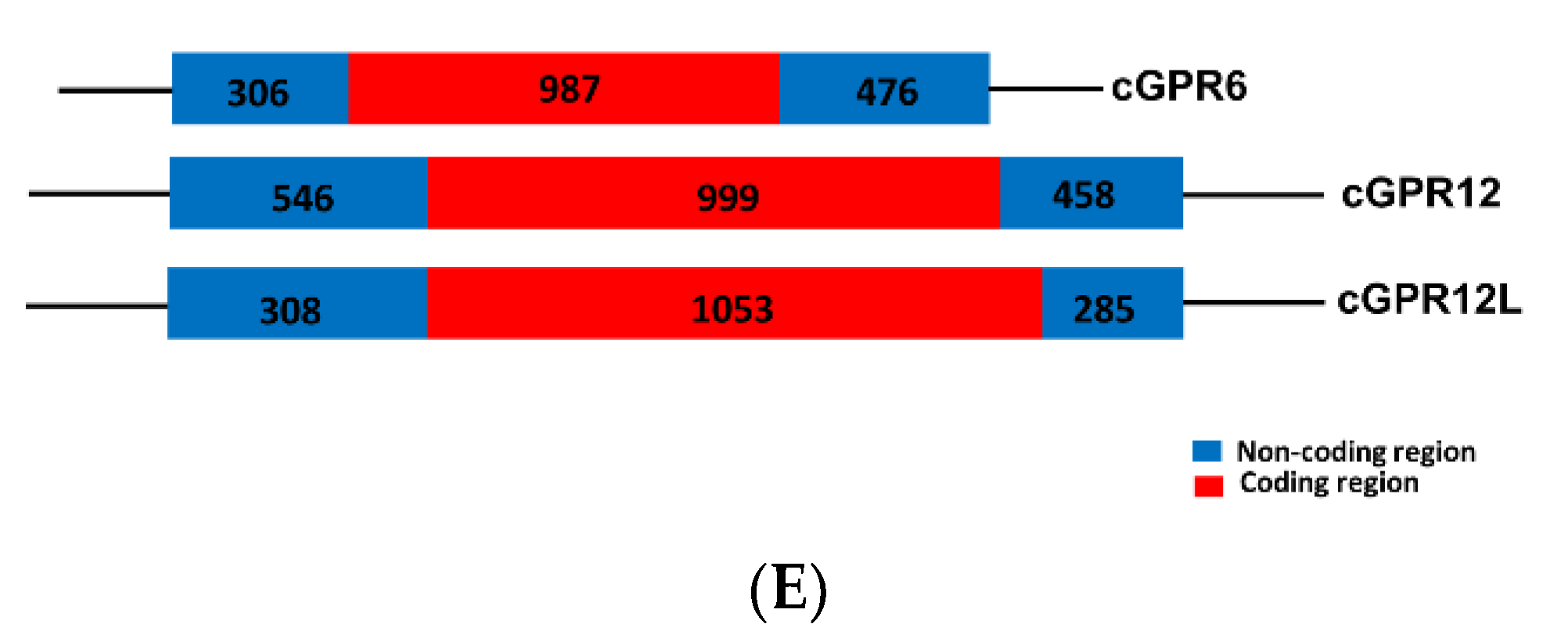
3.1. Cloning the Full-Length cDNAs of GRP3, GPR6, GPR12, and GPR12L in Chickens and Ducks
3.2. GPR3, GPR6, GPR12, and GPR12L in Birds and Other Vertebrates
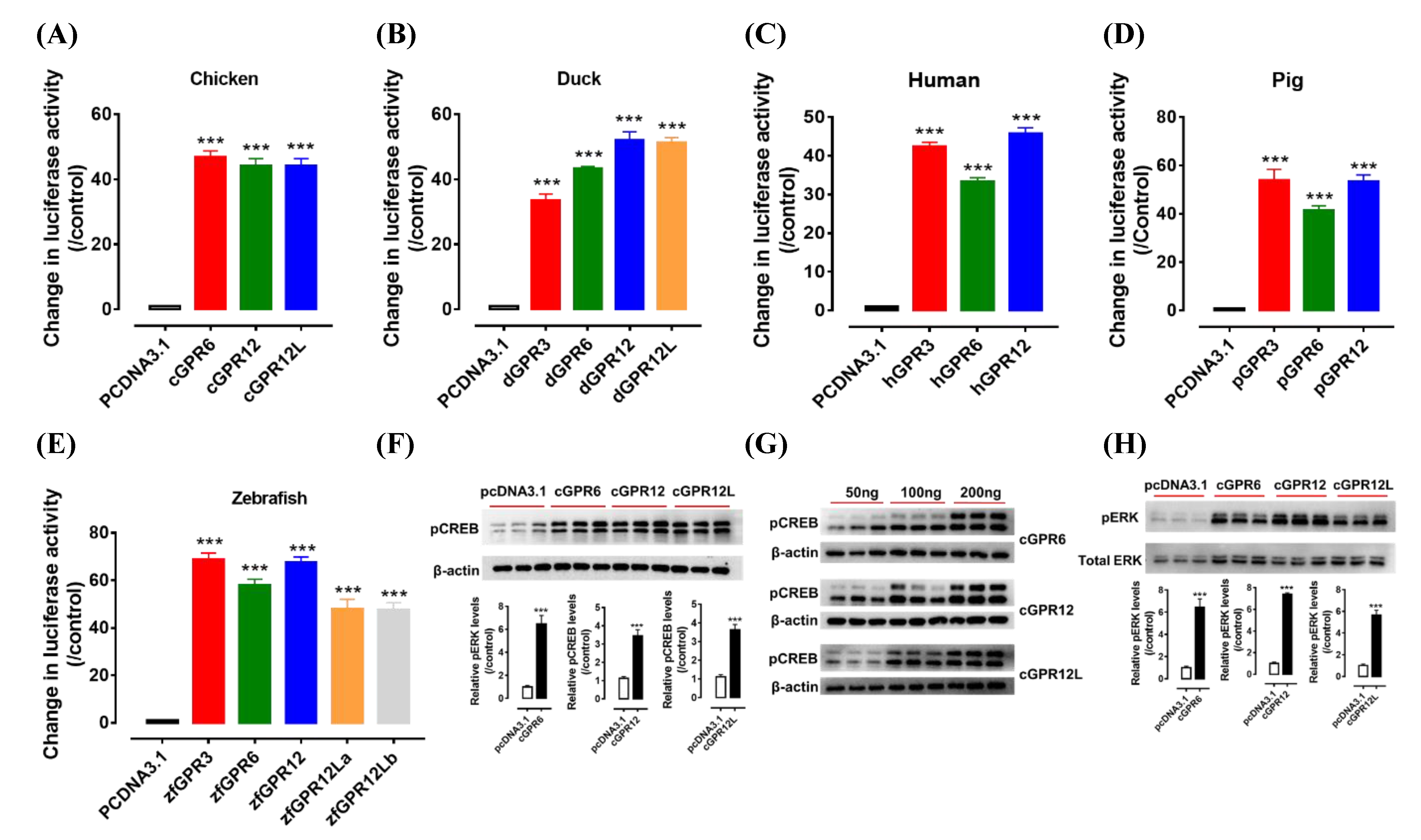
3.3. Detection of the Constitutive Activity of GPR3, GPR6, GPR12 and GPR12L in Birds
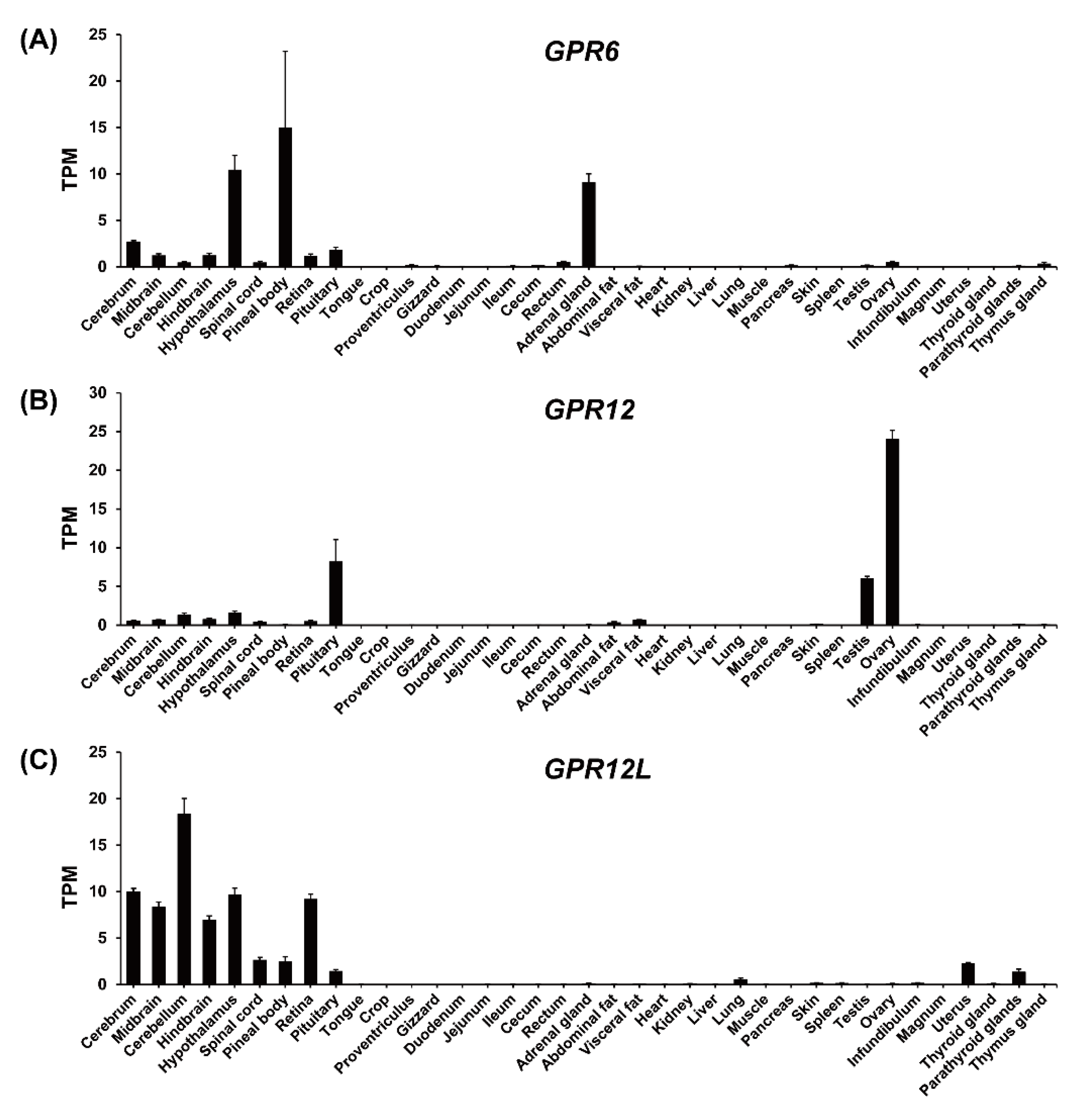
3.4. Tissue Expression of GPR3, GPR6, GPR12, and GPR12L in Chickens and Ducks
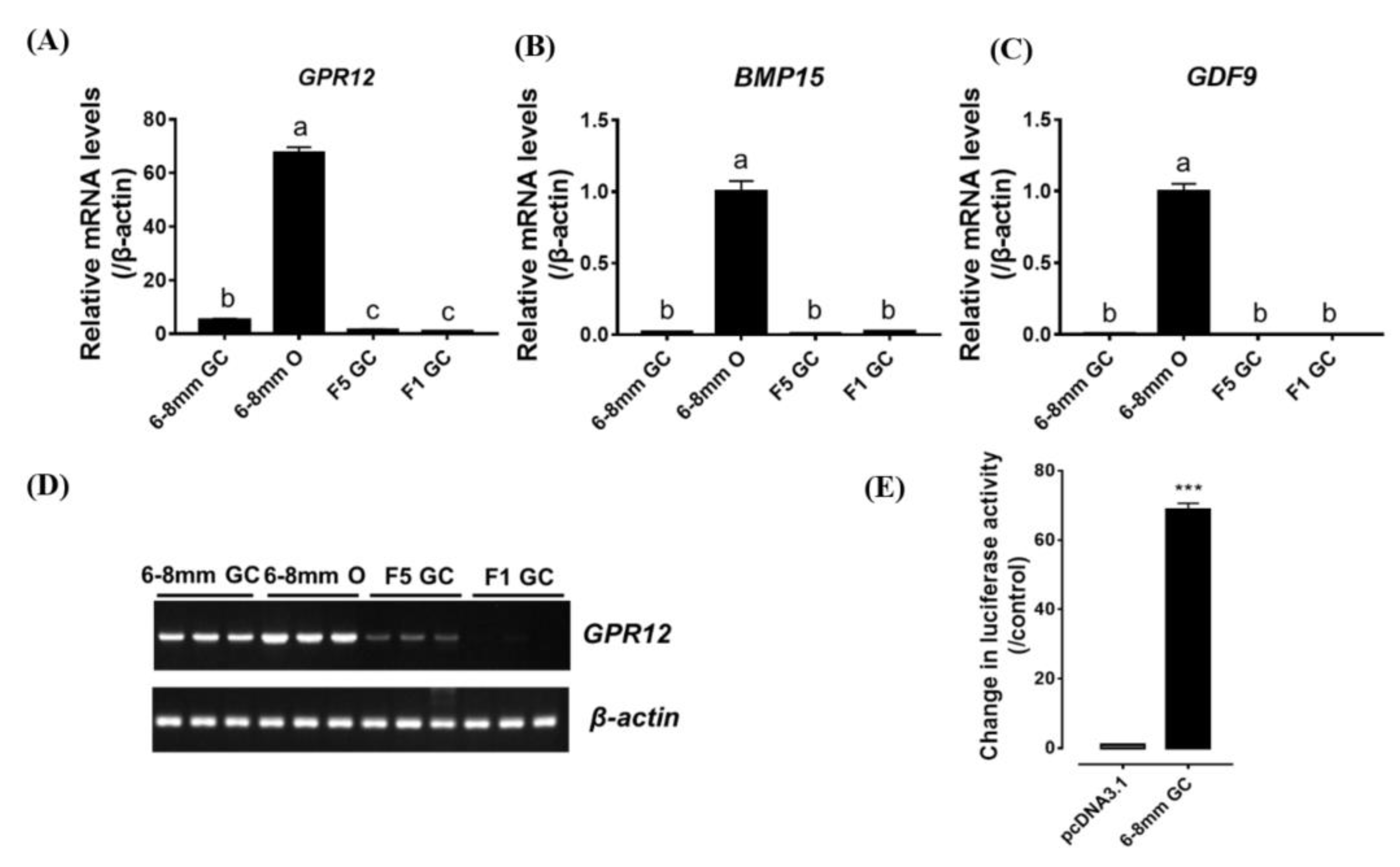
3.5. Regulation of GPR12 Expression in Ovarian Granulosa Cell (GC) by Steroid Hormones
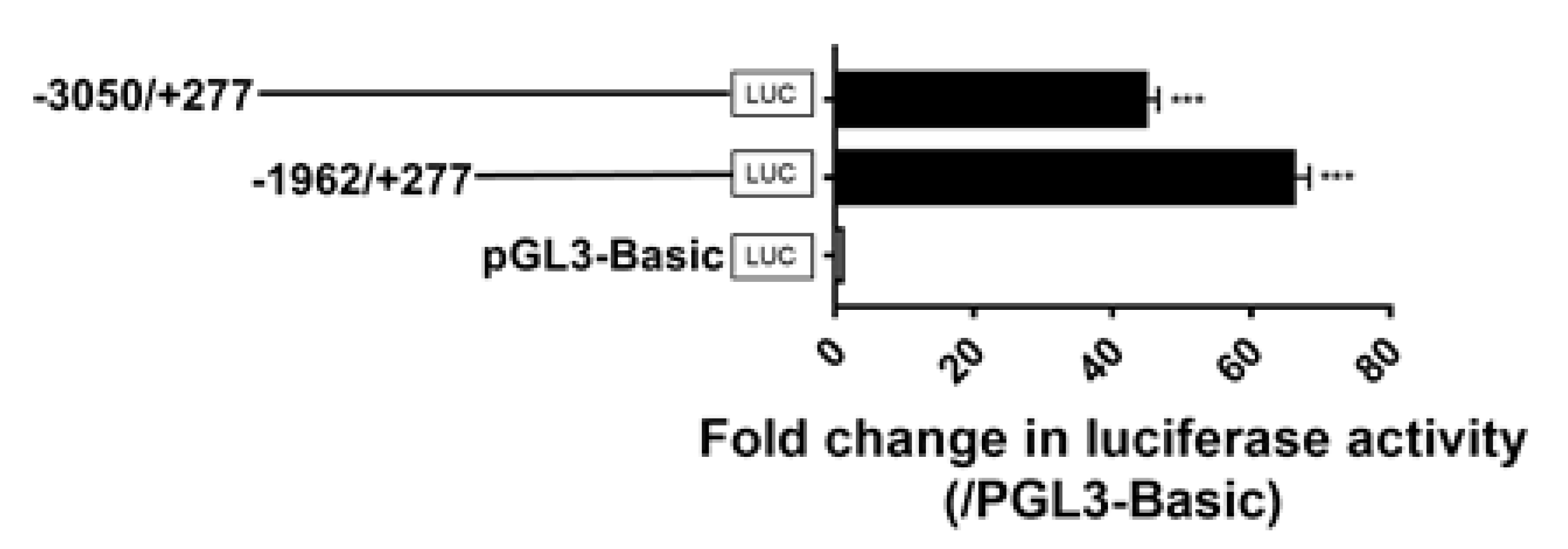
3.6. Identification of the Promoter Regions of Chicken GPR12
4. Discussion
4.1. Identification of GPR3, GPR6, GPR12, and GPR12L in Birds
4.2. Tissue Expression of GPR3, GPR6, GPR12, and GPR12L in Chickens and Ducks
4.3. GPR12 is Expressed in Oocytes and GCs of 6-mm Growing Follicles
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morales, P.; Isawi, I.; Reggio, P.H. Towards a better understanding of the cannabinoid-related orphan receptors GPR3, GPR6, and GPR12. Drug Metab. Rev. 2018, 50, 74–93. [Google Scholar] [CrossRef]
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef]
- Fredriksson, R.; Schioth, H.B. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol. Pharmacol. 2005, 67, 1414–1425. [Google Scholar] [CrossRef]
- Sun, Q.Y.; Miao, Y.L.; Schatten, H. Towards a new understanding on the regulation of mammalian oocyte meiosis resumption. Cell Cycle 2009, 8, 2741–2747. [Google Scholar] [CrossRef]
- Uhlenbrock, K.; Gassenhuber, H.; Kostenis, E. Sphingosine 1-phosphate is a ligand of the human gpr3, gpr6 and gpr12 family of constitutively active G protein-coupled receptors. Cell. Signal. 2002, 14, 941–953. [Google Scholar] [CrossRef]
- Yin, H.; Chu, A.; Li, W.; Wang, B.; Shelton, F.; Otero, F.; Nguyen, D.G.; Caldwell, J.S.; Chen, Y.A. Lipid G protein-coupled receptor ligand identification using β-arrestin PathHunter assay. J. Biol. Chem. 2009, 284, 12328–12338. [Google Scholar] [CrossRef] [PubMed]
- Valverde, O.; Celerier, E.; Baranyi, M.; Vanderhaeghen, P.; Maldonado, R.; Sperlagh, B.; Vassart, G.; Ledent, C. GPR3 receptor, a novel actor in the emotional-like responses. PLoS ONE 2009, 4, e4704. [Google Scholar] [CrossRef]
- Eggerickx, D.; Denef, J.F.; Labbe, O.; Hayashi, Y.; Refetoff, S.; Vassart, G.; Parmentier, M.; Libert, F. Molecular cloning of an orphan G-protein-coupled receptor that constitutively activates adenylate cyclase. Biochem. J. 1995, 309 (Pt 3), 837–843. [Google Scholar] [CrossRef]
- Martin, A.L.; Steurer, M.A.; Aronstam, R.S. Constitutive Activity among Orphan Class-A G Protein Coupled Receptors. PLoS ONE 2015, 10, e0138463. [Google Scholar] [CrossRef]
- Seifert, R.; Wenzel-Seifert, K. Constitutive activity of G-protein-coupled receptors: Cause of disease and common property of wild-type receptors. Naunyn. Schmiedebergs Arch. Pharmacol. 2002, 366, 381–416. [Google Scholar] [CrossRef] [PubMed]
- Godlewski, G.; Jourdan, T.; Szanda, G.; Tam, J.; Cinar, R.; Harvey-White, J.; Liu, J.; Mukhopadhyay, B.; Pacher, P.; Ming Mo, F.; et al. Mice lacking GPR3 receptors display late-onset obese phenotype due to impaired thermogenic function in brown adipose tissue. Sci. Rep. 2015, 5, 14953. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Skwarek-Maruszewska, A.; Horre, K.; Vandewyer, E.; Wolfs, L.; Snellinx, A.; Saito, T.; Radaelli, E.; Corthout, N.; Colombelli, J.; et al. Loss of GPR3 reduces the amyloid plaque burden and improves memory in Alzheimer’s disease mouse models. Sci. Transl. Med. 2015, 7, 309ra164. [Google Scholar] [CrossRef] [PubMed]
- Mehlmann, L.M.; Saeki, Y.; Tanaka, S.; Brennan, T.J.; Evsikov, A.V.; Pendola, F.L.; Knowles, B.B.; Eppig, J.J.; Jaffe, L.A. The Gs-linked receptor GPR3 maintains meiotic arrest in mammalian oocytes. Science 2004, 306, 1947–1950. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Ishii, K.; Kasai, K.; Yoon, S.O.; Saeki, Y. Neural expression of G protein-coupled receptors GPR3, GPR6, and GPR12 up-regulates cyclic AMP levels and promotes neurite outgrowth. J. Biol. Chem. 2007, 282, 10506–10515. [Google Scholar] [CrossRef]
- Oeckl, P.; Hengerer, B.; Ferger, B. G-protein coupled receptor 6 deficiency alters striatal dopamine and cAMP concentrations and reduces dyskinesia in a mouse model of Parkinson’s disease. Exp. Neurol. 2014, 257, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Benoit, M.E.; Hernandez, M.X.; Dinh, M.L.; Benavente, F.; Vasquez, O.; Tenner, A.J. C1q-induced LRP1B and GPR6 proteins expressed early in Alzheimer disease mouse models, are essential for the C1q-mediated protection against amyloid-β neurotoxicity. J. Biol. Chem. 2013, 288, 654–665. [Google Scholar] [CrossRef]
- Han, S.J.; Conti, M. New pathways from PKA to the Cdc2/cyclin B complex in oocytes: Wee1B as a potential PKA substrate. Cell Cycle 2006, 5, 227–231. [Google Scholar] [CrossRef]
- Oh, J.S.; Han, S.J.; Conti, M. Wee1B, Myt1, and Cdc25 function in distinct compartments of the mouse oocyte to control meiotic resumption. J. Cell Biol. 2010, 188, 199–207. [Google Scholar] [CrossRef]
- Hinckley, M.; Vaccari, S.; Horner, K.; Chen, R.; Conti, M. The G-protein-coupled receptors GPR3 and GPR12 are involved in cAMP signaling and maintenance of meiotic arrest in rodent oocytes. Dev. Biol. 2005, 287, 249–261. [Google Scholar] [CrossRef]
- Deng, J.; Lang, S.; Wylie, C.; Hammes, S.R. The Xenopus laevis isoform of G protein-coupled receptor 3 (GPR3) is a constitutively active cell surface receptor that participates in maintaining meiotic arrest in X. laevis oocytes. Mol. Endocrinol. 2008, 22, 1853–1865. [Google Scholar] [CrossRef]
- Yang, C.R.; Wei, Y.; Qi, S.T.; Chen, L.; Zhang, Q.H.; Ma, J.Y.; Luo, Y.B.; Wang, Y.P.; Hou, Y.; Schatten, H.; et al. The G protein coupled receptor 3 is involved in cAMP and cGMP signaling and maintenance of meiotic arrest in porcine oocytes. PLoS ONE 2012, 7, e38807. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Fang, C.; Mo, C.; Wang, Y.; Huang, Y.; Li, J. Transcriptomic analysis of granulosa cell populations proximal and distal to the germinal disc of chicken preovulatory follicles. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, A.; Evans, A.; Perry, M.; Davidson, M. A method for separating the granulosa cells, the basal lamina and the theca of the preovulatory ovarian follicle of the domestic fowl (Gallus domesticus). Reproduction 1977, 50, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Mo, C.; Huang, L.; Li, J.; Wang, Y. Characterization of the Two CART Genes (CART1 and CART2) in Chickens (Gallus gallus). PLoS ONE 2015, 10, e0127107. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Zhou, Y.; Cui, L.; Li, J.; Wu, C.; Wan, Y.; Li, J.; Wang, Y. The interaction of MC3R and MC4R with MRAP2, ACTH, α-MSH and AgRP in chickens. J. Endocrinol. 2017, 234, 155–174. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.; Wu, C.; Hu, Z.; An, L.; Wan, Y.; Fang, C.; Zhang, X.; Li, J.; Wang, Y. The Asp298Asn polymorphism of melanocortin-4 receptor (MC4R) in pigs: Evidence for its potential effects on MC4R constitutive activity and cell surface expression. Anim. Genet. 2020, 51, 694–706. [Google Scholar] [CrossRef]
- He, C.; Zhang, J.; Gao, S.; Meng, F.; Bu, G.; Li, J.; Wang, Y. Molecular characterization of three NPY receptors (Y2, Y5 and Y7) in chickens: Gene structure, tissue expression, promoter identification, and functional analysis. Gen. Comp. Endocrinol. 2016, 236, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Ying Wang, C.; Yan Kwok, A.H.; Leung, F.C. Epidermal growth factor (EGF) receptor ligands in the chicken ovary: I. Evidence for heparin-binding EGF-like growth factor (HB-EGF) as a potential oocyte-derived signal to control granulosa cell proliferation and HB-EGF and kit ligand expression. Endocrinology 2007, 148, 3426–3440. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Yan Kwok, A.H.; Ge, W.; Leung, F.C. A novel prolactin-like protein (PRL-L) gene in chickens and zebrafish: Cloning and characterization of its tissue expression. Gen. Comp. Endocrinol. 2010, 166, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Morales, P.; Hurst, D.P.; Reggio, P.H. Methods for the Development of In Silico GPCR Models. Methods Enzymol. 2017, 593, 405–448. [Google Scholar] [CrossRef]
- Cheng, Z.; Garvin, D.; Paguio, A.; Stecha, P.; Wood, K.; Fan, F. Luciferase Reporter Assay System for Deciphering GPCR Pathways. Curr. Chem. Genom. 2010, 4, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Miao, Y.-R.; Jia, L.-H.; Yu, Q.-Y.; Zhang, Q.; Guo, A.-Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019, 47, D33–D38. [Google Scholar] [CrossRef]
- Olender, T.; Jones, T.E.; Bruford, E.; Lancet, D. A unified nomenclature for vertebrate olfactory receptors. BMC Evol. Biol. 2020, 20, 42. [Google Scholar] [CrossRef] [PubMed]
- Ignatov, A.; Lintzel, J.; Kreienkamp, H.J.; Schaller, H.C. Sphingosine-1-phosphate is a high-affinity ligand for the G protein-coupled receptor GPR6 from mouse and induces intracellular Ca2+ release by activating the sphingosine-kinase pathway. Biochem. Biophys. Res. Commun. 2003, 311, 329–336. [Google Scholar] [CrossRef]
- Tanaka, S.; Miyagi, T.; Dohi, E.; Seki, T.; Hide, I.; Sotomaru, Y.; Saeki, Y.; Antonio Chiocca, E.; Matsumoto, M.; Sakai, N. Developmental expression of GPR3 in rodent cerebellar granule neurons is associated with cell survival and protects neurons from various apoptotic stimuli. Neurobiol. Dis. 2014, 68, 215–227. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, N.; Dong, S.; Hu, Y. Involvement of GPR12 in the induction of neurite outgrowth in PC12 cells. Brain Res. Bull. 2012, 87, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Lubrano-Berthelier, C.; Govaerts, C.; Picard, F.; Santiago, P.; Conklin, B.R.; Vaisse, C. Constitutive activity of the melanocortin-4 receptor is maintained by its N-terminal domain and plays a role in energy homeostasis in humans. J. Clin. Invest. 2004, 114, 1158–1164. [Google Scholar] [CrossRef]
- Song, Z.H.; Young, W.S., 3rd; Brownstein, M.J.; Bonner, T.I. Molecular cloning of a novel candidate G protein-coupled receptor from rat brain. FEBS Lett. 1994, 351, 375–379. [Google Scholar] [CrossRef]
- Heiber, M.; Docherty, J.M.; Shah, G.; Nguyen, T.; Cheng, R.; Heng, H.H.; Marchese, A.; Tsui, L.C.; Shi, X.; George, S.R.; et al. Isolation of three novel human genes encoding G protein-coupled receptors. DNA Cell Biol. 1995, 14, 25–35. [Google Scholar] [CrossRef]
- Saeki, Y.; Ueno, S.; Mizuno, R.; Nishimura, T.; Fujimura, H.; Nagai, Y.; Yanagihara, T. Molecular-Cloning of a Novel Putative G-Protein-Coupled Receptor (Gpcr21) Which Is Expressed Predominantly in Mouse Central-Nervous-System. FEBS Lett. 1993, 336, 317–322. [Google Scholar] [CrossRef]
- Eidne, K.A.; Zabavnik, J.; Peters, T.; Yoshida, S.; Anderson, L.; Taylor, P.L. Cloning, sequencing and tissue distribution of a candidate G protein-coupled receptor from rat pituitary gland. FEBS Lett. 1991, 292, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ding, J.; Li, Y.; Wang, J.; Zhao, Y.; Wang, W.; Shi, S.; Dong, F.; Zhang, Z.; Shi, F.; et al. The porcine Gpr3 gene: Molecular cloning, characterization and expression level in tissues and cumulus-oocyte complexes during in vitro maturation. Mol. Biol. Rep. 2012, 39, 5831–5839. [Google Scholar] [CrossRef] [PubMed]
- Iismaa, T.P.; Kiefer, J.; Liu, M.L.; Baker, E.; Sutherland, G.R.; Shine, J. Isolation and chromosomal localization of a novel human G-protein-coupled receptor (GPR3) expressed predominantly in the central nervous system. Genomics 1994, 24, 391–394. [Google Scholar] [CrossRef]
- Cho, W.K.; Stern, S.; Biggers, J.D. Inhibitory effect of dibutyryl cAMP on mouse oocyte maturation in vitro. J. Exp. Zool. 1974, 187, 383–386. [Google Scholar] [CrossRef]
- Dekel, N.; Aberdam, E.; Sherizly, I. Spontaneous maturation in vitro of cumulus-enclosed rat oocytes is inhibited by forskolin. Biol. Reprod. 1984, 31, 244–250. [Google Scholar] [CrossRef]
- Bornslaeger, E.A.; Schultz, R.M. Adenylate cyclase activity in zona-free mouse oocytes. Exp. Cell Res. 1985, 156, 277–281. [Google Scholar] [CrossRef]
- Kalinowski, R.R.; Berlot, C.H.; Jones, T.L.; Ross, L.F.; Jaffe, L.A.; Mehlmann, L.M. Maintenance of meiotic prophase arrest in vertebrate oocytes by a Gs protein-mediated pathway. Dev. Biol. 2004, 267, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Webb, R.J.; Marshall, F.; Swann, K.; Carroll, J. Follicle-stimulating hormone induces a gap junction-dependent dynamic change in [cAMP] and protein kinase a in mammalian oocytes. Dev. Biol. 2002, 246, 441–454. [Google Scholar] [CrossRef]
- Dekel, N.; Lawrence, T.S.; Gilula, N.B.; Beers, W.H. Modulation of cell-to-cell communication in the cumulus-oocyte complex and the regulation of oocyte maturation by LH. Dev. Biol. 1981, 86, 356–362. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Jiang, B.; Cao, B.; Zhang, Z.; Zhang, J.; Li, J.; Huang, Y.; Wang, Y. Characterization of Four Orphan Receptors (GPR3, GPR6, GPR12 and GPR12L) in Chickens and Ducks and Regulation of GPR12 Expression in Ovarian Granulosa Cells by Progesterone. Genes 2021, 12, 489. https://doi.org/10.3390/genes12040489
Li Z, Jiang B, Cao B, Zhang Z, Zhang J, Li J, Huang Y, Wang Y. Characterization of Four Orphan Receptors (GPR3, GPR6, GPR12 and GPR12L) in Chickens and Ducks and Regulation of GPR12 Expression in Ovarian Granulosa Cells by Progesterone. Genes. 2021; 12(4):489. https://doi.org/10.3390/genes12040489
Chicago/Turabian StyleLi, Zejiao, Biying Jiang, Baolong Cao, Zheng Zhang, Jiannan Zhang, Juan Li, Yan Huang, and Yajun Wang. 2021. "Characterization of Four Orphan Receptors (GPR3, GPR6, GPR12 and GPR12L) in Chickens and Ducks and Regulation of GPR12 Expression in Ovarian Granulosa Cells by Progesterone" Genes 12, no. 4: 489. https://doi.org/10.3390/genes12040489
APA StyleLi, Z., Jiang, B., Cao, B., Zhang, Z., Zhang, J., Li, J., Huang, Y., & Wang, Y. (2021). Characterization of Four Orphan Receptors (GPR3, GPR6, GPR12 and GPR12L) in Chickens and Ducks and Regulation of GPR12 Expression in Ovarian Granulosa Cells by Progesterone. Genes, 12(4), 489. https://doi.org/10.3390/genes12040489






