Cloning, Characterization, and RNA Interference Effect of the UDP-N-Acetylglucosamine Pyrophosphorylase Gene in Cnaphalocrocis medinalis
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Sample Preparation
2.2. RNA Extraction and cDNA Synthesis
2.3. Cloning CmUAP by Both RT-PCR and RACE
2.4. Bioinformatic Analyses of CmUAP
2.5. Gene Expression Analyses Using ddPCR
2.6. RNA Interference
2.7. Statistical Analysis
3. Results
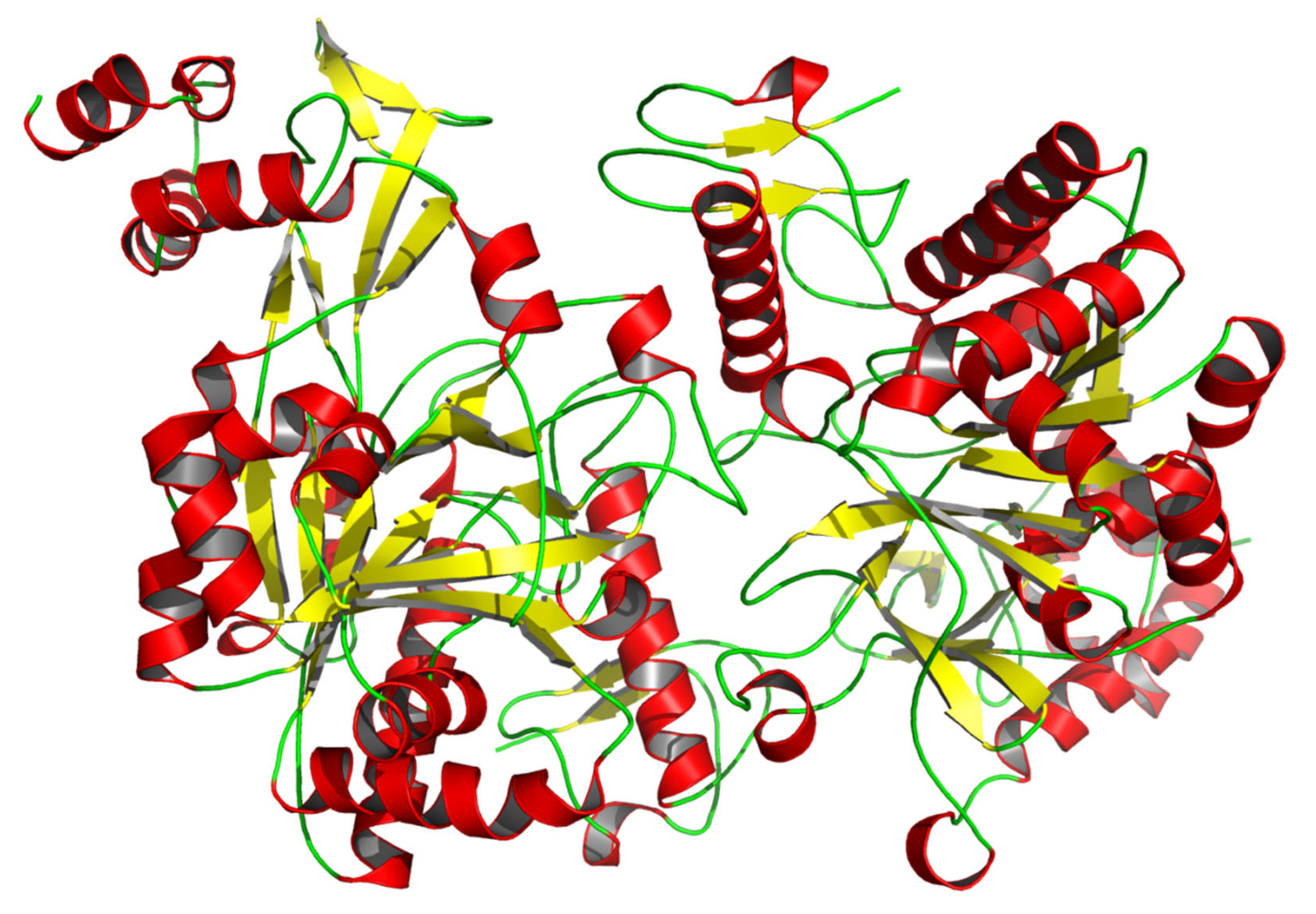
3.1. Characteristic Analyses of CmUAP and the Zymoprotein
3.2. Homology Comparison and Cluster Dendrogram
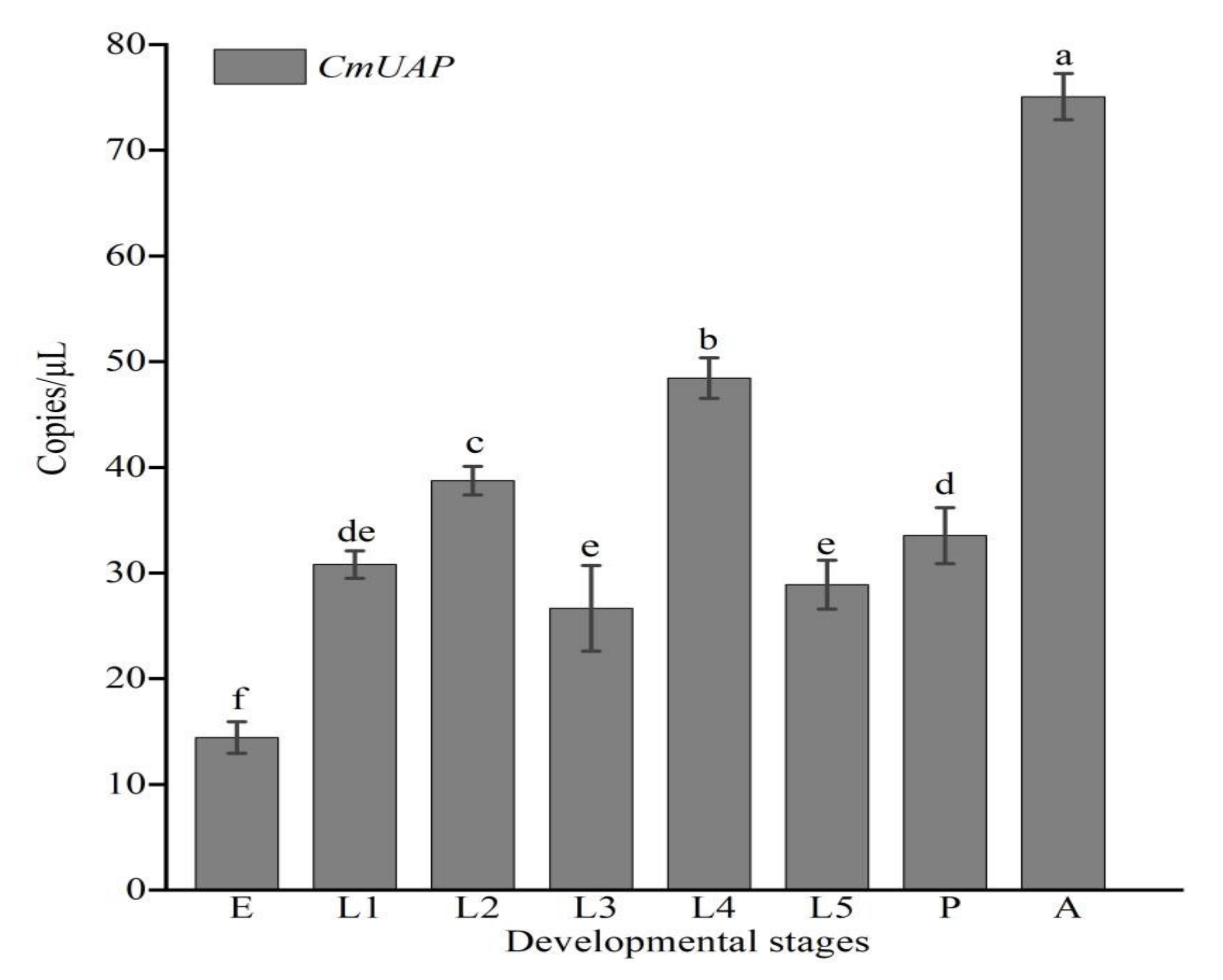
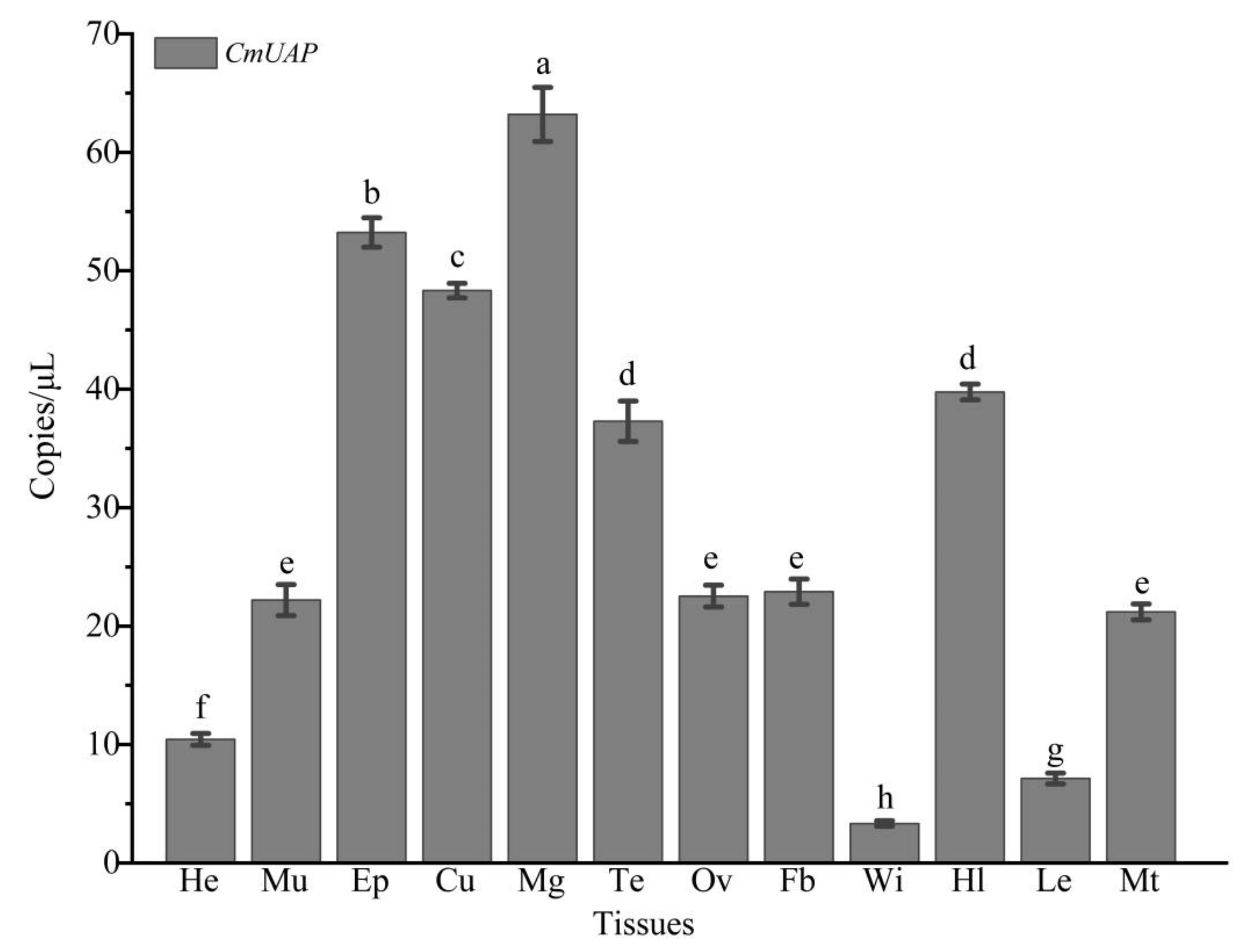
3.3. Gene Expression Profiles
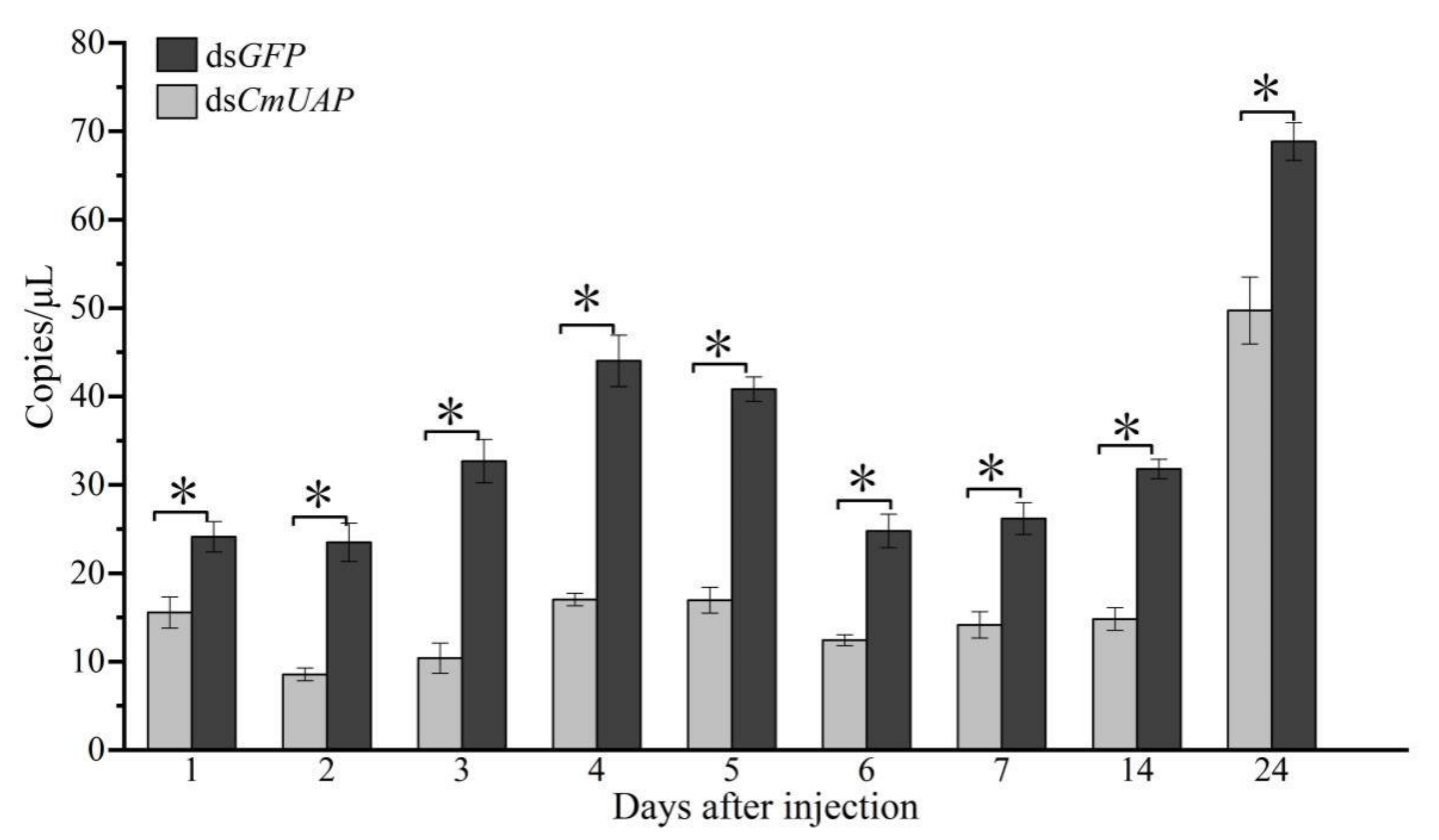
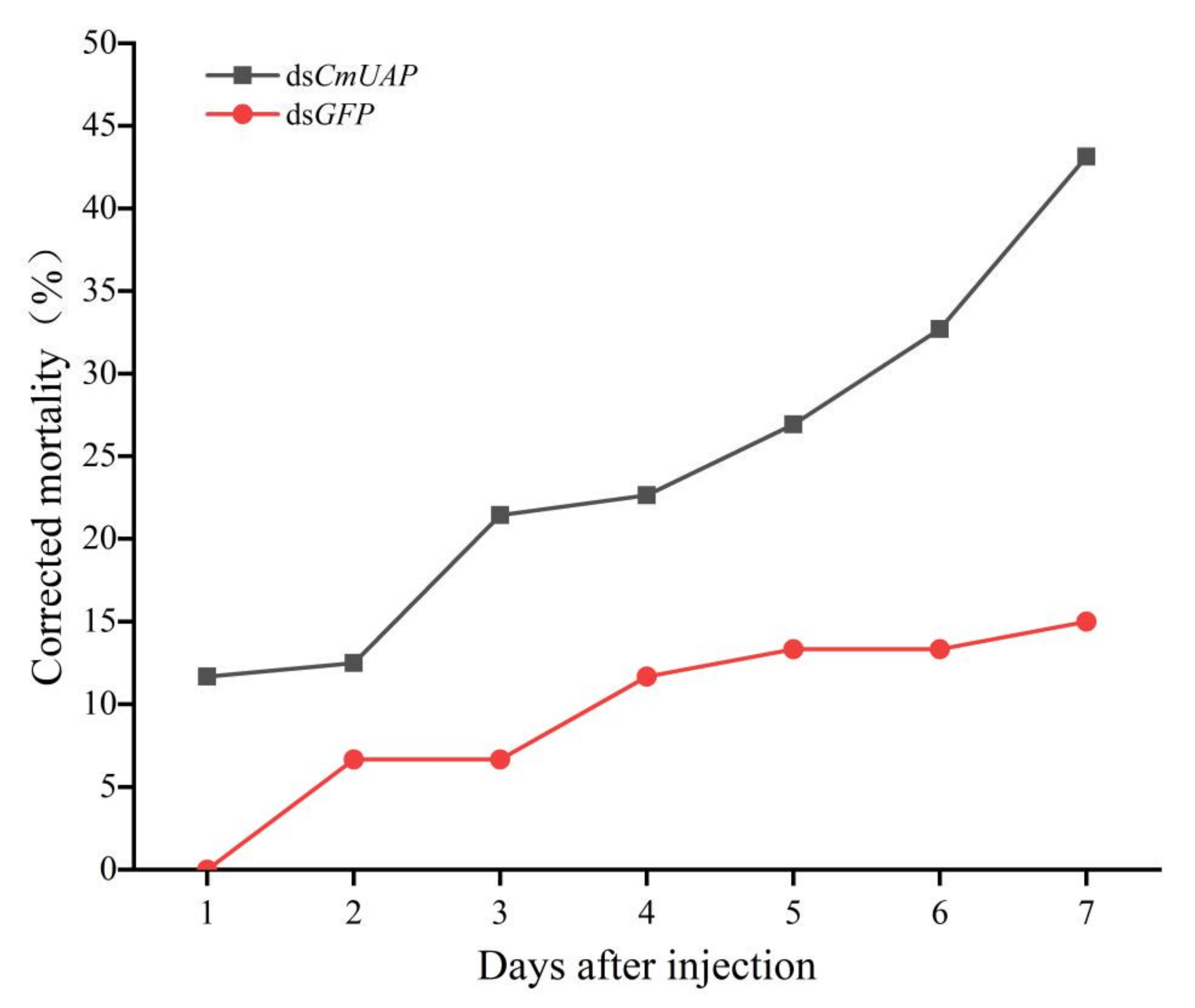
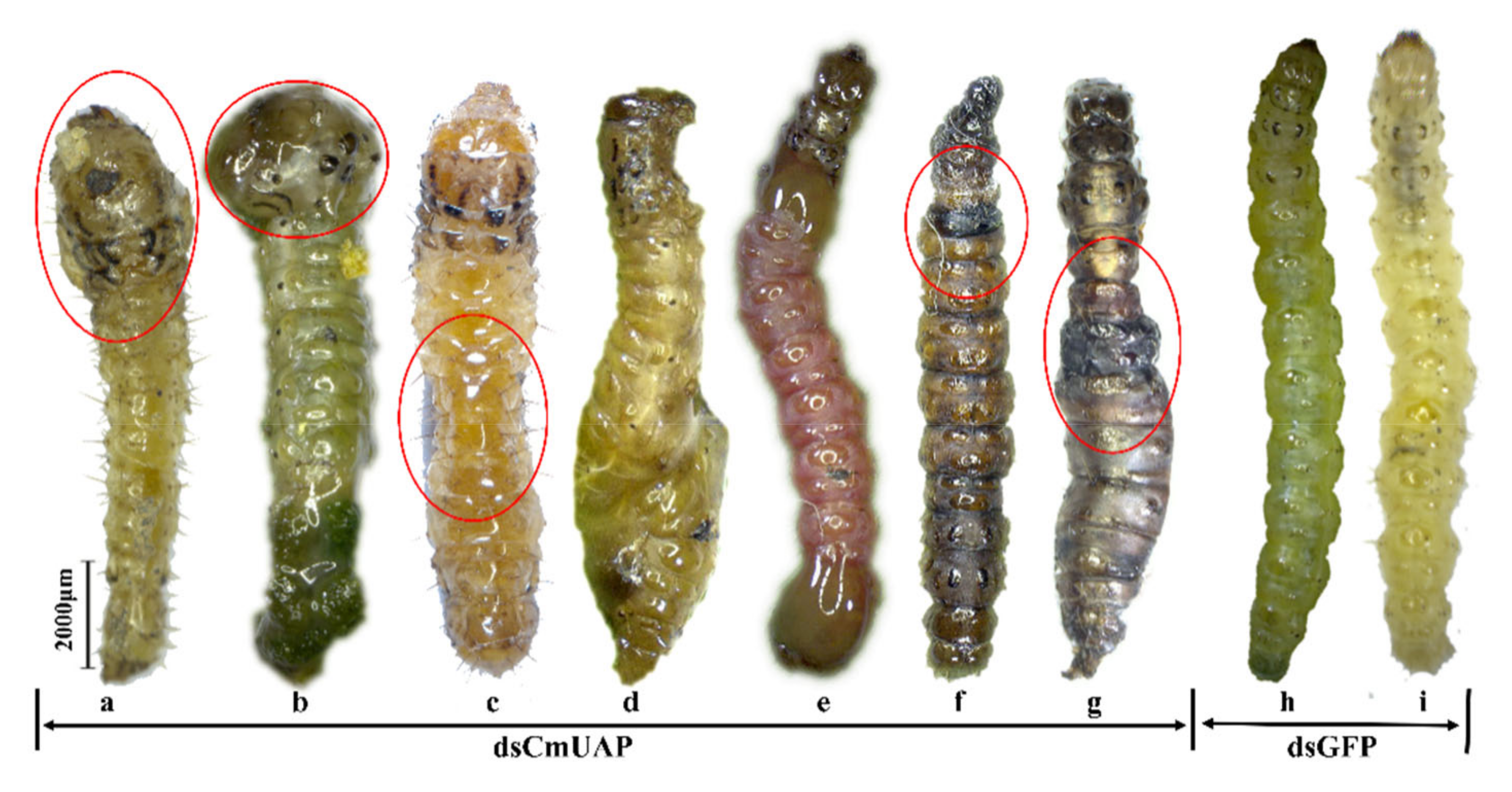
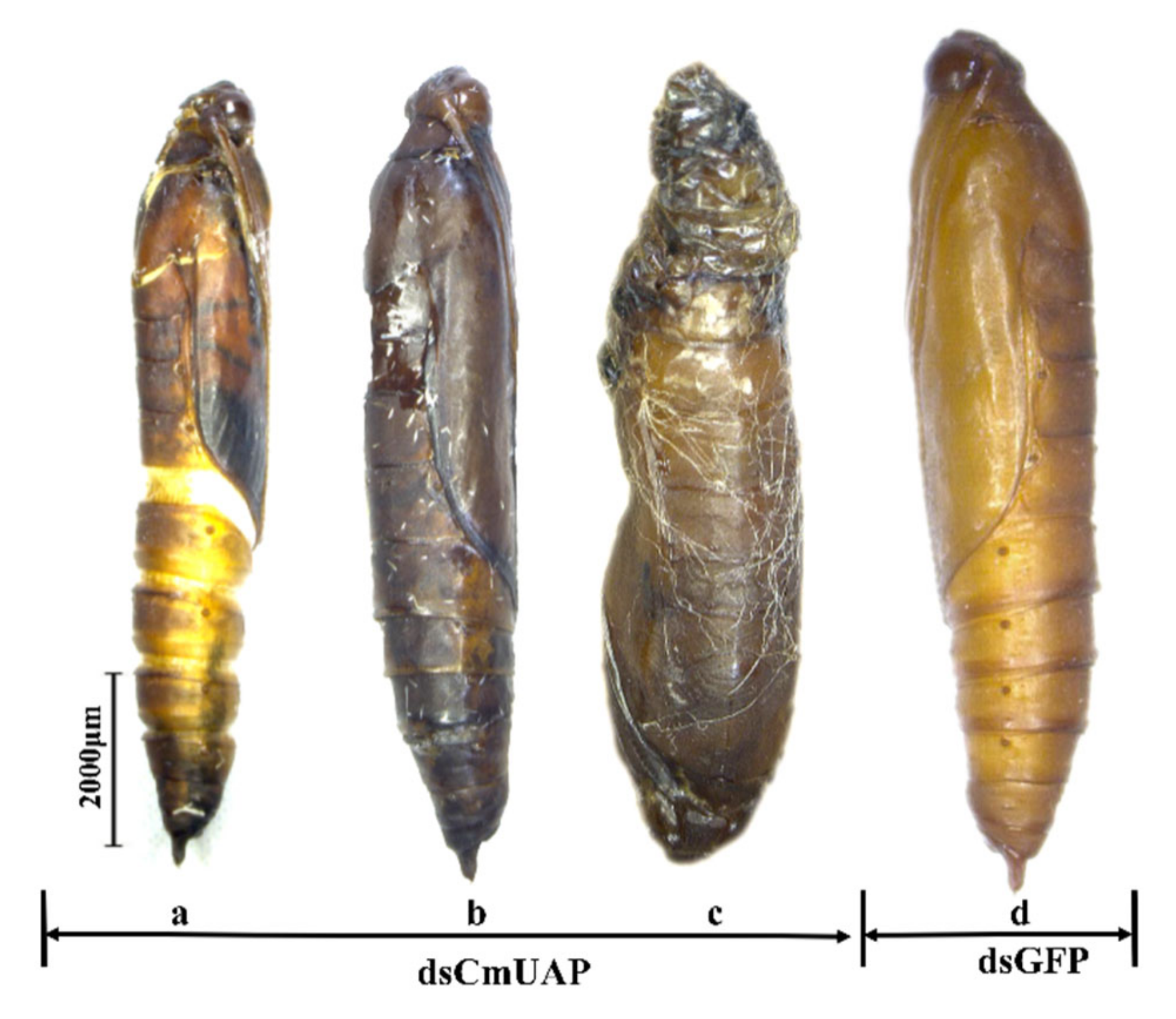
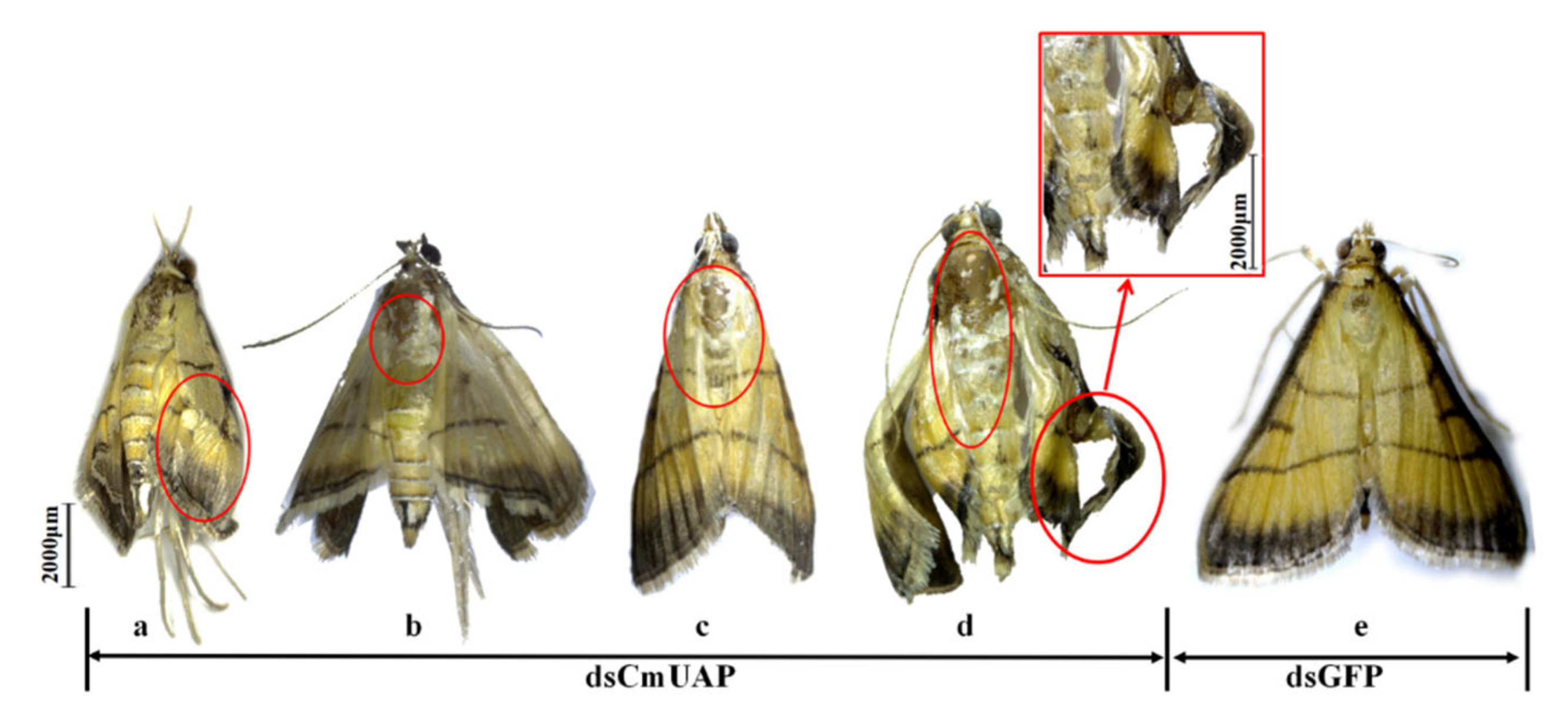
3.4. RNAi Effects of CmUAP
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bharati, L.R.; Om, H.; Kushwaha, K.S. Alternate plant hosts of rice leaffolder. Int. Rice Res. Newsl. 1990, 15, 21–22. [Google Scholar]
- Nathan, S.S.; Chung, P.G.; Murugan, K. Effect of botanical insecticides and bacterial toxins on the gut enzyme of the rice leaffolderCnaphalocrocis medinalis. Phytoparasitica 2004, 32, 433. [Google Scholar] [CrossRef]
- Nathan, S.S. Effects of Melia azedarach on nutritional physiology and enzyme activities of the rice leaffolder Cnaphalocrocis medinalis (Guenée) (Lepidoptera: Pyralidae). Pestic. Biochem. Physiol. 2006, 84, 98–108. [Google Scholar] [CrossRef]
- Zhang, S.-K.; Ren, X.-B.; Wang, Y.-C.; Su, J. Resistance in Cnaphalocrocis medinalis (Lepidoptera: Pyralidae) to new chemistry insecticides. J. Econ. Entomol. 2014, 107, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Fraenkel, G.; Fallil, F. The spinning (stitching) behaviour of the rice leaf folder, Cnaphalocrocis medinalis. Entomol. Exp. Appl. 1981, 29, 138–146. [Google Scholar] [CrossRef]
- Islam, Z.; Karim, A. Leaf folding behaviour of Cnaphalocrocis medinalis (Guenee) and Marasmia patnalis Bradley, and the influence of rice leaf morphology on damage incidence. Crop. Prot. 1997, 16, 215–220. [Google Scholar] [CrossRef]
- Misra, A.K.; Singh, S.P.N.; Parwez, A. Incidence of Cnaphalocrosis medinalis in different cultivars of Boro rice. Ann. Plant Prot. Sci. 2006, 14, 224–226. [Google Scholar]
- Alvi, S.M.; Ali, M.A.; Chaudhary, S.A.N.A.U.L.L.A.H.; Iqbal, S.H.A.H.E.E.N. Population trends and chemical control of rice leaf folder, Cnaphalocrocis medinalis on rice crop. Int. J. Agric. Biol. 2003, 5, 615–617. [Google Scholar]
- Huang, J.; Hu, R.; Pray, C.; Qiao, F.; Rozelle, S. Biotechnology as an alternative to chemical pesticides: A case study of Bt cotton in China. Agric. Econ. 2003, 29, 55–67. [Google Scholar] [CrossRef]
- Matteson, P.C. Insect Pest Management in Tropical Asian Irrigated Rice. Annu. Rev. Èntomol. 2000, 45, 549–574. [Google Scholar] [CrossRef]
- Jinek, M.; Doudna, J.A. A three-dimensional view of the molecular machinery of RNA interference. Nat. Cell Biol. 2008, 457, 405–412. [Google Scholar] [CrossRef]
- Whangbo, J.S.; Hunter, C.P. Environmental RNA interference. Trends Genet. 2008, 24, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, G.; Wang-Pruski, G.; You, M. Phyllotreta striolata (Coleoptera: Chrysomelidae): Arginine kinase cloning and RNAi-based pest control. Eur. J. Èntomol. 2008, 105, 815–822. [Google Scholar] [CrossRef]
- Walshe, D.P.; Lehane, S.M.; Lehane, M.J.; Haines, L.R. Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol. Biol. 2009, 18, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Chandrashekar, K.; Thakur, N.; Verma, P.C.; Borgio, J.F.; Singh, P.K.; Tuli, R. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 2011, 36, 153–161. [Google Scholar] [CrossRef]
- Burand, J.P.; Hunter, W.B. RNAi: Future in insect management. J. Invertebr. Pathol. 2013, 112, S68–S74. [Google Scholar] [CrossRef]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef]
- Tariq, K.; Ali, A.; Davies, T.G.E.; Naz, E.; Naz, L.; Sohail, S.; Hou, M.; Ullah, F. RNA interference-mediated knockdown of voltage-gated sodium channel (MpNav) gene causes mortality in peach-potato aphid, Myzus persicae. Sci. Rep. 2019, 9, 5291. [Google Scholar] [CrossRef]
- Turner, C.T.; Davy, M.W.; MacDiarmid, R.M.; Plummer, K.M.; Birch, N.P.; Newcomb, R.D. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol. Biol. 2006, 15, 383–391. [Google Scholar] [CrossRef]
- Tang, B.; Yang, M.; Shen, Q.; Xu, Y.; Wang, H.; Wang, S. Suppressing the activity of trehalase with validamycin disrupts the trehalose and chitin biosynthesis pathways in the rice brown planthopper, Nilaparvata lugens. Pestic. Biochem. Physiol. 2017, 137, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-W.; Wang, H.-J.; Jin, X.; Pan, B.-Y.; Zeng, B.-P.; Xiao, Z.-J.; Xu, C.-D.; Tang, B. Knockdown of glycogen phosphorylase and glycogen synthase influences expression of chitin synthesis genes of rice brown planthopper Nilaparvata lugens. J. Asia Pacific Èntomol. 2019, 22, 786–794. [Google Scholar] [CrossRef]
- Zhao, L.; Yang, M.; Shen, Q.; Liu, X.; Shi, Z.; Wang, S.; Tang, B. Functional characterization of three trehalase genes regulating the chitin metabolism pathway in rice brown planthopper using RNA interference. Sci. Rep. 2016, 6, 27841. [Google Scholar] [CrossRef]
- Chen, Q.; Jin, S.; Zhang, L.; Shen, Q.; Wei, P.; Wei, Z.; Wang, S.; Tang, B. Regulatory functions of trehalose-6-phosphate synthase in the chitin biosynthesis pathway in Tribolium castaneum (Coleoptera: Tenebrionidae) revealed by RNA interference. Bull. Entomol. Res. 2017, 108, 388–399. [Google Scholar] [CrossRef]
- Candy, D.J.; Kilby, B.A. Studies on chitin synthesis in the desert locust. J. Exp. Biol. 1962, 39, 129–140. [Google Scholar]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, S.; Merzendorfer, H.; Arakane, Y.; Kramer, K.J. Chitin metabolism in insects. In Insect Molecular Biology and Biochemistry; Academic Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Liu, X.; Li, F.; Li, D.; Ma, E.; Zhang, W.; Zhu, K.Y.; Zhang, J. Molecular and Functional Analysis of UDP-N-Acetylglucosamine Pyrophosphorylases from the Migratory Locust, Locusta migratoria. PLoS ONE 2013, 8, e71970. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.F.; Fu, J.; Mu, L.L.; Guo, W.C.; Li, G.Q. Two Leptinotarsa uridine diphosphate N-acetylglucosamine pyrophosphorylases are specialized for chitin synthesis in larval epidermal cuticle and midgut peritrophic matrix. Insect Biochem. Mol. Biol. 2016, 68, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Baguinon, M.C.; Jasrapuria, S.; Chaudhari, S.; Doyungan, A.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Both UDP N-acetylglucosamine pyrophosphorylases of Tribolium castaneum are critical for molting, survival and fecundity. Insect Biochem. Mol. Biol. 2011, 41, 42–50. [Google Scholar] [CrossRef]
- Li, S.-W.; Yang, H.; Liu, Y.-F.; Liao, Q.-R.; Du, J.; Jin, D.-C. Transcriptome and Gene Expression Analysis of the Rice Leaf Folder, Cnaphalocrosis medinalis. PLoS ONE 2012, 7, e47401. [Google Scholar] [CrossRef]
- Gabius, H.-J. The sugar code: Why glycans are so important. Biosystems 2018, 164, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.J.; Hopkins, T.L.; Schaefer, J. Applications of solids NMR to the analysis of insect sclerotized structures. Insect Biochem. Mol. Biol. 1995, 25, 1067–1080. [Google Scholar] [CrossRef]
- Kumar, N.S.; Tang, B.; Chen, X.; Tian, H.; Zhang, W. Molecular cloning, expression pattern and comparative analysis of chitin synthase gene B in Spodoptera exigua. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2008, 149, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.J.; Koga, D. Insect chitin: Physical state, synthesis, degradation and metabolic regulation. Insect Biochem. 1986, 16, 851–877. [Google Scholar] [CrossRef]
- Stokes, M.J.; Güther, M.L.S.; Turnock, D.C.; Prescott, A.R.; Martin, K.L.; Alphey, M.S.; Ferguson, M.A.J. The Synthesis of UDP-N-acetylglucosamine Is Essential for Bloodstream Form Trypanosoma brucei in Vitro and in Vivo and UDP-N-acetylglucosamine Starvation Reveals a Hierarchy in Parasite Protein Glycosylation. J. Biol. Chem. 2008, 283, 16147–16161. [Google Scholar] [CrossRef]
- Zhang, Z.-J.; Xia, L.; Du, J.; Li, S.-W.; Zhao, F. Cloning, characterization, and RNAi effect of the chitin synthase B gene in Cnaphalocrocis medinalis. J. Asia Pacific Entomol. 2020. [Google Scholar] [CrossRef]
- Shakeel, M.; Du, J.; Li, S.-W.; Zhou, Y.-J.; Sarwar, N.; Bukhari, S.A.H. Characterization, Knockdown and Parental Effect of Hexokinase Gene of Cnaphalocrocis medinalis (Lepidoptera: Pyralidae) Revealed by RNA Interference. Genes 2020, 11, 1258. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, X.; Kumar, N.S.; Tang, B.; Sun, X.; Qiu, X.; Hu, J.; Zhang, W. The class A chitin synthase gene of Spodoptera exigua: Molecular cloning and expression patterns. Insect Biochem. Mol. Biol. 2007, 37, 409–417. [Google Scholar] [CrossRef]
- Chen, J.; Yao, Q.; Zhang, W.-Q. Molecular Cloning, Expression Patterns and RNAi of UDP-N-acetylglucosamine Pyrophosphorylase in Spodoptera exigua. Sci. Agric. Sin. 2014, 47, 1351–1361. [Google Scholar]



| Primer Name | Primer Sequence (5′ → 3′) | Primer Usage |
|---|---|---|
| CmUAP-F | ATTTAACCGTGGAGGGAGAAC | RT-PCR |
| CmUAP-R | GTGATTTCCGTTCAGTCCATTC | |
| CmUAP-3F1 | ATACGTAGATGACAGTGGAGTC | RACE-PCR |
| CmUAP-3R1 | GCTGTCAACGATACGCTACGTAAC | |
| CmUAP-3F2 | TCAGAAGCCGACAGAACCAAAT | |
| CmUAP-3R2 | GCTACGTAACGGCATGACAGTG | |
| CmUAP-dF | CGGATGGGAAAGTTGGTGTTT | ddPCR |
| CmUAP-dR | TCTTCTGGATTCGCACAATTCT | |
| CmUAP-iF | TGGAAACATCTGCAACCATTACTTC | dsRNA synthesis |
| CmUAP-iR | CGAGAGTAATGGTGAAATCTCAACG | |
| CmUAP-dsF | taatacgactcactatagggTGGAAACATCTGCAACCATTACTTC | |
| CmUAP-dsR | taatacgactcactatagggCGAGAGTAATGGTGAAATCTCAACG | |
| GFP-iF | GCCAACACTTGTCACTACTT | |
| GFP-iR | GGAGTATTTTGTTGATAATGGTCTG | |
| GFP-dsF | taatacgactcactatagggGCCAACACTTGTCACTACTT | |
| GFP-dsR | taatacgactcactatagggGGAGTATTTTGTTGATAATGGTCTG |
| Component | Volume Per Reaction, μL | Final Concentration | Cycling Step | Temperature, °C | Time | Number of Cycles | Other Reaction Conditions |
|---|---|---|---|---|---|---|---|
| 2 × QX200 ddPCR EvaGreen Supermix | 10 | 1× | Enzyme activation | 95 | 5 min | 1 | Ramp rate: 2 °C/s Lid temperature: 105 °C Set the sample volume to 40 μL |
| Forward primer (2 μM) | 1 | 100 nM | Denaturation | 95 | 30 s | 40 | |
| Reverse primer (2 μM) | 1 | 100 nM | Annealing and extension | 60 | 1 min | 40 | |
| Diluted cDNA template | 2 | 100 ng/μL | Signal stabilization | 4 | 5 min | 1 | |
| DNase-free water | 6 | — | 90 | 5 min | 1 | ||
| Total volume | 20 | — | Hold | 4 | Infinity | 1 |
| Injection Treatment | Total Excretion in 7 Days (g) | Weight on Day 7 (g) | Pupal Weight (g) | Pupal Calendar Period (d) | Eclosion Rate (%) | Adult Life Span (d) | Egg Production | Hatching Rate of Eggs (%) |
|---|---|---|---|---|---|---|---|---|
| dsCmUAP | 1.79 ± 0.03 a | 0.152 ± 0.03 a | 0.144 ± 0.08 a | 9.2 ± 0.6 b | 58.7 ± 8.5 a | 9.8 ± 0.4 a | 145.2 ± 12.9 a | 68.6 ± 3 a |
| dsGFP | 2.25 ± 0.17 b | 0.199 ± 0.03 b | 0.202 ± 0.11 b | 8.2 ± 0.6 b | 86 ± 5 b | 11.1 ± 0.8 a | 86.3 ± 18.1 b | 86.2 ± 1.7 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.-J.; Du, J.; Li, S.-W.; Shakeel, M.; Li, J.-J.; Meng, X.-G. Cloning, Characterization, and RNA Interference Effect of the UDP-N-Acetylglucosamine Pyrophosphorylase Gene in Cnaphalocrocis medinalis. Genes 2021, 12, 464. https://doi.org/10.3390/genes12040464
Zhou Y-J, Du J, Li S-W, Shakeel M, Li J-J, Meng X-G. Cloning, Characterization, and RNA Interference Effect of the UDP-N-Acetylglucosamine Pyrophosphorylase Gene in Cnaphalocrocis medinalis. Genes. 2021; 12(4):464. https://doi.org/10.3390/genes12040464
Chicago/Turabian StyleZhou, Yuan-Jin, Juan Du, Shang-Wei Li, Muhammad Shakeel, Jia-Jing Li, and Xiao-Gui Meng. 2021. "Cloning, Characterization, and RNA Interference Effect of the UDP-N-Acetylglucosamine Pyrophosphorylase Gene in Cnaphalocrocis medinalis" Genes 12, no. 4: 464. https://doi.org/10.3390/genes12040464
APA StyleZhou, Y.-J., Du, J., Li, S.-W., Shakeel, M., Li, J.-J., & Meng, X.-G. (2021). Cloning, Characterization, and RNA Interference Effect of the UDP-N-Acetylglucosamine Pyrophosphorylase Gene in Cnaphalocrocis medinalis. Genes, 12(4), 464. https://doi.org/10.3390/genes12040464





