Characterization of the Ghd8 Flowering Time Gene in a Mini-Core Collection of Miscanthus sinensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Genomic DNA Extraction and Isolation of Ghd8 in Miscanthus
2.3. RNA Isolation and Quantitative Reverse Transcription-PCR Analysis
2.4. Data Analysis
3. Results
3.1. Characterization of M. sinensis Ghd8
3.2. Geographical Distribution of Naturally Occurring MsiGhd8 Protein Variants
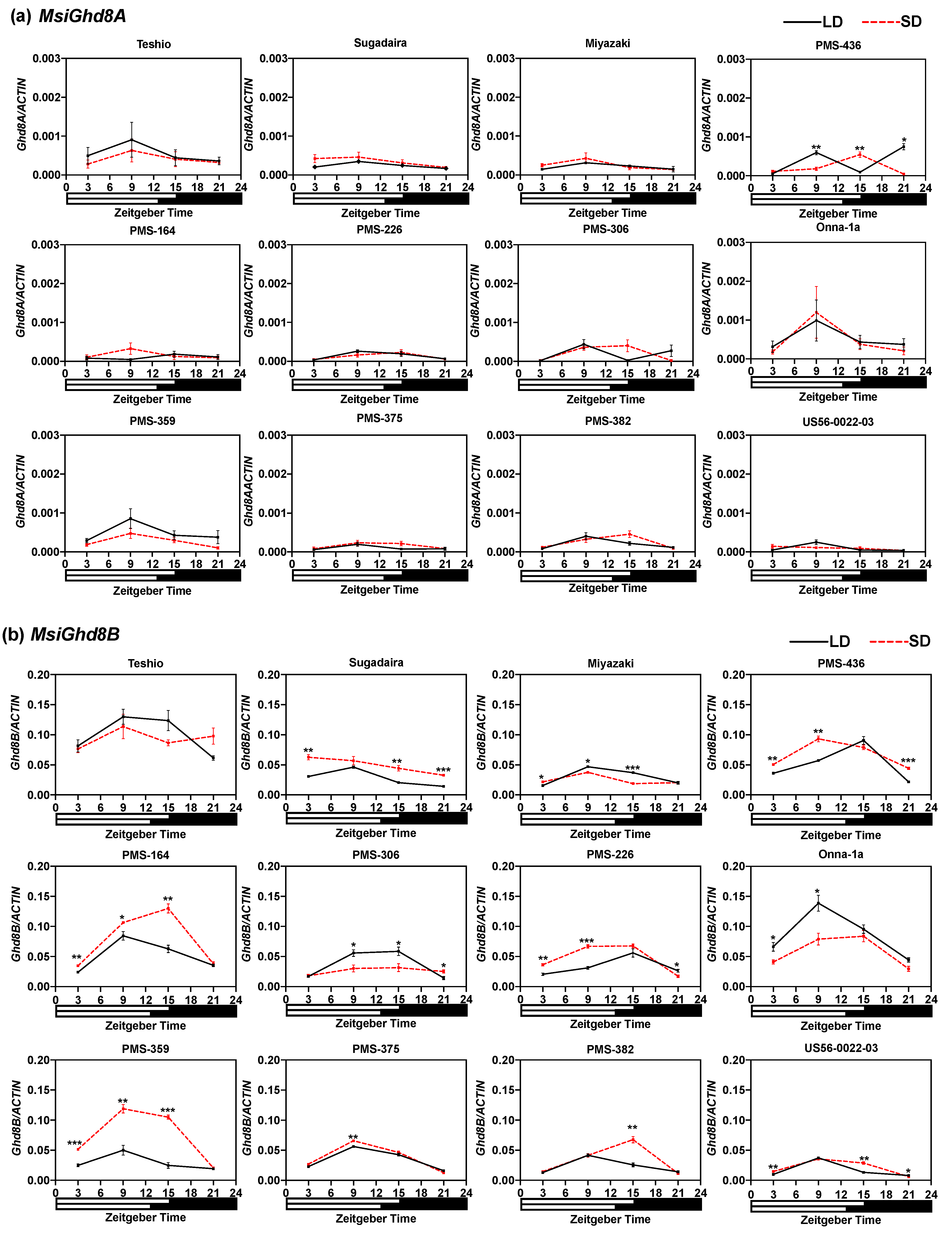
3.3. Expressions Patterns of M. sinensis Ghd8
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hodkinson, T.R.; Klaas, M.; Jones, M.B.; Prickett, R.; Barth, S. Miscanthus: A case study for the utilization of natural genetic variation. Plant Genet. Resour. Charact. Util. 2015, 13, 219–237. [Google Scholar] [CrossRef]
- Greef, J.; Deuter, M. Syntaxonomy of Miscanthus × giganteus GREEF et DEU. Angew. Bot. 1993, 67, 87–90. [Google Scholar]
- Heaton, E.A.; Dohleman, F.G.; Long, S.P. Meeting US biofuel goals with less land: The potential of Miscanthus. Glob. Chang. Biol. 2008, 14, 2000–2014. [Google Scholar] [CrossRef]
- Jensen, E.; Robson, P.; Norris, J.; Cookson, A.; Farrar, K.; Donnison, I.; Clifton-Brown, J. Flowering induction in the bioenergy grass Miscanthus sacchariflorus is a quantitative short-day response, whilst delayed flowering under long days increases biomass accumulation. J. Exp. Bot. 2013, 64, 541–552. [Google Scholar] [CrossRef][Green Version]
- Robson, P.; Jensen, E.; Hawkins, S.; White, S.R.; Kenobi, K.; Clifton-Brown, J.; Donnison, I.; Farrar, K. Accelerating the domestication of a bioenergy crop: Identifying and modelling morphological targets for sustainable yield increase in Miscanthus. J. Exp. Bot. 2013, 64, 4143–4155. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Y.; Kiesel, A.; Wagner, M.; Nunn, C.; Kalinina, O.; Hastings, A.F.S.J.; Clifton-Brown, J.C.; Lewandowski, I. Harvest time optimization for combustion quality of different Miscanthus genotypes across Europe. Front. Plant Sci. 2017, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Clark, L.V.; Jin, X.; Anzoua, K.; Bagmet, L.; Chebukin, P.; Dzyubenko, E.; Dzyubenko, N.; Ghimire, B.K.; Heo, K.; et al. Managing flowering time in Miscanthus and sugarcane to facilitate intra- and intergeneric crosses. PLoS ONE 2021, 16, e0240390. [Google Scholar] [CrossRef]
- Jensen, E.; Farrar, K.; Thomas-Jones, S.; Hastings, A.; Donnison, I.; Clifton-Brown, J. Characterization of flowering time diversity in Miscanthus species. GCB Bioenergy 2011, 3, 387–400. [Google Scholar] [CrossRef]
- Hastings, A.; Mos, M.; Yesufu, J.A.; McCalmont, J.; Schwarz, K.; Shafei, R.; Ashman, C.; Nunn, C.; Schuele, H.; Cosentino, S.; et al. Economic and environmental assessment of seed and rhizome propagated Miscanthus in the UK. Front. Plant Sci. 2017, 8, 1058. [Google Scholar] [CrossRef]
- Clifton-Brown, J.; Harfouche, A.; Casler, M.D.; Dylan Jones, H.; Macalpine, W.J.; Murphy-Bokern, D.; Smart, L.B.; Adler, A.; Ashman, C.; Awty-Carroll, D.; et al. Breeding progress and preparedness for mass-scale deployment of perennial lignocellulosic biomass crops switchgrass, Miscanthus, willow and poplar. GCB Bioenergy 2019, 11, 118–151. [Google Scholar] [CrossRef] [PubMed]
- Deuter, M. Breeding approaches to improvement of yield and quality in Miscanthus grown in Europe. In European Miscanthus Improvement (FAIR3 CT-96-1392) Final Report; Lewandowski, I., Clifton-Brown, J., Eds.; Institute of Crop Production and Grassland Research, University of Hohenheim: Stuttgart, Germany, 2000; pp. 28–52. [Google Scholar]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef]
- Yang, S.; Weers, B.D.; Morishige, D.T.; Mullet, J.E. CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Goretti, D.; Martignago, D.; Landini, M.; Brambilla, V.; Gómez-Ariza, J.; Gnesutta, N.; Galbiati, F.; Collani, S.; Takagi, H.; Terauchi, R.; et al. Transcriptional and post-transcriptional mechanisms limit Heading Date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 2017, 13, e1006530. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyam, R.; Sawad, Y.; Yano, M.; Hirai, M.Y.; et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef]
- Murphy, R.L.; Morishige, D.T.; Brady, J.A.; Rooney, W.L.; Yang, S.; Klein, P.E.; Mullet, J.E. Ghd7 (Ma6) represses sorghum flowering in long days: Ghd7 alleles enhance biomass accumulation and grain production. Plant Genome 2014, 7, 1–10. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Yang, Q.; Li, Z.; Li, W.; Ku, L.; Wang, C.; Ye, J.; Li, K.; Yang, N.; Li, Y.; Zhong, T.; et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 2013, 110, 16969–16974. [Google Scholar] [CrossRef]
- Itoh, H.; Nonoue, Y.; Yano, M.; Izawa, T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010, 42, 635–638. [Google Scholar] [CrossRef]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef]
- Yan, W.; Wang, P.; Chen, H.; Zhou, H.; Li, Q.; Wang, C.; Ding, Z.; Zhang, Y.; Yu, S.; Xing, Y.; et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef]
- Gómez-Ariza, J.; Galbiati, F.; Goretti, D.; Brambilla, V.; Shrestha, R.; Pappolla, A.; Courtois, B.; Fornara, F. Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 2015, 66, 2027–2039. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jin, M.; Zheng, X.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Murphy, R.L.; Morishige, D.T.; Klein, P.E.; Rooney, W.L.; Mullet, J.E. Sorghum Phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS ONE 2014, 9, e105352. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.L.; Klein, R.R.; Morishige, D.T.; Brady, J.A.; Rooney, W.L.; Miller, F.R.; Dugas, D.V.; Klein, P.E.; Mullet, J.E. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA 2011, 108, 16469–16474. [Google Scholar] [CrossRef] [PubMed]
- Casto, A.L.; Mattison, A.J.; Olson, S.N.; Thakran, M.; Rooney, W.L.; Mullet, J.E. Maturity2, a novel regulator of flowering time in Sorghum bicolor, increases expression of SbPRR37 and SbCO in long days delaying flowering. PLoS ONE 2019, 14, e0212154. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Liu, S.; Clark, L.V.; Sharma, S.; Gifford, J.M.; Juvik, J.A.; Lipka, A.E.; Sacks, E.J. Genetic mapping of biomass yield in three interconnected Miscanthus populations. GCB Bioenergy 2018, 10, 165–185. [Google Scholar] [CrossRef]
- Gifford, J.M.; Chae, W.B.; Swaminathan, K.; Moose, S.P.; Juvik, J.A. Mapping the genome of Miscanthus sinensis for QTL associated with biomass productivity. GCB Bioenergy 2015, 7, 797–810. [Google Scholar] [CrossRef]
- Jensen, E.; Shafiei, R.; Ma, X.; Serba, D.D.; Smith, D.P.; Slavov, G.T.; Robson, P.; Farrar, K.; Thomas Jones, S.; Swaller, T.; et al. Linkage mapping evidence for a syntenic QTL associated with flowering time in perennial C4 rhizomatous grasses Miscanthus and switchgrass. GCB Bioenergy 2021, 13, 98–111. [Google Scholar] [CrossRef]
- Chalfun-Junior, A.; Franken, J.; Mes, J.J.; Marsch-Martinez, N.; Pereira, A.; Angenent, G.C. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol. Biol. 2005, 57, 559–575. [Google Scholar] [CrossRef]
- Nagano, H.; Clark, L.V.; Zhao, H.; Peng, J.; Yoo, J.H.; Heo, K.; Yu, C.Y.; Anzoua, K.G.; Matsuo, T.; Sacks, E.J.; et al. Contrasting allelic distribution of CO/Hd1 homologues in Miscanthus sinensis from the East Asian mainland and the Japanese archipelago. J. Exp. Bot. 2015, 66, 4227–4237. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Gong, R.; Yang, Y.; Yu, S. Ghd8 controls rice photoperiod sensitivity by forming a complex that interacts with Ghd7. BMC Plant Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef]
- Cai, X.; Ballif, J.; Endo, S.; Davis, E.; Liang, M.; Chen, D.; Dewald, D.; Kreps, J.; Zhu, T.; Wu, Y. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 2007, 145, 98–105. [Google Scholar] [CrossRef]
- Li, X.; Liu, H.; Wang, M.; Liu, H.; Tian, X.; Zhou, W.; Lü, T.; Wang, Z.; Chu, C.; Fang, J.; et al. Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang Province, northern limit of China. J. Integr. Plant Biol. 2015, 57, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Yamanouchi, U.; Yano, M. Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor. Appl. Genet. 2013, 126, 611–618. [Google Scholar] [CrossRef]
- Gale, M.D.; Devos, K.M. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 1998, 95, 1971–1974. [Google Scholar] [CrossRef]
- Glémin, S.; Bataillon, T. A comparative view of the evolution of grasses under domestication: Tansley review. New Phytol. 2009, 183, 273–290. [Google Scholar] [CrossRef] [PubMed]
- Salse, J.; Abrouk, M.; Bolot, S.; Guilhot, N.; Courcelle, E.; Faraut, T.; Waugh, R.; Close, T.J.; Messing, J.; Feuillet, C. Reconstruction of monocotelydoneous proto-chromosomes reveals faster evolution in plants than in animals. Proc. Natl. Acad. Sci. USA 2009, 106, 14908–14913. [Google Scholar] [CrossRef] [PubMed]
- Mitros, T.; Session, A.M.; James, B.T.; Wu, G.A.; Belaffif, M.B.; Clark, L.V.; Shu, S.; Dong, H.; Barling, A.; Holmes, J.R.; et al. Genome biology of the paleotetraploid perennial biomass crop Miscanthus. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Kim, C.; Zhang, D.; Auckland, S.A.; Rainville, L.K.; Jakob, K.; Kronmiller, B.; Sacks, E.J.; Deuter, M.; Paterson, A.H. SSR-based genetic maps of Miscanthus sinensis and M. sacchariflorus, and their comparison to sorghum. Theor. Appl. Genet. 2012, 124, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, K.; Chae, W.B.; Mitros, T.; Varala, K.; Xie, L.; Barling, A.; Glowacka, K.; Hall, M.; Jezowski, S.; Ming, R.; et al. A framework genetic map for Miscanthus sinensis from RNAseq-based markers shows recent tetraploidy. BMC Genomics 2012, 13, 142. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, T.; Li, B.; Sui, Y.; Chen, J.; Shi, J.; Wing, R.A.; Chen, M. Comparative sequence analysis of the Ghd7 orthologous regions revealed movement of Ghd7 in the grass genomes. PLoS ONE 2012, 7, e50236. [Google Scholar] [CrossRef]
- Ming, R.; Del Monte, T.A.; Hernandez, E.; Moore, P.H.; Irvine, J.E.; Paterson, A.H. Comparative analysis of QTLs affecting plant height and flowering among closely-related diploid and polyploid genomes. Genome 2002, 45, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Tao, D.; Sacks, E.; Fu, B.; Xu, P.; Li, J.; Yang, Y.; McNally, K.; Khush, G.S.; Paterson, A.H.; et al. Convergent evolution of perenniality in rice and sorghum. Proc. Natl. Acad. Sci. USA 2003, 100, 4050–4054. [Google Scholar] [CrossRef] [PubMed]
- Van der Weijde, T.; Kamei, C.L.A.; Severing, E.I.; Torres, A.F.; Gomez, L.D.; Dolstra, O.; Maliepaard, C.A.; McQueen-Mason, S.J.; Visser, R.G.F.; Trindade, L.M. Genetic complexity of Miscanthus cell wall composition and biomass quality for biofuels. BMC Genomics 2017, 18, 406. [Google Scholar] [CrossRef]
- Liu, H.; Liu, H.; Zhou, L.; Zhang, Z.; Zhang, X.; Wang, M.; Li, H.; Lin, Z. Parallel domestication of the heading date 1 gene in cereals. Mol. Biol. Evol. 2015, 32, 2726–2737. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.V.; Brummer, J.E.; Głowacka, K.; Hall, M.C.; Heo, K.; Peng, J.; Yamada, T.; Yoo, J.H.; Yu, C.Y.; Zhao, H.; et al. A footprint of past climate change on the diversity and population structure of Miscanthus sinensis. Ann. Bot. 2014, 114, 97–107. [Google Scholar] [CrossRef]
- Clark, L.V.; Stewart, J.R.; Nishiwaki, A.; Toma, Y.; Kjeldsen, J.B.; Jørgensen, U.; Zhao, H.; Peng, J.; Yoo, J.H.; Heo, K.; et al. Genetic structure of Miscanthus sinensis and Miscanthus sacchariflorus in Japan indicates a gradient of bidirectional but asymmetric introgression. J. Exp. Bot. 2015, 66, 4213–4225. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Dwiyanti, M.S.; Yamada, T.; Sato, M.; Abe, J.; Kitamura, K. Genetic variation of γ-tocopherol methyltransferase gene contributes to elevated α-tocopherol content in soybean seeds. BMC Plant Biol. 2011, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Bookout, A.L.; Mangelsdorf, D.J. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal. 2003, 1, nrs.01012. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Vernon, B., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–166. ISBN 9781483227344. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Lallemand, T.; Leduc, M.; Landès, C.; Rizzon, C.; Lerat, E. An overview of duplicated gene detection methods: Why the duplication mechanism has to be accounted for in their choice. Genes 2020, 11, 1046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, X.; Yan, W.; Zhang, Z.; Lu, L.; Han, Z.; Zhao, H.; Liu, H.; Song, P.; Hu, Y.; et al. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 2015, 208, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.C.; Boe, A.; Lee, D.K. A simple system for promoting flowering of upland switchgrass in the greenhouse. Crop Sci. 2011, 51, 2607–2614. [Google Scholar] [CrossRef]
- McMillan, C. The role of ecotypic variation in the distribution of the central grassland of North America. Ecol. Monogr. 1959, 29, 285–308. [Google Scholar] [CrossRef]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef]
- Ben-Naim, O.; Eshed, R.; Parnis, A.; Teper-Bamnolker, P.; Shalit, A.; Coupland, G.; Samach, A.; Lifschitz, E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006, 46, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Ren, D.; Huang, M.; Sun, K.; Feng, J.; Zhao, J.; Xiao, D.; Xie, W.; Liu, S.; Zhang, H.; et al. Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading. New Phytol. 2021, 229, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Du, A.; Tian, W.; Wei, M.; Yan, W.; He, H.; Zhou, D.; Huang, X.; Li, S.; Ouyang, X. The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol. Plant 2017, 10, 948–961. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, H.; Qi, F.; Zhang, Z.; Li, Q.; Han, Z.; Xing, Y. Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice. Rice 2019, 12, 48. [Google Scholar] [CrossRef]
- Liu, J.; Gong, J.; Wei, X.; Yang, S.; Huang, X.; Li, C.; Zhou, X. Dominance complementation of Hd1 and Ghd8 contributes to extremely late flowering in two rice hybrids. Mol. Breed. 2020, 40, 1–10. [Google Scholar] [CrossRef]
- Nemoto, Y.; Nonoue, Y.; Yano, M.; Izawa, T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 2016, 86, 221–233. [Google Scholar] [CrossRef]
- Wang, P.; Xiong, Y.; Gong, R.; Yang, Y.; Fan, K.; Yu, S. A key variant in the cis-regulatory element of flowering gene Ghd8 associated with cold tolerance in rice. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Adachi, S.; Yoshikawa, K.; Yamanouchi, U.; Tanabata, T.; Sun, J.; Ookawa, T.; Yamamoto, T.; Sage, R.F.; Hirasawa, T.; Yonemaru, J. Fine mapping of Carbon assimilation rate 8, a quantitative trait locus for flag leaf nitrogen content, stomatal conductance and photosynthesis in rice. Front. Plant Sci. 2017, 8, 60. [Google Scholar] [CrossRef] [PubMed]
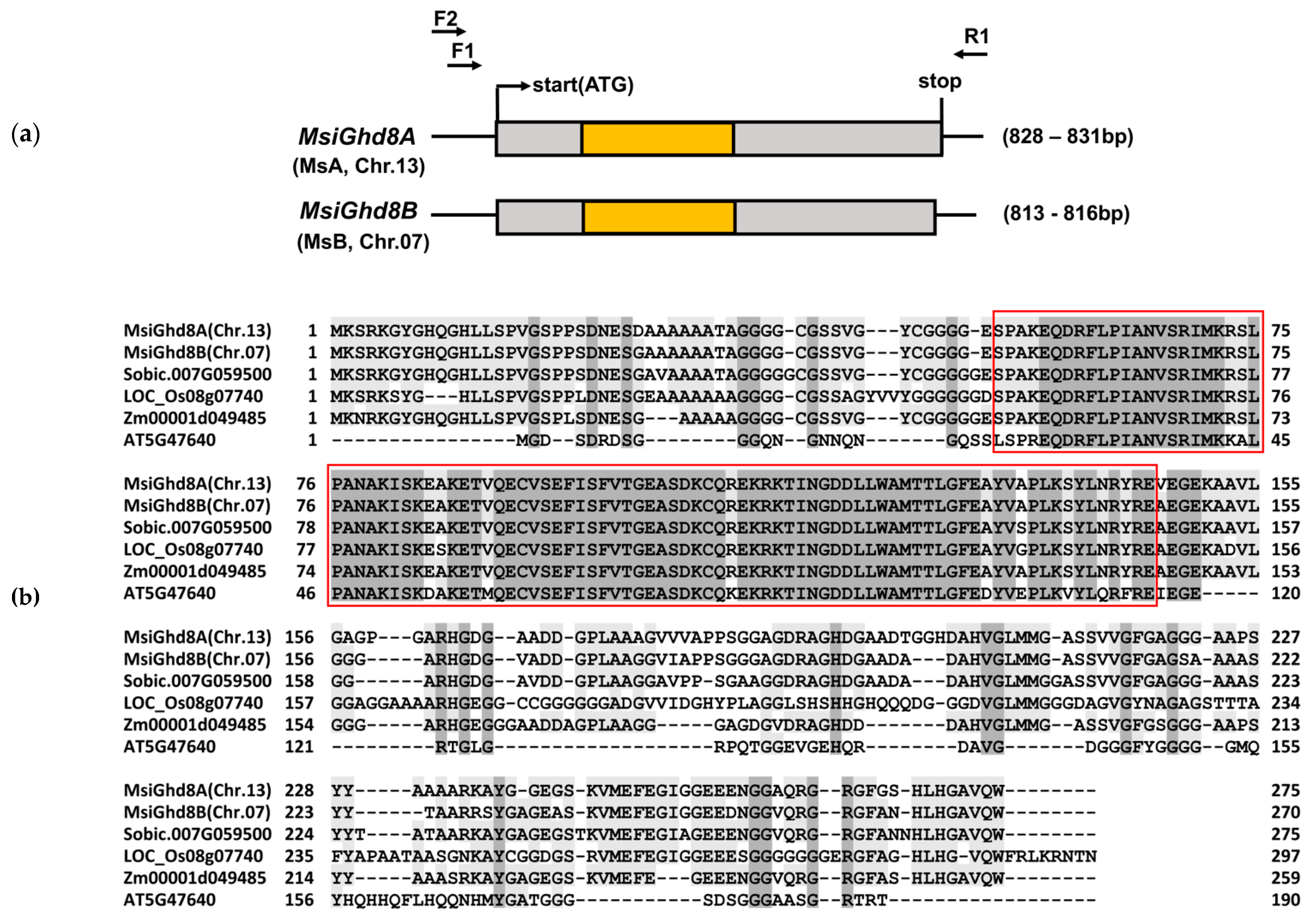
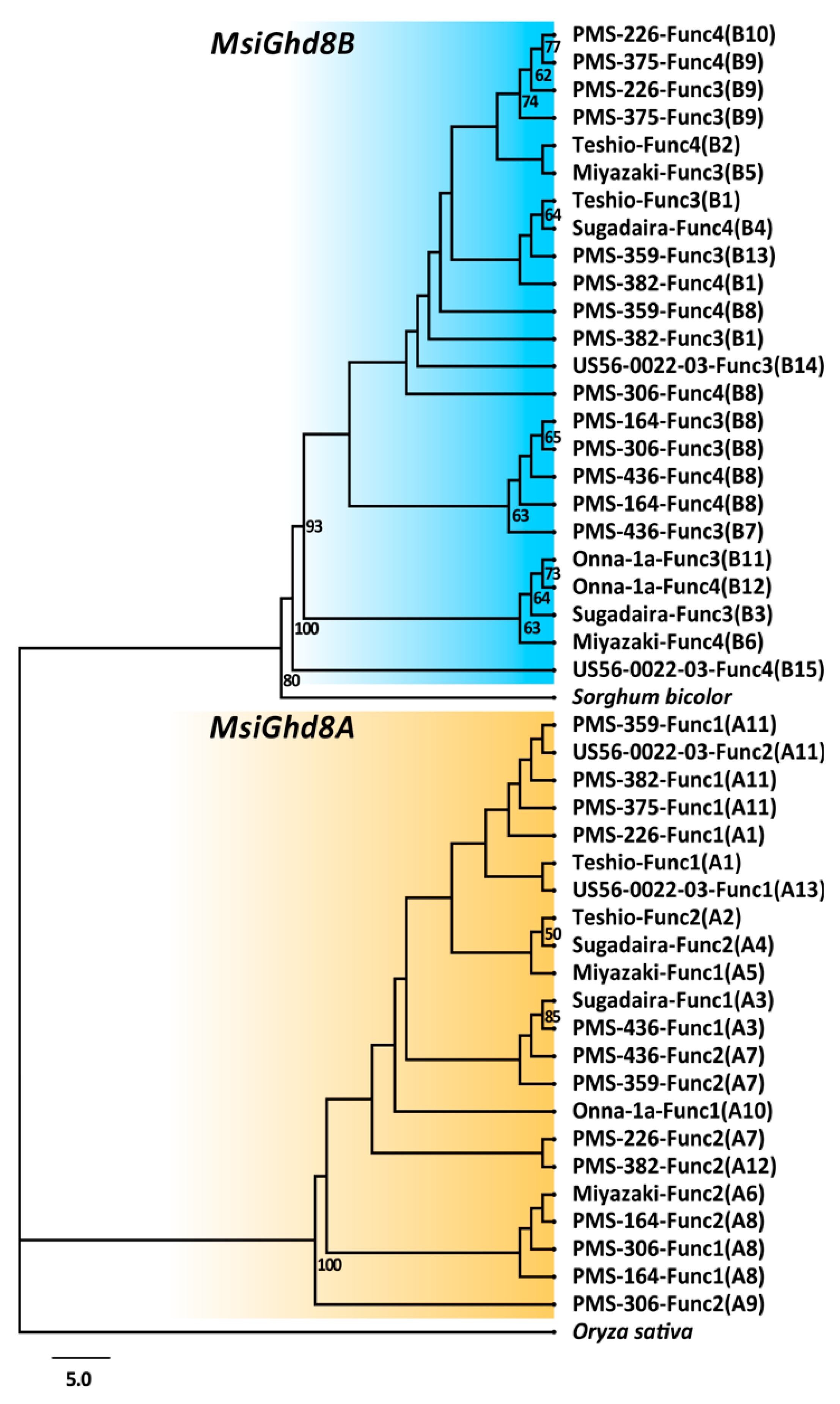
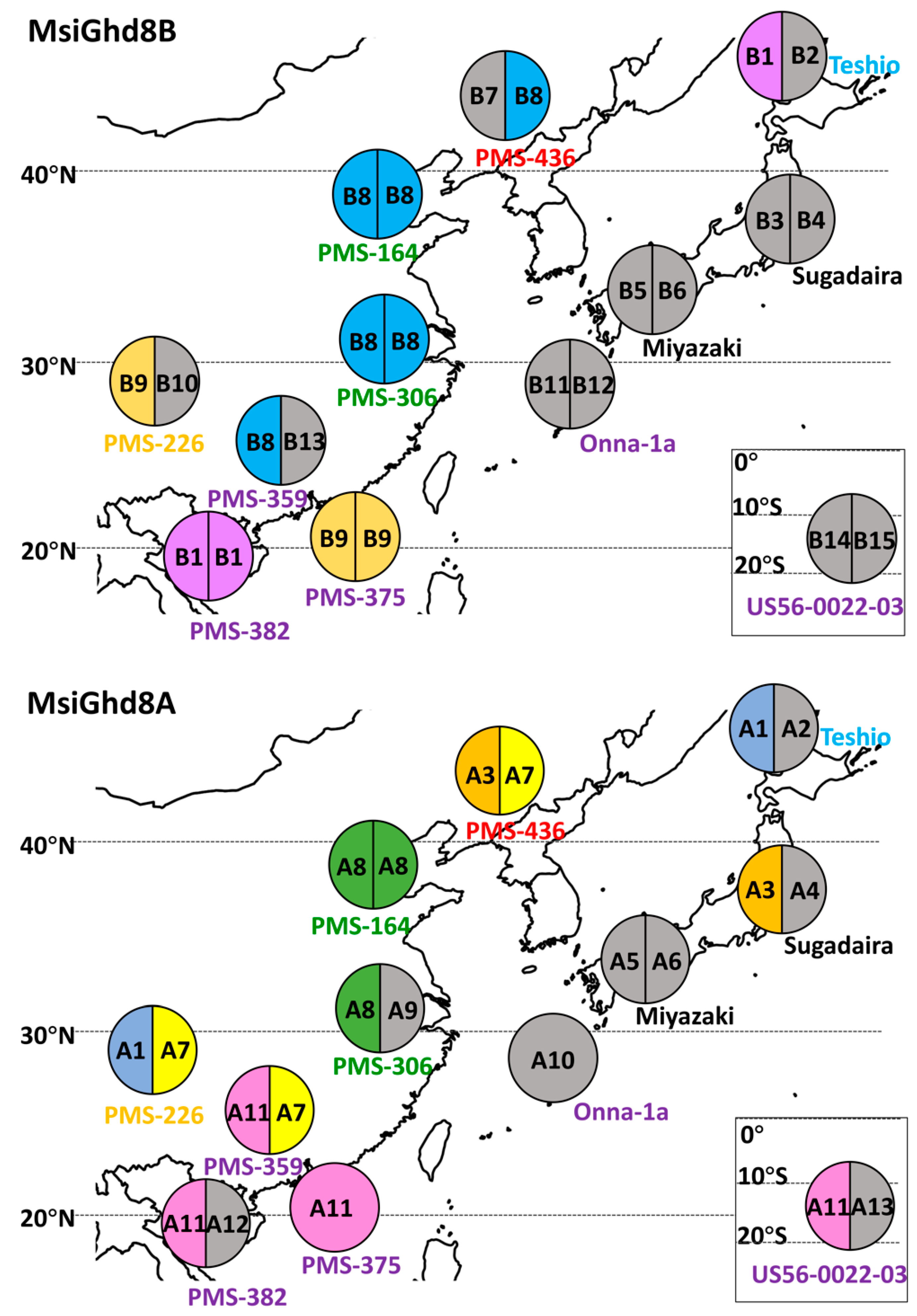

| Genotypes | Ploidy | Lat | Long | Genetic Group † | Genetic Group Color Code † | Days to First Flowering ‡ | Variant Types Classified by Predicted Protein in Ghd8 Homoeologs | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12.5 h | 15 h | MsiGhd8A | MsiGhd8B | ||||||||
| M. sinensis “Teshio” | 2x | 44.9 | 141.9 | Northern Japan | Blue | 66 | A1 | A2 | B1 | B2 | |
| M. sinensis “Sugadaira” | 2x | 36.0 | 138.1 | Southern Japan | Yellow | 96 | A3 | A4 | B3 | B4 | |
| M. sinensis “Miyazaki” | 2x | 31.8 | 131.4 | Southern Japan | Yellow | 61 | 167 | A5 | A6 | B5 | B6 |
| M. sinensis “PMS-436” | 2x | 41.3 | 123.7 | Korea/North China | Red | 115 | A3 | A7 | B7 | B8 | |
| M. sinensis “PMS-164” | 2x | 37.3 | 114.3 | Yangtze-Qinling | Green | 130 | A8 | A8 | B8 | B8 | |
| M. sinensis “PMS-306” | 2x | 29.9 | 118.8 | Yangtze-Qinling | Green | 84 | 173 | A8 | A9 | B8 | B8 |
| M. sinensis “PMS-226” | 2x | 26.6 | 106.8 | Sichuan Basin | Orange | 76 | 189 | A1 | A7 | B9 | B10 |
| M. sinensis “Onna-1a” | 2x | 26.5 | 126.8 | SE China plus tropical | Purple | 274 | A10 | B11 | B12 | ||
| M. sinensis “PMS-359” | 2x | 22.9 | 112.3 | SE China plus tropical | Purple | 81 | 179 | A11 | A7 | B8 | B13 |
| M. sinensis “PMS-375” | 2x | 19.6 | 110.3 | SE China plus tropical | Purple | 142 | A11 | B9 | B9 | ||
| M. sinensis “PMS-382” | 2x | 18.9 | 109.5 | SE China plus tropical | Purple | 184 | A11 | A12 | B1 | B1 | |
| M. floridulus “US56-0022-03” | 2x | −20.9 | 165.3 | SE China plus tropical | Purple | 114 | A11 | A13 | B14 | B15 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.; Xu, M.; Nagano, H.; Clark, L.V.; Sacks, E.J.; Yamada, T. Characterization of the Ghd8 Flowering Time Gene in a Mini-Core Collection of Miscanthus sinensis. Genes 2021, 12, 288. https://doi.org/10.3390/genes12020288
Guo Z, Xu M, Nagano H, Clark LV, Sacks EJ, Yamada T. Characterization of the Ghd8 Flowering Time Gene in a Mini-Core Collection of Miscanthus sinensis. Genes. 2021; 12(2):288. https://doi.org/10.3390/genes12020288
Chicago/Turabian StyleGuo, Zhihui, Meilan Xu, Hironori Nagano, Lindsay V. Clark, Erik J. Sacks, and Toshihiko Yamada. 2021. "Characterization of the Ghd8 Flowering Time Gene in a Mini-Core Collection of Miscanthus sinensis" Genes 12, no. 2: 288. https://doi.org/10.3390/genes12020288
APA StyleGuo, Z., Xu, M., Nagano, H., Clark, L. V., Sacks, E. J., & Yamada, T. (2021). Characterization of the Ghd8 Flowering Time Gene in a Mini-Core Collection of Miscanthus sinensis. Genes, 12(2), 288. https://doi.org/10.3390/genes12020288







