The Transcriptional Network in the Arabidopsis Circadian Clock System
Abstract
1. Introduction
2. Transcriptional Networks under Clock Regulation
2.1. The Transcriptional Network of CCA1 and LHY
2.2. The Transcriptional Network of LNK1 and RVE8
2.3. The Transcriptional Network of PRR9, PRR7, PRR5 and TOC1
2.4. The Transcriptional Network of ELF3, ELF4 and LUX
2.5. The Transcriptional Network of GI
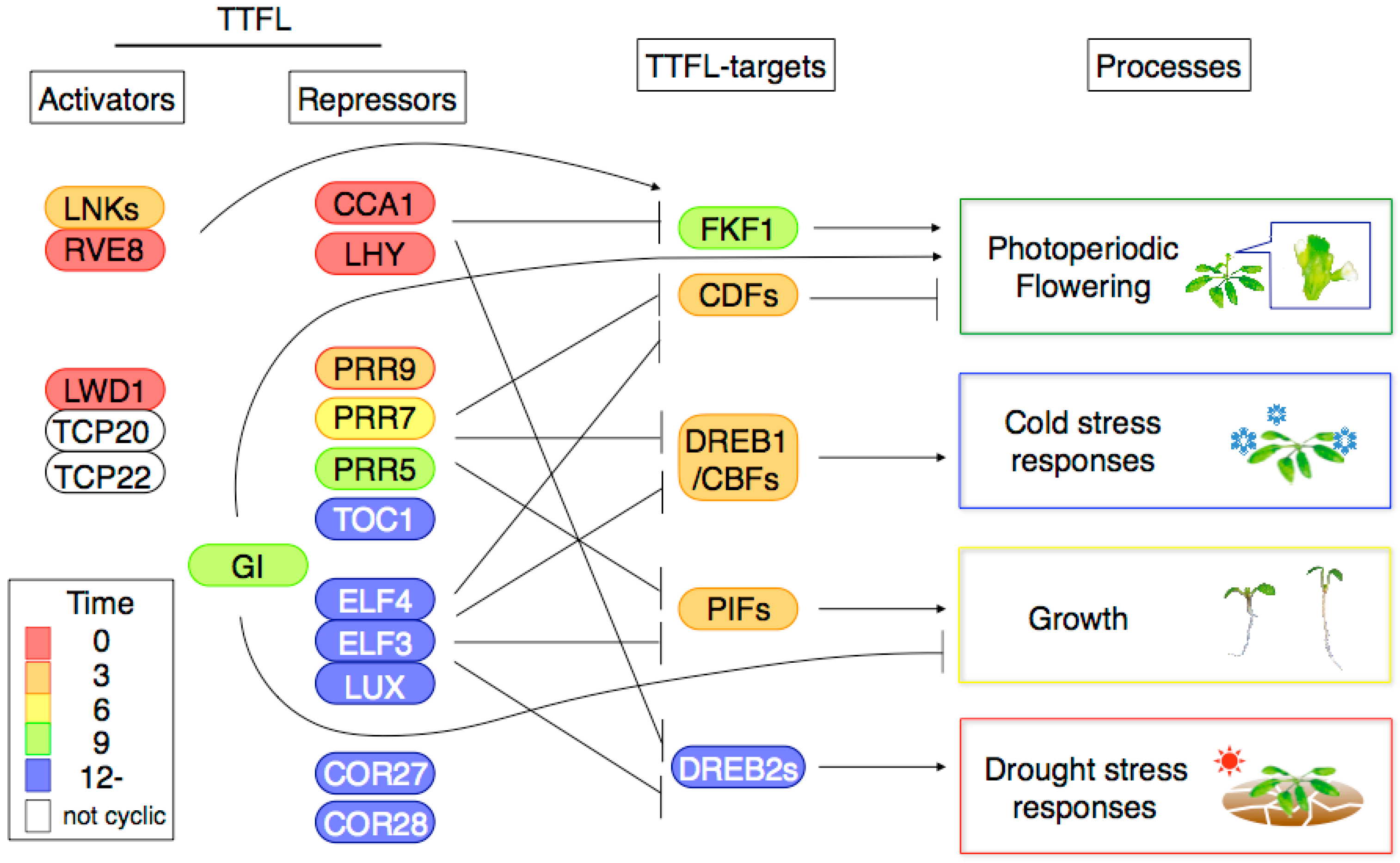
2.6. Possible Interactions among Clock Transcription Factors for Regulating Gene Expression
3. An Evolutionary View of Clock-Dependent Transcriptional Networks in Plants
3.1. The Transcriptional Network under Clock Control in Angiosperms
3.2. The Transcriptional Network under Clock Control in the Plant Lineage
4. Conclusions
Funding
Conflicts of Interest
References
- McClung, C.R. The Plant Circadian Oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef]
- Barak, S.; Tobin, E.M.; Andronis, C.; Sugano, S.; Green, R.M. All in good time: The Arabidopsis circadian clock. Trends Plant Sci. 2000, 5, 517–522. [Google Scholar] [CrossRef]
- McClung, C.R. Plant circadian rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef]
- Kim, H.Y.; Cote, G.G.; Crain, R.C. Potassium Channels in Samanea saman Protoplasts Controlled by Phytochrome and the Biological Clock. Science 1993, 260, 960–962. [Google Scholar] [CrossRef]
- Niinuma, K.; Someya, N.; Kimura, M.; Yamaguchi, I.; Hamamoto, H. Circadian rhythm of circumnutation in inflorescence stems of Arabidopsis. Plant Cell Physiol. 2005, 46, 1423–1427. [Google Scholar] [CrossRef]
- Salome, P.A.; Michael, T.P.; Kearns, E.V.; Fett-Neto, A.G.; Sharrock, R.A.; McClung, C.R. The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 2002, 129, 1674–1685. [Google Scholar] [CrossRef]
- Wang, W.; Barnaby, J.Y.; Tada, Y.; Li, H.; Tor, M.; Caldelari, D.; Lee, D.U.; Fu, X.D.; Dong, X. Timing of plant immune responses by a central circadian regulator. Nature 2011, 470, 110–114. [Google Scholar] [CrossRef]
- Goodspeed, D.; Chehab, E.W.; Min-Venditti, A.; Braam, J.; Covington, M.F. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 2012, 109, 4674–4677. [Google Scholar] [CrossRef]
- Graf, A.; Schlereth, A.; Stitt, M.; Smith, A.M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 2010, 107, 9458–9463. [Google Scholar] [CrossRef]
- Webb, A.A.R.; Seki, M.; Satake, A.; Caldana, C. Continuous dynamic adjustment of the plant circadian oscillator. Nat. Commun. 2019, 10, 550. [Google Scholar] [CrossRef]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kevei, E.; Toth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef]
- Green, R.M.; Tingay, S.; Wang, Z.Y.; Tobin, E.M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002, 129, 576–584. [Google Scholar] [CrossRef]
- Yerushalmi, S.; Yakir, E.; Green, R.M. Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 2011, 20, 1155–1165. [Google Scholar] [CrossRef]
- Rubin, M.J.; Brock, M.T.; Davis, A.M.; German, Z.M.; Knapp, M.; Welch, S.M.; Harmer, S.L.; Maloof, J.N.; Davis, S.J.; Weinig, C. Circadian rhythms vary over the growing season and correlate with fitness components. Mol. Ecol. 2017, 26, 5528–5540. [Google Scholar] [CrossRef]
- Rubin, M.J.; Brock, M.T.; Baker, R.L.; Wilcox, S.; Anderson, K.; Davis, S.J.; Weinig, C. Circadian rhythms are associated with shoot architecture in natural settings. New Phytol. 2018, 219, 246–258. [Google Scholar] [CrossRef]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef]
- Edwards, K.D.; Anderson, P.E.; Hall, A.; Salathia, N.S.; Locke, J.C.; Lynn, J.R.; Straume, M.; Smith, J.Q.; Millar, A.J. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 2006, 18, 639–650. [Google Scholar] [CrossRef]
- Blasing, O.E.; Gibon, Y.; Gunther, M.; Hohne, M.; Morcuende, R.; Osuna, D.; Thimm, O.; Usadel, B.; Scheible, W.R.; Stitt, M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 2005, 17, 3257–3281. [Google Scholar] [CrossRef]
- Michael, T.P.; Mockler, T.C.; Breton, G.; McEntee, C.; Byer, A.; Trout, J.D.; Hazen, S.P.; Shen, R.; Priest, H.D.; Sullivan, C.M.; et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008, 4, e14. [Google Scholar] [CrossRef]
- Covington, M.F.; Harmer, S.L. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007, 5, e222. [Google Scholar] [CrossRef]
- Fowler, S.G.; Cook, D.; Thomashow, M.F. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005, 137, 961–968. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef]
- Kidokoro, S.; Yoneda, K.; Takasaki, H.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Different Cold-Signaling Pathways Function in the Responses to Rapid and Gradual Decreases in Temperature. Plant Cell 2017, 29, 760–774. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyama, T.; Kinoshita, T.; Nakamichi, N. Direct Repression of Evening Genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis Circadian Clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.H.; Doherty, C.J.; Pruneda-Paz, J.L.; Schmitz, R.J.; Ecker, J.R.; Kay, S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E4802–E4810. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Manfield, I.; Stockley, P.; Carre, I.A. Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLoS ONE 2015, 10, e0143943. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carre, I.A. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018, 220, 893–907. [Google Scholar] [CrossRef]
- Li, G.; Siddiqui, H.; Teng, Y.; Lin, R.; Wan, X.Y.; Li, J.; Lau, O.S.; Ouyang, X.; Dai, M.; Wan, J.; et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 2011, 13, 616–622. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farre, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013, 76, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Newton, L.; Liu, M.J.; Shiu, S.H.; Farre, E.M. A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 2016, 170, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Perez-Garcia, P.; Pokhilko, A.; Millar, A.J.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 2012, 336, 75–79. [Google Scholar] [CrossRef]
- Dixon, L.E.; Knox, K.; Kozma-Bognar, L.; Southern, M.M.; Pokhilko, A.; Millar, A.J. Temporal Repression of Core Circadian Genes Is Mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011, 21, 120–125. [Google Scholar] [CrossRef]
- Helfer, A.; Nusinow, D.A.; Chow, B.Y.; Gehrke, A.R.; Bulyk, M.L.; Kay, S.A. LUX ARRHYTHMO Encodes a Nighttime Repressor of Circadian Gene Expression in the Arabidopsis Core Clock. Curr. Biol. 2011, 21, 126–133. [Google Scholar] [CrossRef]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farre, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef]
- Ma, Y.; Gil, S.; Grasser, K.D.; Mas, P. Targeted Recruitment of the Basal Transcriptional Machinery by LNK Clock Components Controls the Circadian Rhythms of Nascent RNAs in Arabidopsis. Plant Cell 2018, 30, 907–924. [Google Scholar] [CrossRef]
- Li, X.; Ma, D.; Lu, S.X.; Hu, X.; Huang, R.; Liang, T.; Xu, T.; Tobin, E.M.; Liu, H. Blue Light- and Low Temperature-Regulated COR27 and COR28 Play Roles in the Arabidopsis Circadian Clock. Plant Cell 2016, 28, 2755–2769. [Google Scholar] [CrossRef]
- Wu, J.F.; Tsai, H.L.; Joanito, I.; Wu, Y.C.; Chang, C.W.; Li, Y.H.; Wang, Y.; Hong, J.C.; Chu, J.W.; Hsu, C.P.; et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181. [Google Scholar] [CrossRef] [PubMed]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009, 323, 1481–1485. [Google Scholar] [CrossRef] [PubMed]
- Flis, A.; Sulpice, R.; Seaton, D.D.; Ivakov, A.A.; Liput, M.; Abel, C.; Millar, A.J.; Stitt, M. Photoperiod-dependent changes in the phase of core clock transcripts and global transcriptional outputs at dawn and dusk in Arabidopsis. Plant Cell Environ. 2016, 39, 1955–1981. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.J. The Intracellular Dynamics of Circadian Clocks Reach for the Light of Ecology and Evolution. Annu. Rev. Plant Biol. 2016, 67, 595–618. [Google Scholar] [CrossRef]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Tobin, E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar] [CrossRef]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carre, I.A.; Coupland, G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93, 1219–1229. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.R.; Carre, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar] [CrossRef]
- Lai, A.G.; Doherty, C.J.; Mueller-Roeber, B.; Kay, S.A.; Schippers, J.H.; Dijkwel, P.P. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 2012, 109, 17129–17134. [Google Scholar] [CrossRef]
- Alabadi, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Mas, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Hsu, P.Y.; Devisetty, U.K.; Harmer, S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife 2013, 2, e00473. [Google Scholar] [CrossRef] [PubMed]
- Rugnone, M.L.; Faigon Soverna, A.; Sanchez, S.E.; Schlaen, R.G.; Hernando, C.E.; Seymour, D.K.; Mancini, E.; Chernomoretz, A.; Weigel, D.; Mas, P.; et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 2013, 110, 12120–12125. [Google Scholar] [CrossRef]
- Xie, Q.; Wang, P.; Liu, X.; Yuan, L.; Wang, L.; Zhang, C.; Li, Y.; Xing, H.; Zhi, L.; Yue, Z.; et al. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 2014, 26, 2843–2857. [Google Scholar] [CrossRef]
- De Leone, M.J.; Hernando, C.E.; Romanowski, A.; Garcia-Hourquet, M.; Careno, D.; Casal, J.; Rugnone, M.; Mora-Garcia, S.; Yanovsky, M.J. The LNK Gene Family: At the Crossroad between Light Signaling and the Circadian Clock. Genes 2019, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Shalit-Kaneh, A.; Kumimoto, R.W.; Filkov, V.; Harmer, S.L. Multiple feedback loops of the Arabidopsis circadian clock provide rhythmic robustness across environmental conditions. Proc. Natl. Acad. Sci. USA 2018, 115, 7147–7152. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef]
- Mas, P.; Kim, W.Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Ito, S.; Yamashino, T.; Mizuno, T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 686–698. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Sato, E.; Shimizu, T.; Nakamich, N.; Sato, S.; Kato, T.; Tabata, S.; Nagatani, A.; Yamashino, T.; Mizuno, T. Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol. 2003, 44, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Niwa, Y.; Nakamichi, N.; Kawamura, H.; Yamashino, T.; Mizuno, T. Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Murakami, M.; Nakamura, Y.; Ito, S.; Nakamichi, N.; Yamashino, T.; Mizuno, T. Mutants of circadian-associated PRR genes display a novel and visible phenotype as to light responses during de-etiolation of Arabidopsis thaliana seedlings. Biosci. Biotechnol. Biochem. 2007, 71, 834–839. [Google Scholar] [CrossRef]
- Fukushima, A.; Kusano, M.; Nakamichi, N.; Kobayashi, M.; Hayashi, N.; Sakakibara, H.; Mizuno, T.; Saito, K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. USA 2009, 106, 7251–7256. [Google Scholar] [CrossRef] [PubMed]
- Yamashino, T.; Ito, S.; Niwa, Y.; Kunihiro, A.; Nakamichi, N.; Mizuno, T. Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol. 2008, 49, 1839–1850. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.E.; Webb, A.A.; Pearson, M.; Kay, S.A. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 1998, 125, 485–494. [Google Scholar] [PubMed]
- Yanovsky, M.J.; Kay, S.A. Molecular basis of seasonal time measurement in Arabidopsis. Nature 2002, 419, 308–312. [Google Scholar] [CrossRef]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.L.; Maloof, J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, Y.; He, Y.; Wang, Y.; Wang, L. Pseudo Response Regulators Regulate Photoperiodic Hypocotyl Growth by Repressing PIF4/5 Transcription. Plant Physiol. 2020, 183, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef]
- Wang, F.; Han, T.; Song, Q.; Ye, W.; Song, X.; Chu, J.; Li, J.; Chen, Z.J. Rice Circadian Clock Regulates Tiller Growth and Panicle Development Through Strigolactone Signaling and Sugar Sensing. Plant Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Matsushika, A.; Kojima, M.; Yamashino, T.; Mizuno, T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002, 43, 58–69. [Google Scholar] [CrossRef]
- Para, A.; Farre, E.M.; Imaizumi, T.; Pruneda-Paz, J.L.; Harmon, F.G.; Kay, S.A. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 2007, 19, 3462–3473. [Google Scholar] [CrossRef]
- Fujiwara, S.; Wang, L.; Han, L.; Suh, S.S.; Salome, P.A.; McClung, C.R.; Somers, D.E. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008, 283, 23073–23083. [Google Scholar] [CrossRef]
- Hayama, R.; Sarid-Krebs, L.; Richter, R.; Fernandez, V.; Jang, S.; Coupland, G. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 2017, 36, 904–918. [Google Scholar] [CrossRef]
- Martin, G.; Rovira, A.; Veciana, N.; Soy, J.; Toledo-Ortiz, G.; Gommers, C.M.M.; Boix, M.; Henriques, R.; Minguet, E.G.; Alabadi, D.; et al. Circadian Waves of Transcriptional Repression Shape PIF-Regulated Photoperiod-Responsive Growth in Arabidopsis. Curr. Biol. 2018, 28, 311–318. [Google Scholar] [CrossRef]
- Zhang, Y.; Pfeiffer, A.; Tepperman, J.M.; Dalton-Roesler, J.; Leivar, P.; Grandio, E.G.; Quail, P.H. Central clock components modulate plant shade avoidance by directly repressing transcriptional activation activity of PIF proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 3261–3269. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Niinuma, K.; Ito, S.; Yamashino, T.; Mizoguchi, T.; Mizuno, T. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007, 48, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.R.; Davis, S.J.; Bastow, R.M.; McWatters, H.G.; Kozma-Bognar, L.; Nagy, F.; Millar, A.J.; Amasino, R.M. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002, 419, 74–77. [Google Scholar] [CrossRef]
- Kolmos, E.; Nowak, M.; Werner, M.; Fischer, K.; Schwarz, G.; Mathews, S.; Schoof, H.; Nagy, F.; Bujnicki, J.M.; Davis, S.J. Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 2009, 3, 350–366. [Google Scholar] [CrossRef]
- Zagotta, M.T.; Hicks, K.A.; Jacobs, C.I.; Young, J.C.; Hangarter, R.P.; Meeks-Wagner, D.R. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996, 10, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Panda, S.; Liu, X.L.; Strayer, C.A.; Wagner, D.R.; Kay, S.A. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 2001, 13, 1305–1315. [Google Scholar] [CrossRef]
- Liu, X.L.; Covington, M.F.; Fankhauser, C.; Chory, J.; Wagner, D.R. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 2001, 13, 1293–1304. [Google Scholar]
- Hazen, S.P.; Schultz, T.F.; Pruneda-Paz, J.L.; Borevitz, J.O.; Ecker, J.R.; Kay, S.A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 2005, 102, 10387–10392. [Google Scholar] [CrossRef]
- Onai, K.; Ishiura, M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 2005, 10, 963–972. [Google Scholar] [CrossRef]
- Ezer, D.; Jung, J.H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stockle, D.; et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Lee, K.; Ezer, D.; Cortijo, S.; Jung, J.; Charoensawan, V.; Box, M.S.; Jaeger, K.; Takahashi, N.; Mas, P.; et al. The Evening Complex establishes repressive chromatin domains via H2A.Z deposition. Plant Physiol. 2020, 182, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Kitayama, M.; Oka, H.; Tsubouchi, M.; Takayama, C.; Nomoto, Y.; Yamashino, T. The EC night-time repressor plays a crucial role in modulating circadian clock transcriptional circuitry by conservatively double-checking both warm-night and night-time-light signals in a synergistic manner in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 2139–2151. [Google Scholar] [CrossRef]
- Thines, B.; Harmon, F.G. Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2010, 107, 3257–3262. [Google Scholar] [CrossRef]
- Silva, C.S.; Nayak, A.; Lai, X.; Hutin, S.; Hugouvieux, V.; Jung, J.H.; Lopez-Vidriero, I.; Franco-Zorrilla, J.M.; Panigrahi, K.C.S.; Nanao, M.H.; et al. Molecular mechanisms of Evening Complex activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 6901–6909. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Takahashi, N.; Hirata, Y.; Ronald, J.; Porco, S.; Davis, S.J.; Nusinow, D.A.; Kay, S.A.; Mas, P. A mobile ELF4 delivers circadian temperature information from shoots to roots. Nat. Plants 2020, 6, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Park, D.H.; Somers, D.E.; Kim, Y.S.; Choy, Y.H.; Lim, H.K.; Soh, M.S.; Kim, H.J.; Kay, S.A.; Nam, H.G. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 1999, 285, 1579–1582. [Google Scholar] [CrossRef]
- Cha, J.Y.; Kim, J.; Kim, T.S.; Zeng, Q.; Wang, L.; Lee, S.Y.; Kim, W.Y.; Somers, D.E. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Kiba, T.; Henriques, R.; Sakakibara, H.; Chua, N.H. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 2007, 19, 2516–2530. [Google Scholar] [CrossRef]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef]
- Nohales, M.A.; Liu, W.; Duffy, T.; Nozue, K.; Sawa, M.; Pruneda-Paz, J.L.; Maloof, J.N.; Jacobsen, S.E.; Kay, S.A. Multi-level Modulation of Light Signaling by GIGANTEA Regulates Both the Output and Pace of the Circadian Clock. Dev. Cell 2019, 49, 840–851.e8. [Google Scholar] [CrossRef]
- Mishra, P.; Panigrahi, K.C. GIGANTEA—an emerging story. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Kay, S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, Y.H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef]
- Tseng, T.S.; Salome, P.A.; McClung, C.R.; Olszewski, N.E. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 2004, 16, 1550–1563. [Google Scholar] [CrossRef]
- Mockler, T.C.; Michael, T.P.; Priest, H.D.; Shen, R.; Sullivan, C.M.; Givan, S.A.; McEntee, C.; Kay, S.A.; Chory, J. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Breton, G.; Priest, H.D.; Dharmawardhana, P.; Jaiswal, P.; Fox, S.E.; Michael, T.P.; Chory, J.; Kay, S.A.; Mockler, T.C. Global Profiling of Rice and Poplar Transcriptomes Highlights Key Conserved Circadian-Controlled Pathways and cis-Regulatory Modules. PLoS ONE 2011, 6, e16907. [Google Scholar] [CrossRef]
- Toda, Y.; Kudo, T.; Kinoshita, T.; Nakamichi, N. Evolutionary Insight into the Clock-Associated PRR5 Transcriptional Network of Flowering Plants. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Oikawa, T.; Ishimaru, Y.; Munemasa, S.; Takeuchi, Y.; Washiyama, K.; Hamamoto, S.; Yoshikawa, N.; Mutara, Y.; Uozumi, N.; Ueda, M. Ion Channels Regulate Nyctinastic Leaf Opening in Samanea saman. Curr. Biol. 2018, 28, 2230–2238. [Google Scholar] [CrossRef]
- Atamian, H.S.; Creux, N.M.; Brown, E.A.; Garner, A.G.; Blackman, B.K.; Harmer, S.L. Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science 2016, 353, 587–590. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, G.; Kim, S.; Thi, T.N.; Kim, H.; Jeong, J.; Kim, J.; Kim, J.; Choi, G.; Oh, E. The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Shimizu, H.; Katayama, K.; Koto, T.; Torii, K.; Araki, T.; Endo, M. Decentralized circadian clocks process thermal and photoperiodic cues in specific tissues. Nat. Plants 2015, 1, 15163. [Google Scholar] [CrossRef]
- Endo, M.; Shimizu, H.; Nohales, M.A.; Araki, T.; Kay, S.A. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 2014, 515, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Hirata, Y.; Aihara, K.; Mas, P. A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell 2015, 163, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Gould, P.D.; Domijan, M.; Greenwood, M.; Tokuda, I.T.; Rees, H.; Kozma-Bognar, L.; Hall, A.J.; Locke, J.C. Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Elife 2018, 7. [Google Scholar] [CrossRef]
- Muranaka, T.; Oyama, T. Heterogeneity of cellular circadian clocks in intact plants and its correction under light-dark cycles. Sci. Adv. 2016, 2, e1600500. [Google Scholar] [CrossRef]
- Fukuda, H.; Nakamichi, N.; Hisatsune, M.; Murase, H.; Mizuno, T. Synchronization of plant circadian oscillators with a phase delay effect of the vein network. Phys. Rev. Lett. 2007, 99, 098102. [Google Scholar] [CrossRef]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyama, R.; Sawada, Y.; Yano, M.; Hirai, M.Y.; et al. Os-GIGANTEA Confers Robust Diurnal Rhythms on the Global Transcriptome of Rice in the Field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef]
- Nakamichi, N. Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol. 2015, 56, 594–604. [Google Scholar] [CrossRef]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian Clock Genes Universally Control Key Agricultural Traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef]
- Li, M.W.; Liu, W.; Lam, H.M.; Gendron, J.M. Characterization of Two Growth Period QTLs Reveals Modification of PRR3 Genes During Soybean Domestication. Plant Cell Physiol. 2019, 60, 407–420. [Google Scholar] [CrossRef]
- Ferrari, C.; Proost, S.; Janowski, M.; Becker, J.; Nikoloski, Z.; Bhattacharya, D.; Price, D.; Tohge, T.; Bar-Even, A.; Fernie, A.; et al. Kingdom-wide comparison reveals the evolution of diurnal gene expression in Archaeplastida. Nat. Commun. 2019, 10, 737. [Google Scholar] [CrossRef] [PubMed]
- Linde, A.M.; Eklund, D.M.; Kubota, A.; Pederson, E.R.A.; Holm, K.; Gyllenstrand, N.; Nishihama, R.; Cronberg, N.; Muranaka, T.; Oyama, T.; et al. Early evolution of the land plant circadian clock. New Phytol. 2017, 216, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Kubota, A.; Kita, S.; Ishizaki, K.; Nishihama, R.; Yamato, K.T.; Kohchi, T. Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Corellou, F.; Schwartz, C.; Motta, J.P.; Djouani-Tahri el, B.; Sanchez, F.; Bouget, F.Y. Clocks in the green lineage: Comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell 2009, 21, 3436–3449. [Google Scholar] [CrossRef]
- Matsuo, T.; Iida, T.; Ohmura, A.; Gururaj, M.; Kato, D.; Mutoh, R.; Ihara, K.; Ishiura, M. The role of ROC75 as a daytime component of the circadian oscillator in Chlamydomonas reinhardtii. Plos Genet. 2020, 16. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamichi, N. The Transcriptional Network in the Arabidopsis Circadian Clock System. Genes 2020, 11, 1284. https://doi.org/10.3390/genes11111284
Nakamichi N. The Transcriptional Network in the Arabidopsis Circadian Clock System. Genes. 2020; 11(11):1284. https://doi.org/10.3390/genes11111284
Chicago/Turabian StyleNakamichi, Norihito. 2020. "The Transcriptional Network in the Arabidopsis Circadian Clock System" Genes 11, no. 11: 1284. https://doi.org/10.3390/genes11111284
APA StyleNakamichi, N. (2020). The Transcriptional Network in the Arabidopsis Circadian Clock System. Genes, 11(11), 1284. https://doi.org/10.3390/genes11111284



