Predictive Value of Circulating miRNAs in Lymph Node Metastasis for Colon Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Sample Collection
2.2. Circulating miRNA Extraction
2.3. miRNAs Expression Analysis
2.4. Statistical Analyses
3. Results
3.1. Patient Characteristics
3.2. Discovery of Candidate miRNAs
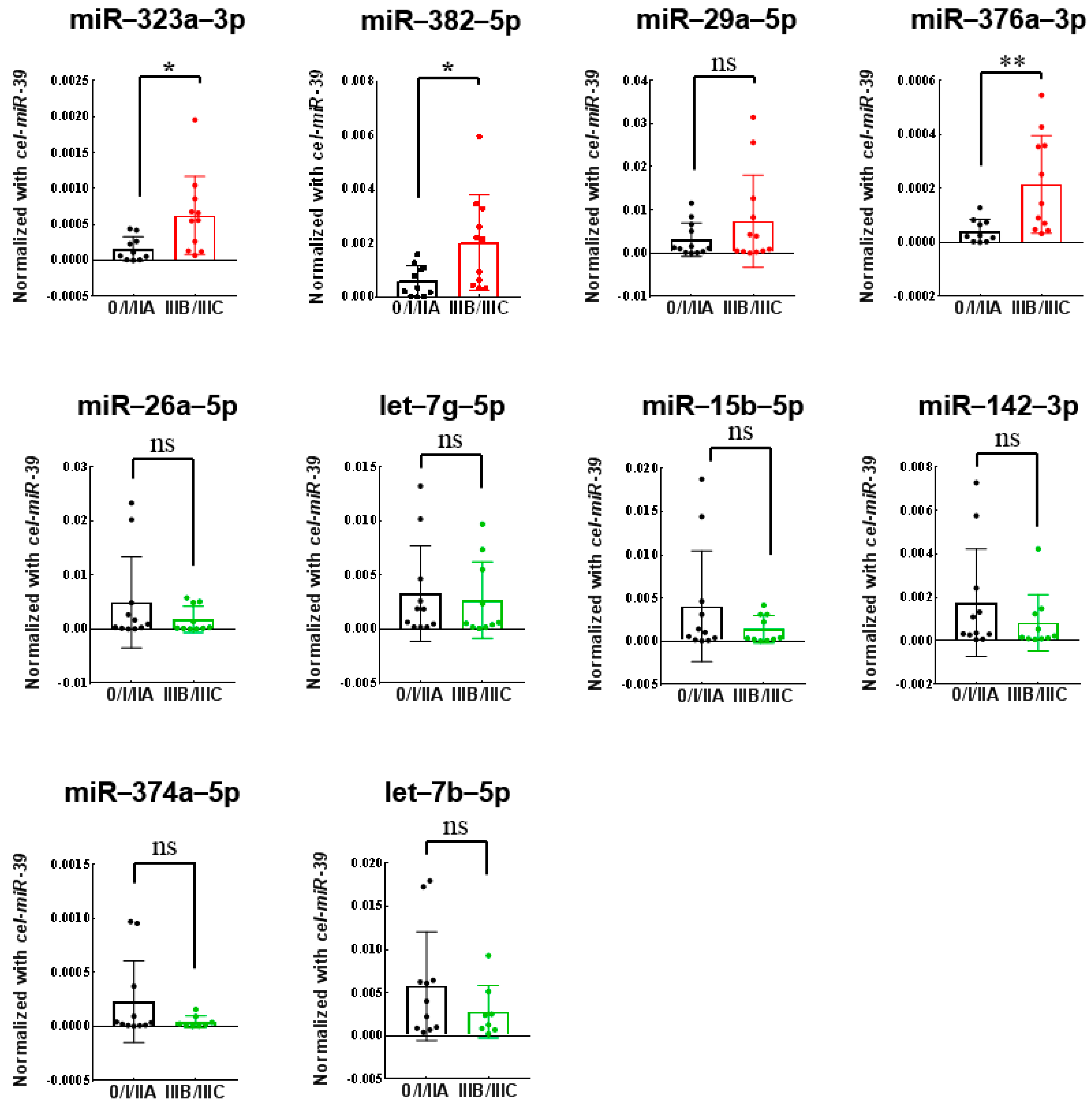
3.3. Validation of Differentially Expressed miRNAs Using Quantitative RT-PCR
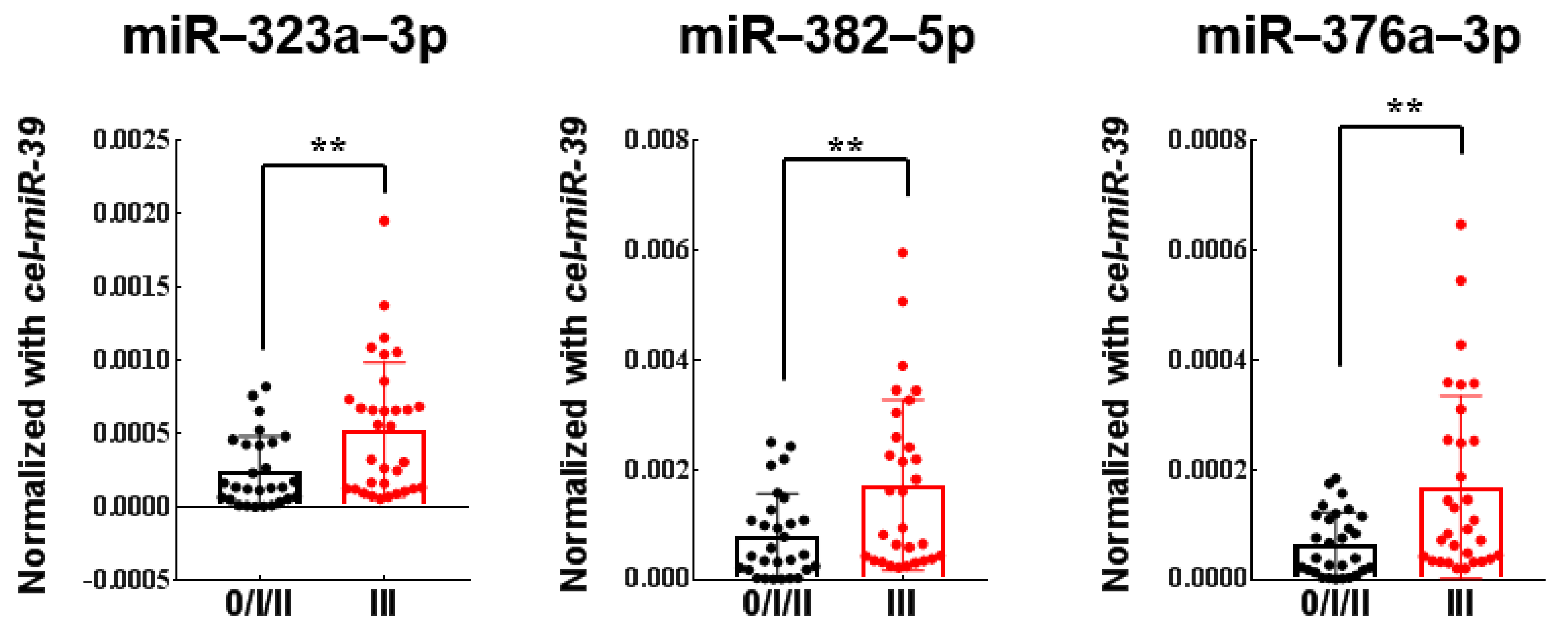
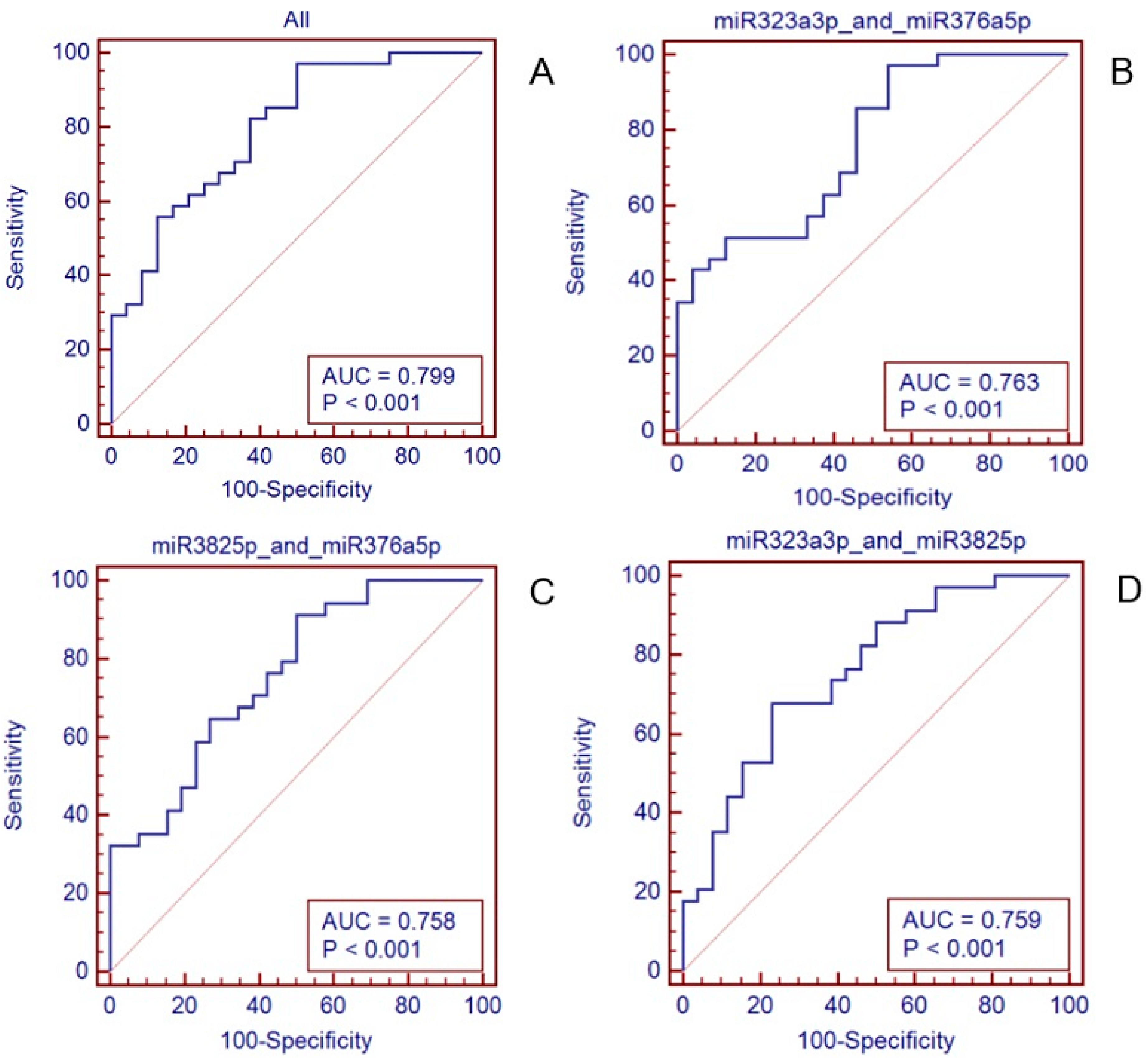
3.4. Establishment and Validation of the 3-miRNAs Signature with Predictive Value
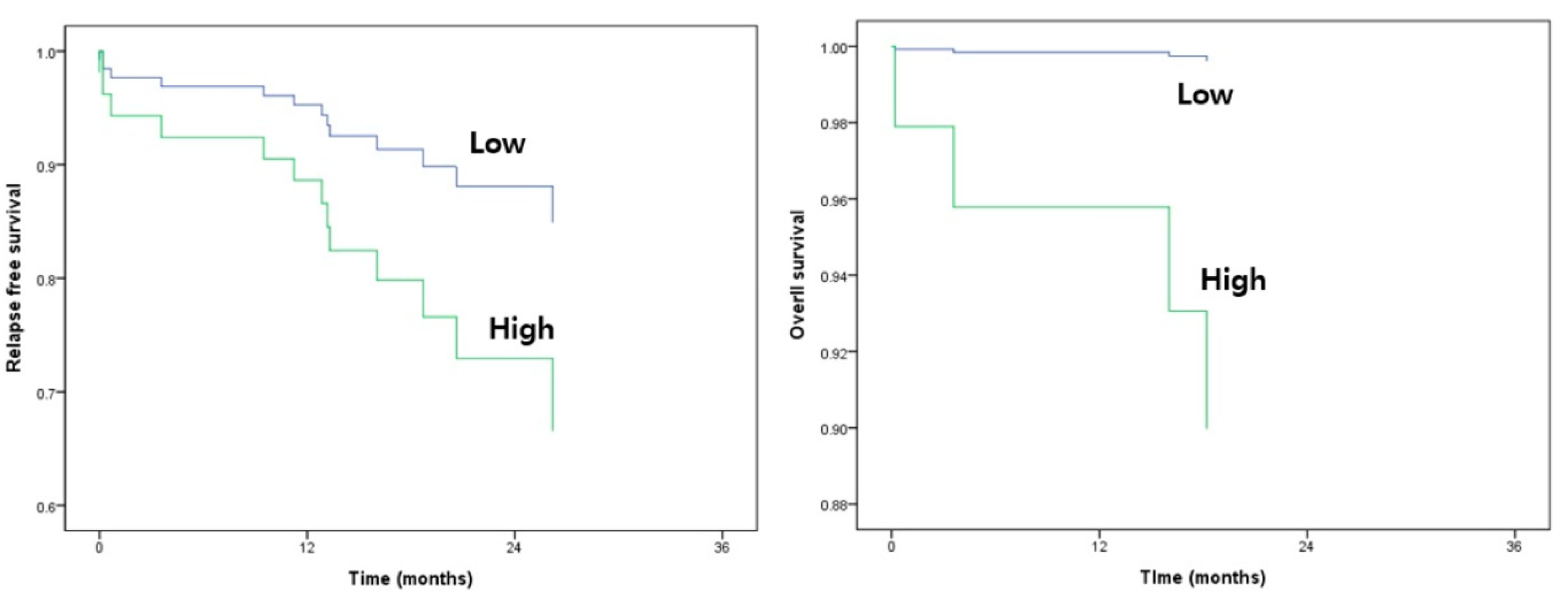
3.5. Survival Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Connell, J.B.; Maggard, M.A.; Ko, C.Y. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J. Nat. Cancer Inst. 2004, 96, 1420–1425. [Google Scholar]
- Nelson, H.; Petrelli, N.; Carlin, A.; Couture, J.; Fleshman, J.; Guillem, J.; Miedema, B.; Ota, D.; Sargent, D. Guidelines 2000 for colon and rectal cancer surgery. J. Nat. Cancer Inst. 2001, 93, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Greene, F.L.; Stewart, A.K.; Norton, H.J. A new TNM staging strategy for node-positive (stage III) colon cancer: An analysis of 50,042 patients. Annal. Surg. 2002, 236, 416–421; discussion 421. [Google Scholar] [CrossRef] [PubMed]
- Gunderson, L.L.; Jessup, J.M.; Sargent, D.J.; Greene, F.L.; Stewart, A.K. Revised TN categorization for colon cancer based on national survival outcomes data. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Drusco, A.; Nuovo, G.J.; Zanesi, N.; Di Leva, G.; Pichiorri, F.; Volinia, S.; Fernandez, C.; Antenucci, A.; Costinean, S.; Bottoni, A. MicroRNA profiles discriminate among colon cancer metastasis. PLoS ONE 2014, 9, e96670. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer–A brief overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Xia, Z.; Deng, Y.N.; Yang, Y.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019, 9, 180212. [Google Scholar] [CrossRef]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225, e1212. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Ma, H.; Chang, L.; Zhou, X.; Wang, N.; Zhao, L.; Zuo, J.; Wang, Y.; Han, J.; Wang, G. Role of microRNA-141 in colorectal cancer with lymph node metastasis. Exper. Ther. Med. 2016, 12, 3405–3410. [Google Scholar] [CrossRef] [PubMed]
- Ju, Q.; Zhao, Y.J.; Dong, Y.; Cheng, C.; Zhang, S.; Yang, Y.; Li, P.; Ge, D.; Sun, B. Identification of a miRNA-mRNA network associated with lymph node metastasis in colorectal cancer. Oncol. Lett. 2019, 18, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Kim, G.; Tak, W.Y.; Jang, S.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Chun, J.M.A.; Park, S.Y.; et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 2019, 144, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.Y.; Hsu, R.J.; Liu, J.M.; Chen, S.C.; Liao, G.S.; Gao, H.W.; Yu, C.P. MicroRNA-382-5p aggravates breast cancer progression by regulating the RERG/Ras/ERK signaling axis. Oncotarget 2017, 8, 22443–22459. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.P.; Xu, K.; Cui, J.; Yuan, D.Y.; Zou, B.; Li, J.; Liu, J.L.; Li, K.Y.; Meng, Z.; Zhang, B. Cancer-associated fibroblast-derived exosomal miR-382-5p promotes the migration and invasion of oral squamous cell carcinoma. Oncol. Rep. 2019, 42, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zheng, J. MicroRNA-323 upregulation promotes prostate cancer growth and docetaxel resistance by repressing p73. Biomed. Pharmacother. 2018, 97, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, J.; Ren, Y.; Zuo, Z.; Ni, S.; Cai, J. MEG3 can affect the proliferation and migration of colorectal cancer cells through regulating miR-376/PRKD1 axis. Am. J. Transl. Res. 2019, 11, 5740–5751. [Google Scholar] [PubMed]
- Chen, X.; Wang, Y.W.; Zhu, W.J.; Li, Y.; Liu, L.; Yin, G.; Gao, P. A 4–MicroRNA signature predicts lymph node metastasis and prognosis in breast cancer. Human Pathol. 2018, 76, 122–132. [Google Scholar] [CrossRef] [PubMed]
| Factors | n = 79 |
|---|---|
| Age | 63 |
| Median, range | (37–86) |
| Age | |
| <65 | 45 (57) |
| ≥65 | 34 (43) |
| Gender | |
| Male | 47 (59.5) |
| Female | 32 (40.5) |
| Tumor location | |
| Colon cancer | 76 (96.2) |
| Rectal cancer | 3 (3.8) |
| Pathologic T stage | |
| Tis | 1 (1.3) |
| T1 | 6 (7.6) |
| T2 | 3 (3.8) |
| T3 | 53 (67.1) |
| T4 | 16 (20.3) |
| Pathologic N stage | |
| N0 | 38 (48.1) |
| N1 | 20 (25.3) |
| N2 | 21 (26.6) |
| Pathologic Stage | |
| I | 9 (4.5) |
| II | 28 (35.4) |
| III | 39 (49.4) |
| IV | 3 (3.8) |
| Differentiation | |
| well | 3 (3.8) |
| moderate | 53 (67.1) |
| poor | 18 (22.8) |
| others | 3 (3.8) |
| KRAS | |
| wild | 42 (53.2) |
| mutant | 30 (38) |
| MSI | |
| low | 69 (87.3) |
| high | 6 (7.6) |
| Histology | |
| adenocarcinoma | 78 (98.7) |
| others | 1 (1.3) |
| Lymphovascular invasion | |
| present | 43 (54.4) |
| absent | 35 (44.3) |
| Venous invasion | |
| present | 43 (54.4) |
| absent | 32 (40.5) |
| Perineural invasion | |
| present | 54 (68.4) |
| absent | 24 (30.4) |
| lymphocyte response | |
| present | 57 (72.2) |
| absent | 18 (22.8) |
| Recurrence | 16 (20.3) |
| Death | 4 (5.1) |
| Probe Name | Fold Change | Pstage IIIC | Pstage 0 + 1 + IIA | p-Value |
|---|---|---|---|---|
| hsa-miR-323a-3p | 2.46 | 50.15 | 20.42 | 0.00616289 |
| hsa-miR-382-5p | 1.74 | 50.05 | 28.74 | 0.03742823 |
| hsa-miR-29a-3p | 1.73 | 63.28 | 36.54 | 0.01971120 |
| hsa-miR-376a-3p | 1.68 | 78.24 | 46.48 | 0.04374451 |
| hsa-let-7b-5p | −1.37 | 215.72 | 294.92 | 0.03424014 |
| hsa-miR-374a-5p | −1.71 | 25.16 | 42.91 | 0.04004743 |
| hsa-miR-142-3p | −1.91 | 52.04 | 99.52 | 0.01377911 |
| hsa-miR-15b-5p | −2.00 | 52.78 | 105.47 | 0.04035384 |
| hsa-let-7g-5p | −2.08 | 41.97 | 87.28 | 0.02688495 |
| hsa-miR-26a-5p | −2.73 | 17.93 | 49.02 | 0.00180513 |
| Relapse-Free Survival | Overall Survival | |||||
|---|---|---|---|---|---|---|
| Variable | Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | ||
| p-Value | HR (95% CI) | p-Value | p-Value | HR (95% CI) | p-Value | |
| Sex (M) | 0.786 | 0.777 | 0.777 | |||
| Age | 0.744 | 0.346 | 0.346 | |||
| Pstage (III and IV) | 0.038 * | 10.859 (0.963–122.475) | 0.317 | 0.317 | ||
| histologic type (2) | 0.733 | 0.862 | 0.862 | |||
| histologic grade (2) | 0.714 | 0.976 | 0.976 | |||
| histologic grade (3) | 0.749 | 0.975 | 0.975 | |||
| histologic grade (4) | 0.984 | 1.000 | 1.000 | |||
| LVI (1) | 0.075 | 0.392 | 0.392 | |||
| Venous invasion (1) | 0.034 * | 2.013 (0.507–7.985) | 0.443 | 0.443 | ||
| PNI (1) | 0.090 | 0.428 | ||||
| miR323a3p×1000 (>0.1326 §) | 0.258 | 0.745 (0.142–3.906) | 0.728 | 0.405 | 487,477.61 (0.000–∞) | 0.974 |
| miR3825p×1000 (>2.1516 §) | 0.132 | 9.399 (0.499–177.143) | 0.135 | 0.961 | 5.016 (0.517–48.628) | 0.164 |
| miR376a5p×1000 (>0.1454 §) | 0.251 | 0.231 (0.010–5.319) | 0.360 | 0.471 | 0.00 (0.000–2.31 × 10302) | 0.969 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, I.H.; Kim, G.; Kwak, S.G.; Baek, D.W.; Kang, B.W.; Kim, H.J.; Park, S.y.; Park, J.S.; Choi, G.-S.; Hur, K.; et al. Predictive Value of Circulating miRNAs in Lymph Node Metastasis for Colon Cancer. Genes 2021, 12, 176. https://doi.org/10.3390/genes12020176
Lee IH, Kim G, Kwak SG, Baek DW, Kang BW, Kim HJ, Park Sy, Park JS, Choi G-S, Hur K, et al. Predictive Value of Circulating miRNAs in Lymph Node Metastasis for Colon Cancer. Genes. 2021; 12(2):176. https://doi.org/10.3390/genes12020176
Chicago/Turabian StyleLee, In Hee, Gyeonghwa Kim, Sang Gyu Kwak, Dong Won Baek, Byung Woog Kang, Hye Jin Kim, Su yeon Park, Jun Seok Park, Gyu-Seog Choi, Keun Hur, and et al. 2021. "Predictive Value of Circulating miRNAs in Lymph Node Metastasis for Colon Cancer" Genes 12, no. 2: 176. https://doi.org/10.3390/genes12020176
APA StyleLee, I. H., Kim, G., Kwak, S. G., Baek, D. W., Kang, B. W., Kim, H. J., Park, S. y., Park, J. S., Choi, G.-S., Hur, K., & Kim, J. G. (2021). Predictive Value of Circulating miRNAs in Lymph Node Metastasis for Colon Cancer. Genes, 12(2), 176. https://doi.org/10.3390/genes12020176







