Comprehensive Genome-Wide Identification and Transcript Profiling of GABA Pathway Gene Family in Apple (Malus domestica)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Classification of the Apple GABA Pathway Genes
2.2. Phylogenetic Analysis of the GABA Pathway Proteins
2.3. Chromosomal Distribution and Synteny Analysis
2.4. Sequences Analysis
2.5. GABA Pathway Genes Expression Profiles
3. Results
3.1. Genome-Wide Identification and Characterization of GABA Pathway Gene Family Members in Apple
3.2. Chromosome Distribution and Duplication of Apple GABA Shunt Gene Family
3.3. Evolutionary Relationships of GABA Shunt Genes between Apple and Arabidopsis
3.4. Gene Structure Analysis and Conserved Motif Identification in Apple GABA Pathway Genes
3.5. Expression Profiles of GABA Pathway Genes in Different Tissues Development, Biological and Abiotic Stress
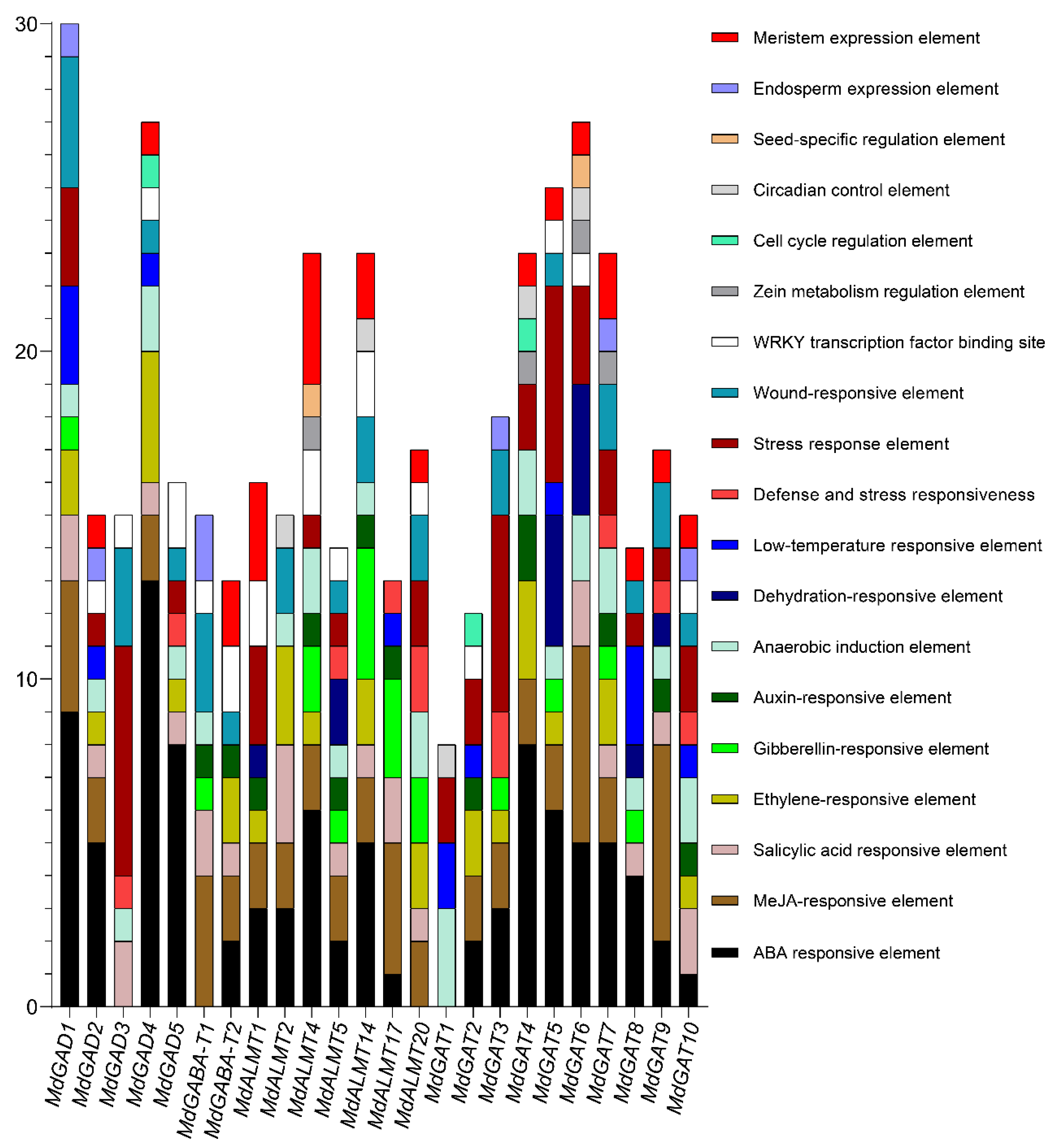
3.6. Cis-Acting Elements in the Promoter of the GABA Pathway Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seifikalhor, M.; Aliniaeifard, S.; Hassani, B.; Niknam, V.; Lastochkina, O. Diverse role of γ-aminobutyric acid in dynamic plant cell responses. Plant Cell Rep. 2019, 38, 847–867. [Google Scholar] [CrossRef]
- Salah, A.; Zhan, M.; Cao, C.; Han, Y.; Ling, L.; Liu, Z.; Li, P.; Ye, M.; Jiang, Y. γ-Aminobutyric Acid Promotes Chloroplast Ultrastructure, Antioxidant Capacity, and Growth of Waterlogged Maize Seedlings. Sci. Rep. 2019, 9, 484. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Tyerman, S.D.; Gilliham, M.; Xu, B. γ-Aminobutyric acid (GABA) signalling in plants. Cell. Mol. Life Sci. 2017, 74, 1577–1603. [Google Scholar] [CrossRef] [PubMed]
- Shelp, B.J.; Bown, A.W.; Faure, D. Extracellular γ-aminobutyrate mediates communication between plants and other organisms. Plant Physiol. 2006, 142, 1350–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renault, H.; El Amrani, A.; Palanivelu, R.; Updegraff, E.P.; Yu, A.; Renou, J.P.; Preuss, D.; Bouchereau, A.; Deleu, C. GABA accumulation causes cell elongation defects and a decrease in expression of genes encoding secreted and cell wall-related proteins in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 894–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renault, H.; El Amrani, A.; Berger, A.; Mouille, G.; Soubigou-Taconnat, L.; Bouchereau, A.; Deleu, C. γ-Aminobutyric acid transaminase deficiency impairs central carbon metabolism and leads to cell wall defects during salt stress in Arabidopsis roots. Plant Cell Environ. 2013, 36, 1009–1018. [Google Scholar] [CrossRef]
- Shelp, B.; Bozzo, G.; Trobacher, C.; Chiu, G.; Bajwa, V. Strategies and tools for studying the metabolism and function of Y-aminobutyrate in plants. I. Pathway structure. Botany 2012, 90, 651–668. [Google Scholar] [CrossRef]
- Zhu, X.; Liao, J.; Xia, X.; Xiong, F.; Li, Y.; Shen, J.; Wen, B.; Ma, Y.; Wang, Y.; Fang, W. Physiological and iTRAQ-based proteomic analyses reveal the function of exogenous γ-aminobutyric acid (GABA) in improving tea plant (Camellia sinensis L.) tolerance at cold temperature. BMC Plant Biol. 2019, 19, 43. [Google Scholar] [CrossRef]
- Vijayakumari, K.; Puthur, J.T. γ-Aminobutyric acid (GABA) priming enhances the osmotic stress tolerance in Piper nigrum Linn. plants subjected to PEG-induced stress. Plant Growth Regul. 2016, 78, 57–67. [Google Scholar] [CrossRef]
- Xing, S.G.; Jun, Y.B.; Hau, Z.W.; Liang, L.Y. Higher accumulation of γ-aminobutyric acid induced by salt stress through stimulating the activity of diamine oxidases in Glycine max (L.) Merr. Roots. Plant Physiol. Bioch. 2007, 45, 560–566. [Google Scholar] [CrossRef]
- Renault, H.; Roussel, V.; El Amrani, A.; Arzel, M.; Renault, D.; Bouchereau, A.; Deleu, C. The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 2010, 10, 20. [Google Scholar] [CrossRef] [Green Version]
- Akcay, N.; Bor, M.; Karabudak, T.; Ozdemir, F.; Turkan, I. Contribution of Gamma amino butyric acid (GABA) to salt stress responses of Nicotiana sylvestris CMSII mutant and wild type plants. J. Plant Physiol. 2012, 169, 452–458. [Google Scholar] [CrossRef]
- Xiang, L.X.; Hu, L.P.; Hu, X.H.; Pan, X.B.; Ren, W.Q. Response of reactive oxygen metabolism in melon chloroplasts to short-term salinity-alkalinity stress regulated by exogenous γ-aminobutyric acid. J. Ecol. 2015, 26, 3746–3752. (In Chinese) [Google Scholar]
- Kalhor, M.S.; Aliniaeifard, S.; Seif, M.; Asayesh, E.J.; Bernard, F.; Hassani, B.; Li, T. Enhanced salt tolerance and photosynthetic performance: Implication of γ-amino butyric acid application in salt-exposed lettuce (Lactuca sativa L.) plants. Plant Physiol. Bioch. 2018, 130, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Tyerman, S.D.; Xu, B.; Bose, J.; Kaur, S.; Conn, V.; Domingos, P.; Ullah, S.; Wege, S.; Shabala, S.; et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat. Commun. 2015, 6, 7879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takayama, M.; Matsukura, C.; Ariizumi, T.; Ezura, H. Activating glutamate decarboxylase activity by removing the autoinhibitory domain leads to hyper γ-aminobutyric acid (GABA) accumulation in tomato fruit. Plant Cell Rep. 2017, 36, 103–116. [Google Scholar] [CrossRef]
- Xie, T.; Ji, J.; Chen, W.; Yue, J.; Du, C.; Sun, J.; Chen, L.; Jiang, Z.; Shi, S. GABA negatively regulates adventitious root development. J. Exp. Bot. 2019, 71, 1459–1474. [Google Scholar] [CrossRef]
- Yue, J.Y.; Du, C.J.; Ji, J.; Xie, T.T.; Chen, W.; Chang, E.M.; Chen, L.Z.; Jiang, Z.P.; Shi, S.Q. Inhibition of α-ketoglutarate dehydrogenase activity affects adventitious root growth in poplar via changes in GABA shunt. Planta 2018, 248, 963–979. [Google Scholar] [CrossRef]
- Bouche, N.; Fromm, H. GABA in plants: Just a metabolite? Trends Plant Sci. 2004, 9, 110–115. [Google Scholar] [CrossRef]
- Fait, A.; Fromm, H.; Walter, D.; Galili, G.; Fernie, A.R. Highway or byway: The metabolic role of the GABA shunt in plants. Trends Plant Sci. 2008, 13, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Kamran, M.; Sullivan, W.; Chirkova, L.; Okamoto, M.; Degryse, F.; McLaughlin, M.; Gilliham, M.; Tyerman, S.D. Aluminum-activated malate transporters can facilitate GABA transport. Plant Cell 2018, 30, 1147–1164. [Google Scholar] [CrossRef] [Green Version]
- Okumoto, S.; Pilot, G. Amino acid export in plants: A missing link in nitrogen cycling. Mol. Plant 2011, 4, 453–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tegeder, M. Transporters for amino acids in plant cells: Some functions and many unknowns. Curr. Opin. Plant Biol. 2012, 15, 315–321. [Google Scholar] [CrossRef]
- Schwacke, R.; Grallath, S.; Breitkreuz, K.E.; Stransky, E.; Stransky, H.; Frommer, W.B.; Rentsch, D. LeProT1, a Transporter for Proline, Glycine Betaine, and γ-Amino Butyric Acid in Tomato Pollen. Plant Cell 1999, 11, 377–391. [Google Scholar] [PubMed] [Green Version]
- Meyer, A.; Eskandari, S.; Grallath, S.; Rentsch, D. AtGAT1, a high affinity transporter for γ-aminobutyric acid in Arabidopsis thaliana. J. Biol. Chemstry 2006, 281, 7197–7204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.; Xu, J.; Shao, X.; Luo, W.; Niu, Z.; Gao, C.; Wan, D. A Novel Gene Coding γ-Aminobutyric Acid Transporter May Improve the Tolerance of Populus euphratica to Adverse Environments. Front. Plant Sci. 2019, 10, 1083. [Google Scholar] [CrossRef]
- Ma, J.; Wan, D.; Duan, B.; Bai, X.; Bai, Q.; Chen, N.; Ma, T. Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 2019, 17, 451–460. [Google Scholar] [CrossRef]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Liu, B.; Qiu, Q.; Wang, Z.; Zhang, J.; et al. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013, 4, 2797. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; VandePeer, Y. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Ma, B.; Yuan, Y.; Gao, M.; Qi, T.; Li, M.; Ma, F. Genome-wide identification, molecular evolution, and expression divergence of aluminum-activated malate transporters in apples. Int. J. Mol. Sci. 2018, 19, 2807. [Google Scholar] [CrossRef] [Green Version]
- Bergmann, R.; Kongsbak, K.; Sorensen, P.L.; Sander, T.; Balle, T. A unified model of the GABAA receptor comprising agonist and benzodiazepine binding sites. PLoS ONE 2013, 8, e52323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, P.S.; Aricescu, A.R. Crystal structure of a human GABAA receptor. Nature 2014, 512, 270–275. [Google Scholar] [CrossRef] [Green Version]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef]
- Da, L.; Liu, Y.; Yang, J.; Tian, T.; She, J.; Ma, X.; Xu, W.; Su, Z. AppleMDO: A Multi-Dimensional Omics Database for Apple Co-Expression Networks and Chromatin States. Front. Plant Sci. 2019, 10, 1333. [Google Scholar] [CrossRef]
- Valentinuzzi, F.; Venuti, S.; Pii, Y.; Marroni, F.; Cesco, S.; Hartmann, F.; Mimmo, T.; Morgante, M.; Pinton, R.; Tomasi, N.; et al. Common and specific responses to iron and phosphorus deficiencies in roots of apple tree (Malus x domestica). Plant Mol. Biol. 2019, 101, 129–148. [Google Scholar] [CrossRef]
- Che-Othman, M.H.; Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Wheat mitochondrial respiration shifts from the tricarboxylic acid cycle to the GABA shunt under salt stress. New Phytol. 2020, 225, 1166–1180. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, X.-M.; Jin, L.-F.; Shi, C.-Y.; Liu, Y.-Z.; Peng, S.-A. Identification and transcript analysis of two glutamate decarboxylase genes, CsGAD1 and CsGAD2, reveal the strong relationship between CsGAD1 and citrate utilization in citrus fruit. Mol. Biol. Rep. 2014, 41, 6253–6262. [Google Scholar] [CrossRef] [PubMed]
- Bouche, N.; Fait, A.; Zik, M.; Fromm, H. The root-specific glutamate decarboxylase (GAD1) is essential for sustaining GABA levels in Arabidopsis. Plant Mol. Biol. 2004, 55, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Ren, G.; He, C.; Li, X.; Meng, Q.; Zhu, C.; Wang, R.; Zhang, J. Cloning and Characterization of a Maize cDNA Encoding Glutamate Decarboxylase. Plant Mol. Biol. Repot. 2010, 28, 620–626. [Google Scholar] [CrossRef]
- Trobacher, C.P.; Zarei, A.; Liu, J.; Clark, S.M.; Bozzo, G.G.; Shelp, B.J. Calmodulin-dependent and calmodulinindependent glutamate decarboxylases in apple fruit. BMC Plant Biol. 2013, 13, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, X.; Chen, Y.; Zhang, L.; Fu, X.; Wei, Q.; Grierson, D.; Zhou, Y.; Huang, Y.; Dong, F.; Yang, Z. Dual mechanisms regulating glutamate decarboxylases and accumulation of γ-aminobutyric acid in tea (Camellia sinensis) leaves exposed to multiple stresses. Sci. Rep. 2016, 6, 23685. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Zheng, L.; Yue, J.; Yao, X.; Chang, E.; Xie, T.; Deng, N.; Chen, L.; Huang, Y.; Jiang, Z.; et al. Identification of two CiGADs from Caragana intermedia and their transcriptional responses to abiotic stresses and exogenous abscisic acid. PeerJ 2017, 5, e3439. [Google Scholar] [CrossRef] [Green Version]
- Akama, K.; Akihiro, T.; Kitagawa, M.; Takaiwa, F. Rice (Oryza sativa) contains a novel isoform of glutamate decarboxylase that lacks an authentic calmodulin-binding domain at the C-terminus. Biochim. Biophys. Acta. 2001, 1522, 143–150. [Google Scholar] [CrossRef]
- Sharma, T.; Dreyer, I.; Kochian, L.; Pineros, M.A. The ALMT Family of Organic Acid Transporters in Plants and Their Involvement in Detoxification and Nutrient Security. Front. Plant Sci. 2016, 7, 1488. [Google Scholar] [CrossRef] [Green Version]
- Hedrich, R.; Salvador-Recatala, V.; Dreyer, I. Electrical Wiring and Long-Distance Plant Communication. Trends Plant Sci. 2016, 21, 376–387. [Google Scholar] [CrossRef]
- Barbier-Brygoo, H.; De Angeli, A.; Filleur, S.; Frachisse, J.M.; Gambale, F.; Thomine, S.; Wege, S. Anion channels/transporters in plants: From molecular bases to regulatory networks. Annu. Rev. Plant Biol. 2011, 62, 25–51. [Google Scholar] [CrossRef] [PubMed]
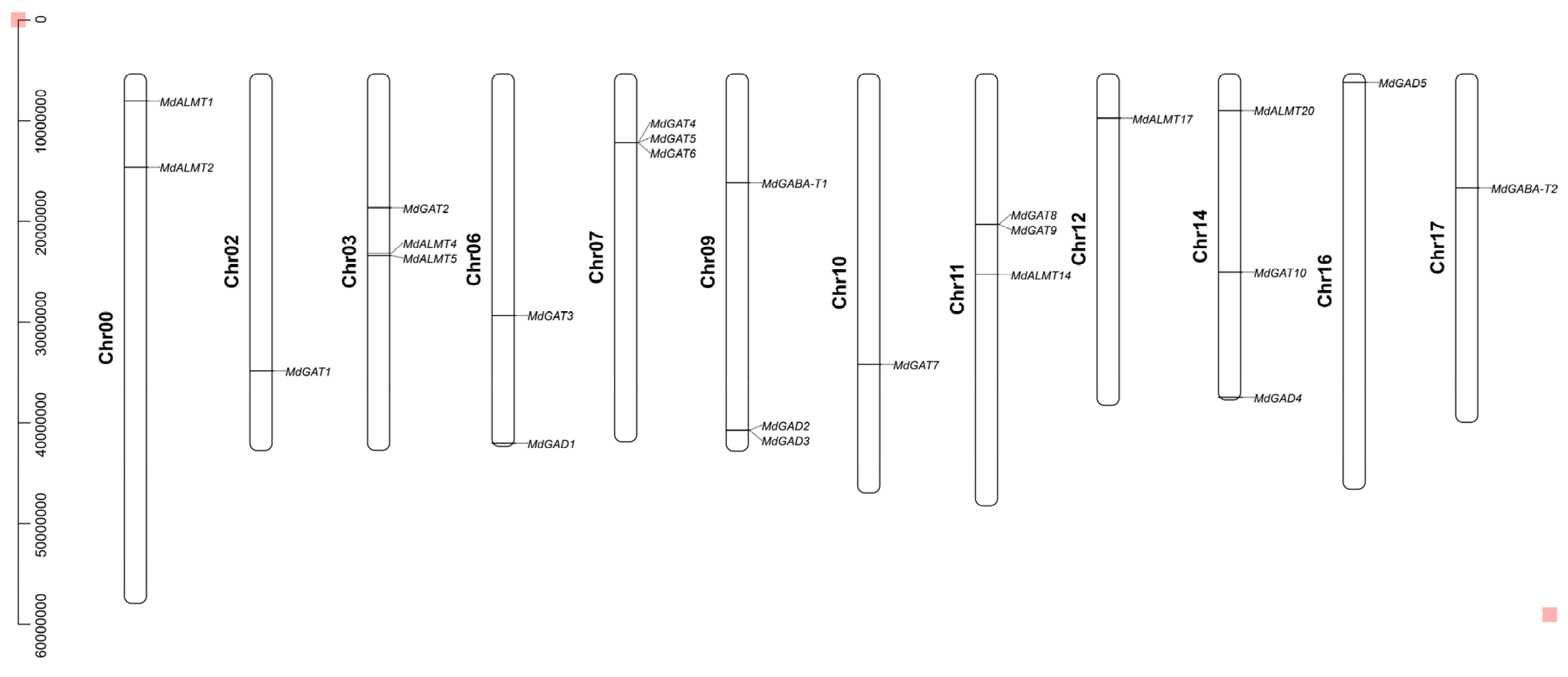
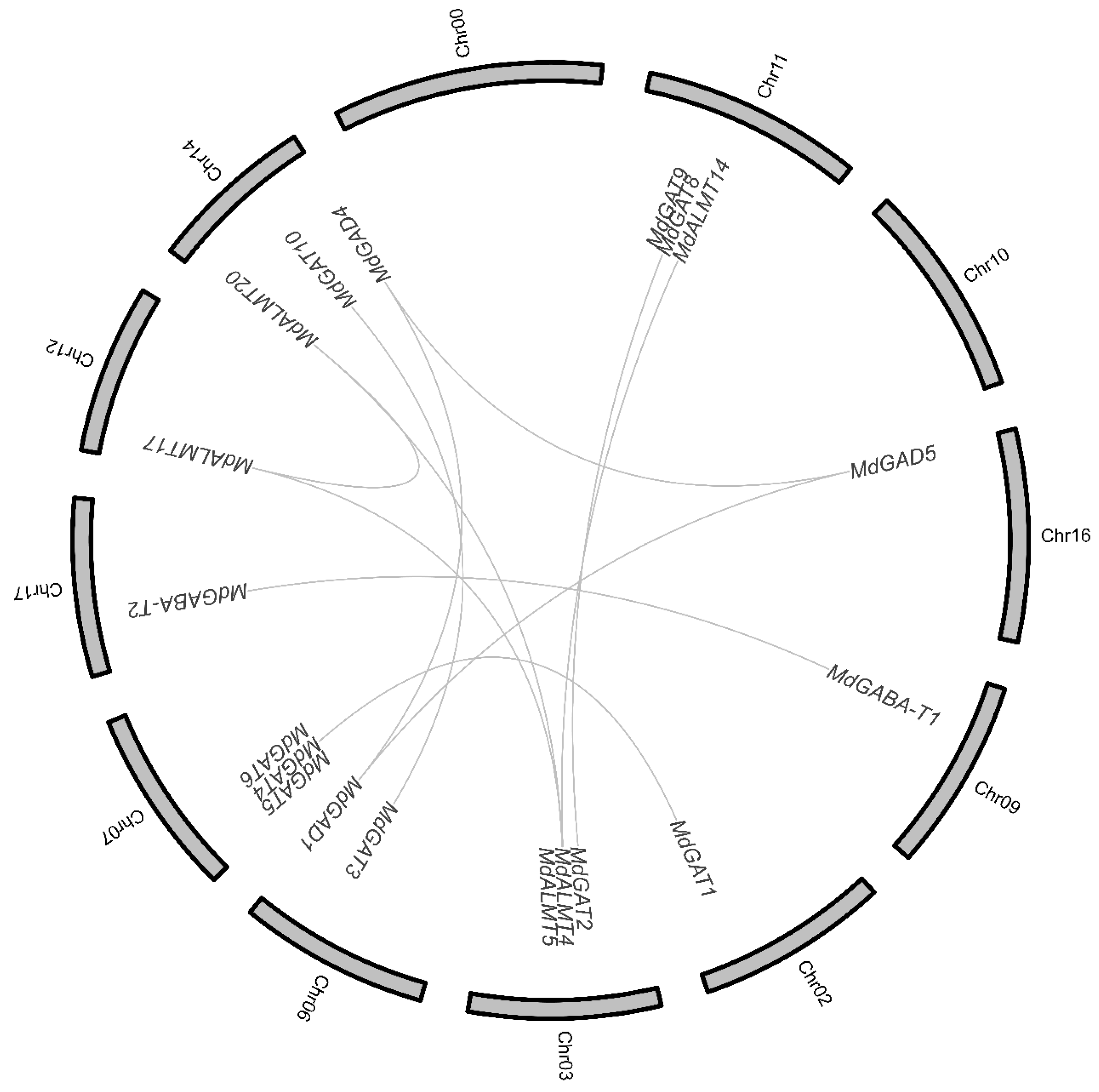


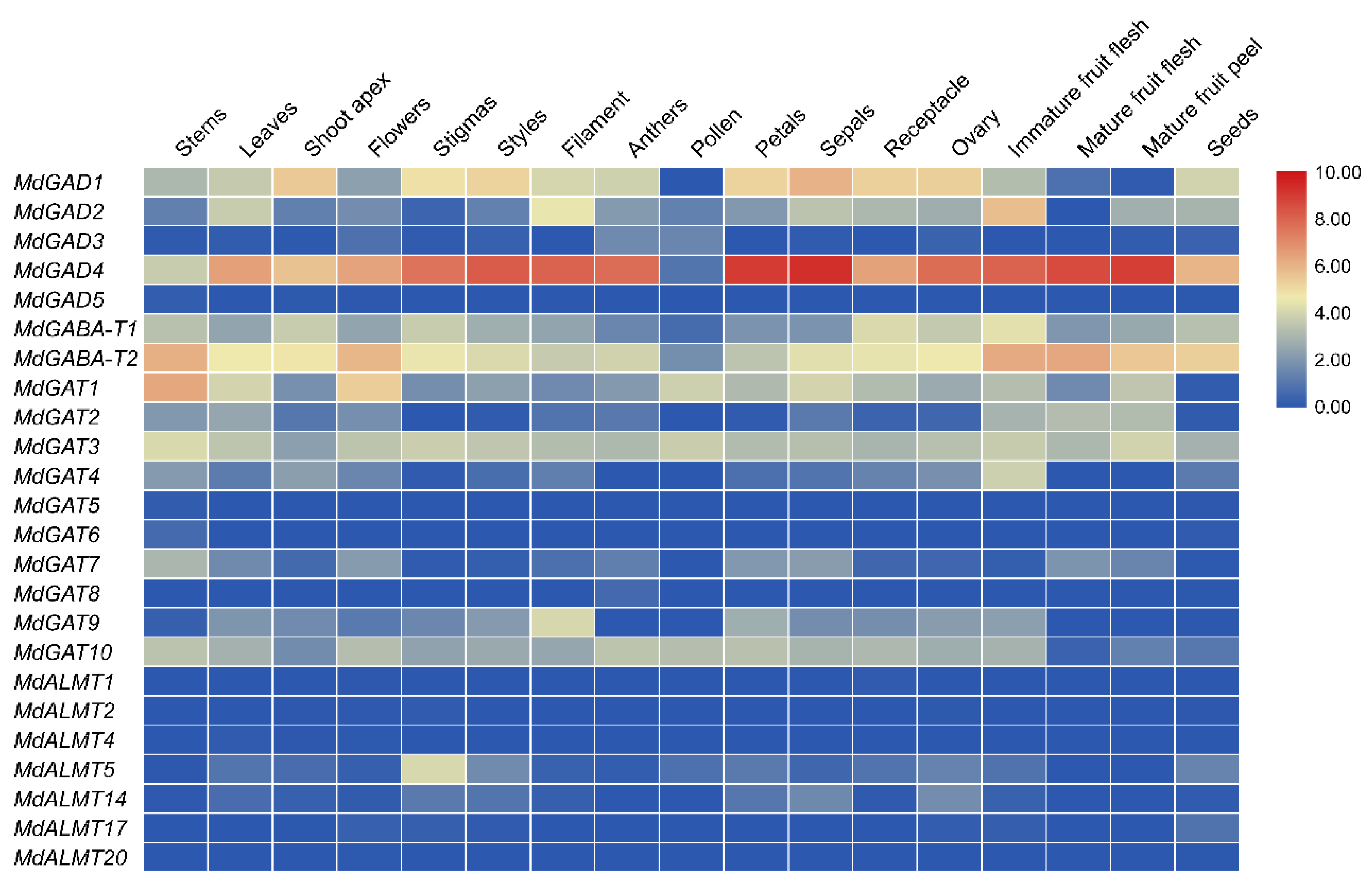
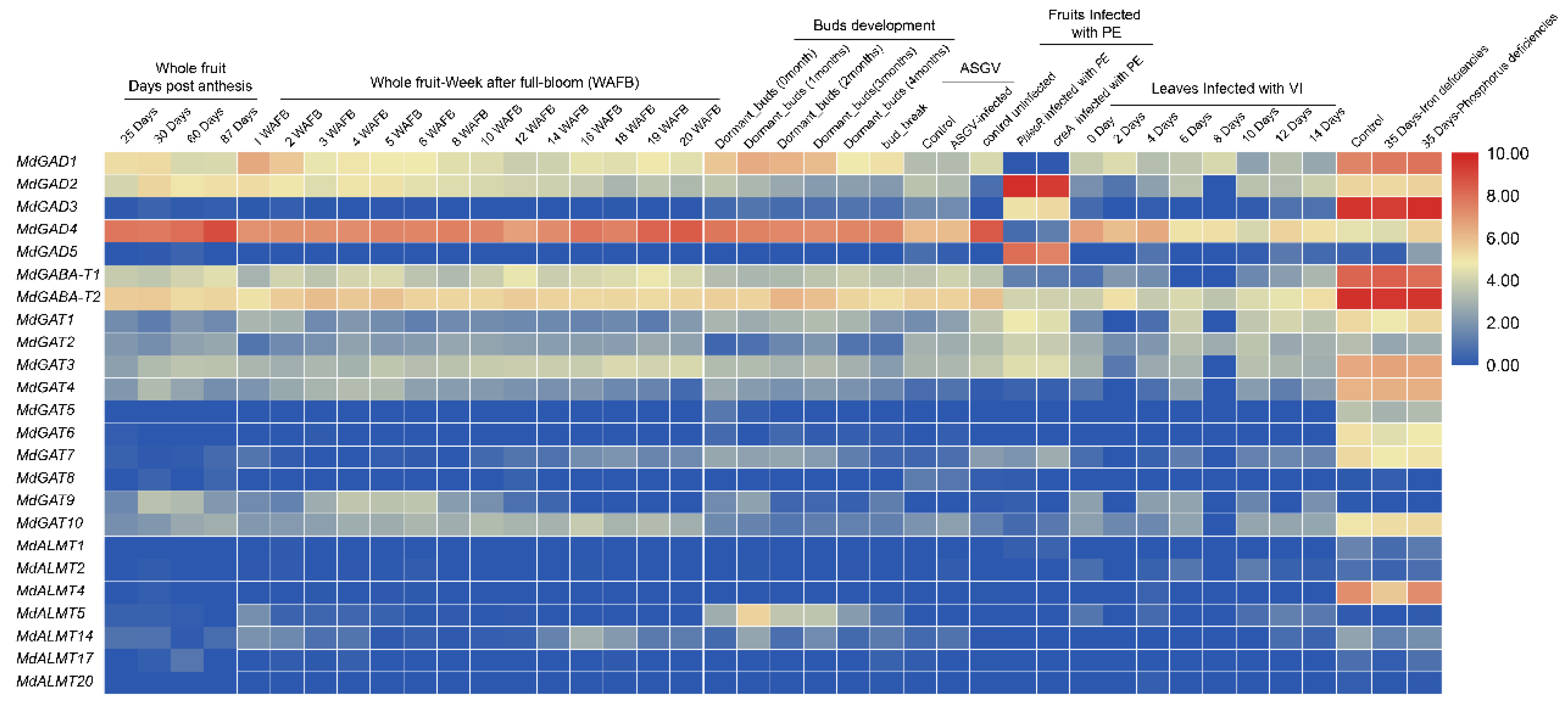
| Gene Name | Gene Name Abbreviation | Locus | Chromosome | Start Position | End Position | Strand | Length (aa) | pI | MW (kD) | Subcellular Localization Predicted | Potential Functions |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Putative glutamate decarboxylase | MdGAD1 | MD06G1235500 | Chr06 | 36665970 | 36669767 | − | 505 | 5.49 | 56.50 | cyto | GABA Biosynthesis |
| MdGAD2 | MD09G1276300 | Chr09 | 35359458 | 35366901 | − | 501 | 5.62 | 56.93 | cyto | ||
| MdGAD3 | MD09G1277000 | Chr09 | 35381876 | 35388878 | − | 501 | 5.71 | 56.85 | cyto | ||
| MdGAD4 | MD14G1242700 | Chr14 | 32108140 | 32110957 | − | 333 | 4.95 | 37.68 | cyto | ||
| MdGAD5 | MD16G1010800 | Chr16 | 826461 | 829289 | + | 510 | 5.65 | 56.95 | chlo | ||
| Putative GABA transaminase | MdGABA-T1 | MD09G1139200 | Chr09 | 10792690 | 10798780 | − | 521 | 6.95 | 57.59 | chlo | GABA Catabolism |
| MdGABA-T2 | MD17G1128700 | Chr17 | 11307951 | 11313609 | − | 514 | 6.95 | 56.69 | chlo | ||
| Putative GABA transporters | MdGAT1 | MD02G1244800 | Chr02 | 29475795 | 29480278 | − | 508 | 9.07 | 56.22 | plas | GABA Transport |
| MdGAT2 | MD03G1133300 | Chr03 | 13280438 | 13300479 | − | 510 | 9.28 | 56.40 | plas | ||
| MdGAT3 | MD06G1103100 | Chr06 | 23991060 | 23995998 | + | 452 | 8.91 | 49.64 | cyto | ||
| MdGAT4 | MD07G1072100 | Chr07 | 6824513 | 6827647 | + | 460 | 8.75 | 50.35 | plas | ||
| MdGAT5 | MD07G1072200 | Chr07 | 6832373 | 6833039 | + | 174 | 4.79 | 19.42 | cyto | ||
| MdGAT6 | MD07G1072300 | Chr07 | 6833186 | 6834458 | + | 299 | 9.22 | 32.55 | chlo | ||
| MdGAT7 | MD10G1191600 | Chr10 | 28831972 | 28834442 | + | 457 | 8.97 | 50.79 | plas | ||
| MdGAT8 | MD11G1155800 | Chr11 | 14931465 | 14932148 | − | 177 | 9.88 | 19.42 | vacu | ||
| MdGAT9 | MD11G1156000 | Chr11 | 14949839 | 14952668 | − | 202 | 7.69 | 22.50 | plas | ||
| MdGAT10 | MD14G1122000 | Chr14 | 19672859 | 19676484 | + | 452 | 8.97 | 49.81 | plas | ||
| Putative GABA receptors | MdALMT1 | MD00G1017600 | Chr00 | 2696703 | 2698931 | − | 472 | 8.21 | 51.81 | plas | GABA Receptors |
| MdALMT2 | MD00G1049200 | Chr00 | 9253069 | 9261444 | + | 485 | 9.28 | 53.69 | plas | ||
| MdALMT4 | MD03G1155200 | Chr03 | 17835181 | 17837669 | + | 476 | 8.73 | 52.20 | plas | ||
| MdALMT5 | MD03G1155400 | Chr03 | 18018465 | 18020999 | + | 484 | 8.83 | 53.34 | plas | ||
| MdALMT14 | MD11G1173000 | Chr11 | 19922876 | 19925870 | + | 485 | 9.39 | 53.75 | plas | ||
| MdALMT17 | MD12G1040500 | Chr12 | 4405915 | 4408221 | − | 493 | 8.30 | 53.72 | plas | ||
| MdALMT20 | MD14G1039500 | Chr14 | 3629641 | 3631853 | − | 494 | 8.84 | 53.85 | plas |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Q.; Su, S.; Wang, Z.; Wang, Y.; Xu, X. Comprehensive Genome-Wide Identification and Transcript Profiling of GABA Pathway Gene Family in Apple (Malus domestica). Genes 2021, 12, 1973. https://doi.org/10.3390/genes12121973
Zheng Q, Su S, Wang Z, Wang Y, Xu X. Comprehensive Genome-Wide Identification and Transcript Profiling of GABA Pathway Gene Family in Apple (Malus domestica). Genes. 2021; 12(12):1973. https://doi.org/10.3390/genes12121973
Chicago/Turabian StyleZheng, Qingbo, Shenghui Su, Zhe Wang, Yongzhang Wang, and Xiaozhao Xu. 2021. "Comprehensive Genome-Wide Identification and Transcript Profiling of GABA Pathway Gene Family in Apple (Malus domestica)" Genes 12, no. 12: 1973. https://doi.org/10.3390/genes12121973
APA StyleZheng, Q., Su, S., Wang, Z., Wang, Y., & Xu, X. (2021). Comprehensive Genome-Wide Identification and Transcript Profiling of GABA Pathway Gene Family in Apple (Malus domestica). Genes, 12(12), 1973. https://doi.org/10.3390/genes12121973





