Concatenation of Transgenic DNA: Random or Orchestrated?
Abstract
1. Historical Overview of the Concatenation Studies
2. Information from the Internal Junctions
3. Palindromes
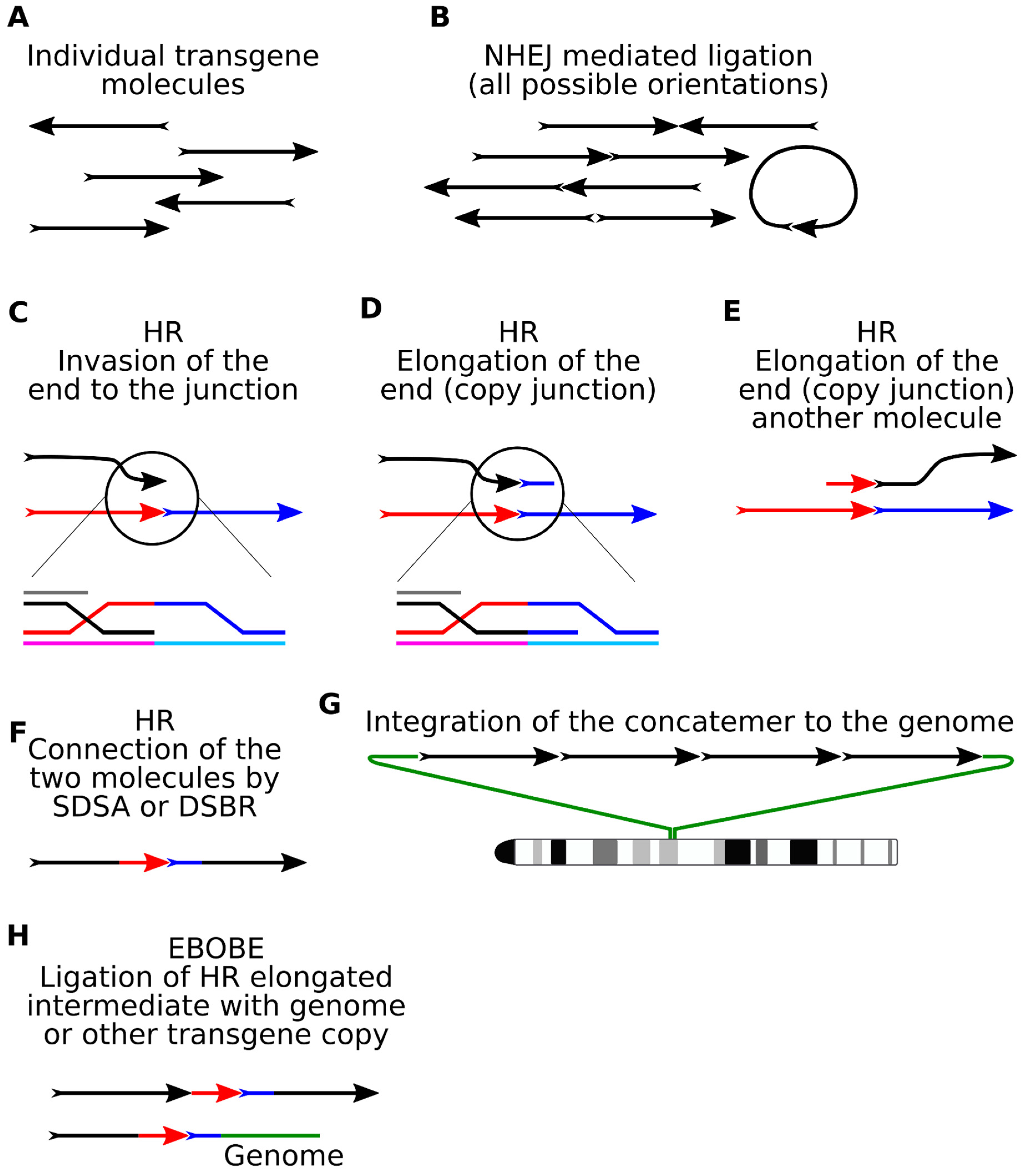
4. Molecular Mechanisms of Concatenation
5. Repeat-Induced Gene Silencing
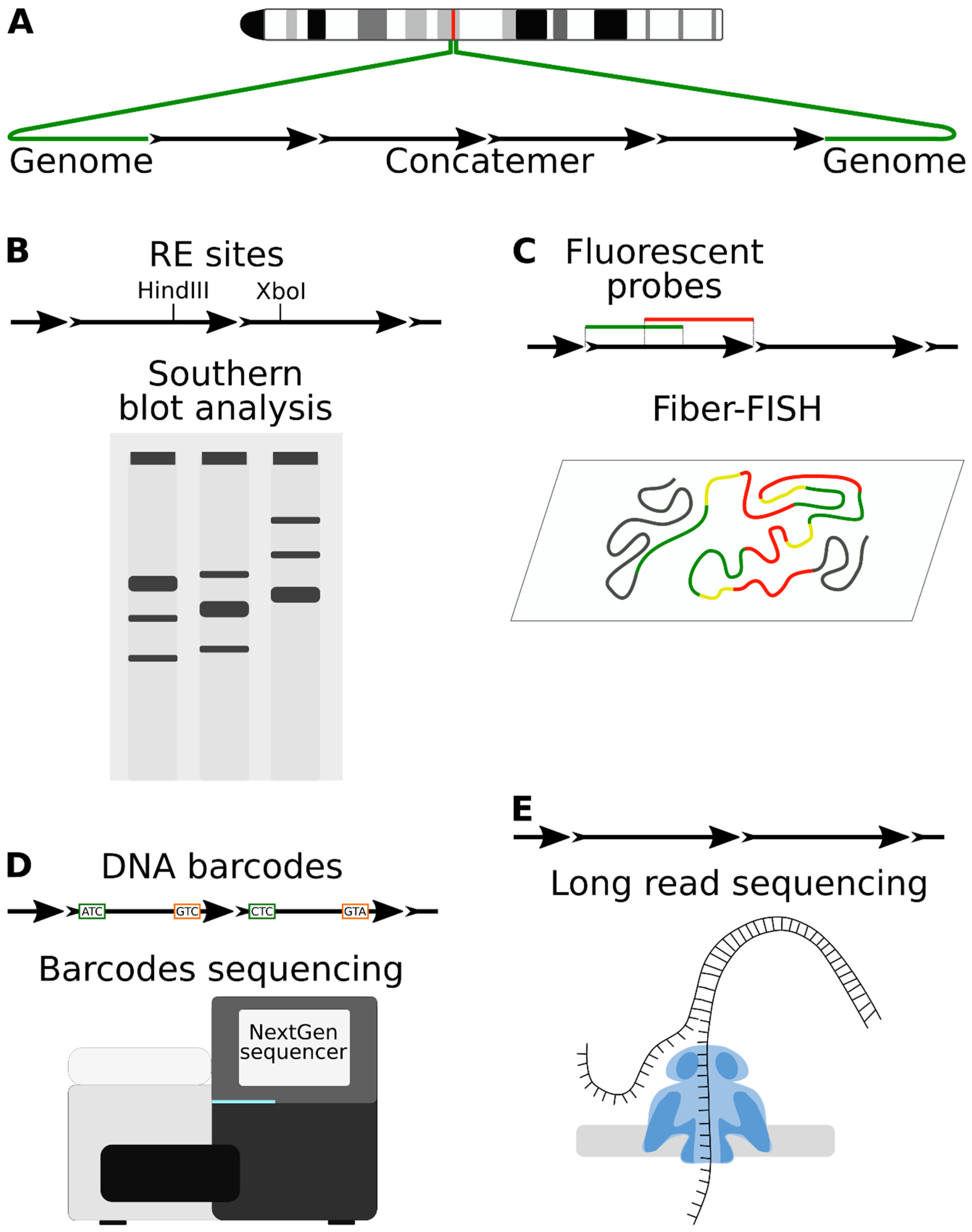
6. Novel Methods for Studying Transgene Concatenation
7. Conclusions and Future Direction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Yang, J.; Zhou, W.; Zhang, Y.; Zidon, T.; Ritchie, T.; Engelhardt, J.F. Concatamerization of adeno-associated virus circular genomes occurs through intermolecular recombination. J. Virol. 1999, 73, 9468–9477. [Google Scholar] [CrossRef]
- Maurer, A.C.; Weitzman, M.D. Adeno-Associated Virus Genome Interactions Important for Vector Production and Transduction. Hum. Gene Ther. 2020, 31, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Clark-Walker, G.D. Unveiling the mystery of mitochondrial DNA replication in yeasts. Mitochondrion 2018, 38, 17–22. [Google Scholar] [CrossRef]
- Tomaska, L.; Nosek, J.; Kar, A.; Willcox, S.; Griffith, J.D. A New View of the T-Loop Junction: Implications for Self-Primed Telomere Extension, Expansion of Disease-Related Nucleotide Repeat Blocks, and Telomere Evolution. Front. Genet. 2019, 10, 792. [Google Scholar] [CrossRef]
- Pesenti, E.; Liskovykh, M.; Okazaki, K.; Mallozzi, A.; Reid, C.; Abad, M.A.; Jeyaprakash, A.A.; Kouprina, N.; Larionov, V.; Masumoto, H.; et al. Analysis of Complex DNA Rearrangements during Early Stages of HAC Formation. ACS Synth. Biol. 2020, 9, 3267–3287. [Google Scholar] [CrossRef]
- Shimizu, N. Gene Amplification and the Extrachromosomal Circular DNA. Genes 2021, 12, 1533. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.O. Chromosomal insertion of foreign DNA. Reprod. Nutr. Dev. 1996, 36, 607–618. [Google Scholar]
- Svitashev, S.K.; Pawlowski, W.P.; Makarevitch, I.; Plank, D.W.; Somers, D.A. Complex transgene locus structures implicate multiple mechanisms for plant transgene rearrangement. Plant J. 2002, 32, 433–445. [Google Scholar] [CrossRef]
- Jupe, F.; Rivkin, A.C.; Michael, T.P.; Zander, M.; Motley, S.T.; Sandoval, J.P.; Keith Slotkin, R.; Chen, H.; Castanon, R.; Nery, J.R.; et al. The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet. 2019, 15, e1007819. [Google Scholar] [CrossRef] [PubMed]
- Mello, C.C.; Kramer, J.M.; Stinchcomb, D.; Ambros, V. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991, 10, 3959–3970. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Chang, T.; Huang, H.; Yee, J.-K. Integration-defective lentiviral vector mediates efficient gene editing through homology-directed repair in human embryonic stem cells. Nucleic Acids Res. 2017, 45, e29. [Google Scholar] [CrossRef]
- Chandler, K.J.; Chandler, R.L.; Broeckelmann, E.M.; Hou, Y.; Southard-Smith, E.M.; Mortlock, D.P. Relevance of BAC transgene copy number in mice: Transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm. Genome 2007, 18, 693–708. [Google Scholar] [CrossRef]
- Le Saux, A.; Houdebine, L.-M.; Jolivet, G. Chromosome integration of BAC (bacterial artificial chromosome): Evidence of multiple rearrangements. Transgenic Res. 2010, 19, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, S.; Balasubramanian, S.; Webster, L.; Lee, M.; Vavilala, D.; Kulikov, N.; Choi, J.; Tang, C.; Hunter, M.; Wang, R.; et al. Accelerating and de-risking CMC development with transposon-derived manufacturing cell lines. Biotechnol. Bioeng. 2021, 118, 2301–2311. [Google Scholar] [CrossRef] [PubMed]
- Danner, E.; Lebedin, M.; de la Rosa, K.; Kühn, R. A homology independent sequence replacement strategy in human cells using a CRISPR nuclease: Replace Targeting. Open Biol. 2021, 11, 200283. [Google Scholar] [CrossRef]
- Boel, A.; De Saffel, H.; Steyaert, W.; Callewaert, B.; De Paepe, A.; Coucke, P.J.; Willaert, A. CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis. Model. Mech. 2018, 11, dmm035352. [Google Scholar] [CrossRef] [PubMed]
- Skryabin, B.V.; Kummerfeld, D.-M.; Gubar, L.; Seeger, B.; Kaiser, H.; Stegemann, A.; Roth, J.; Meuth, S.G.; Pavenstädt, H.; Sherwood, J.; et al. Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9–mediated genome editing events. Sci. Adv. 2020, 6, eaax2941. [Google Scholar] [CrossRef]
- Pu, X.; Young, A.P.; Kubisch, H.M. Production of transgenic mice by pronuclear microinjection. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; Volume 1874, pp. 17–41. [Google Scholar]
- Gordon, J.W.; Scangos, G.A.; Plotkin, D.J.; Barbosa, J.A.; Ruddle, F.H. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 7380–7384. [Google Scholar] [CrossRef] [PubMed]
- Brinster, R.L.; Chen, H.Y.; Trumbauer, M.; Senear, A.W.; Warren, R.; Palmiter, R.D. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell 1981, 27, 223–231. [Google Scholar] [CrossRef]
- Smith, K. Theoretical mechanisms in targeted and random integration of transgene DNA. Reprod. Nutr. Dev. 2001, 41, 465–485. [Google Scholar] [CrossRef][Green Version]
- Nakanishi, T.; Kuroiwa, A.; Yamada, S.; Isotani, A.; Yamashita, A.; Tairaka, A.; Hayashi, T.; Takagi, T.; Ikawa, M.; Matsuda, Y.; et al. Fish analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics 2002, 80, 564–574. [Google Scholar] [CrossRef]
- Palmiter, R.D.; Chen, H.Y.; Brinster, R.L. Differential regulation of metallothionein-thymidine kinase fusion genes in transgenic mice and their offspring. Cell 1982, 29, 701–710. [Google Scholar] [CrossRef]
- Brinster, R.L.; Ritchie, K.A.; Hammer, R.E.; O’Brien, R.L.; Arp, B.; Storb, U. Expression of a microinjected immunoglobulin gene in the spleen of transgenic mice. Nature 1983, 306, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Khillan, J.S.; Overbeek, P.A.; Westphal, H. Drosophila P element integration in the mouse. Dev. Biol. 1985, 109, 247–250. [Google Scholar] [CrossRef]
- Overbeek, P.A.; Lai, S.-P.; Van Quill, K.R.; Westphal, H. Tissue-Specific Expression in Transgenic Mice of a Fused Gene Containing RSV Terminal Sequences. Science 1986, 231, 1574–1577. [Google Scholar] [CrossRef]
- Folger, K.R.; Wong, E.A.; Wahl, G.; Capecchi, M.R. Patterns of integration of DNA microinjected into cultured mammalian cells: Evidence for homologous recombination between injected plasmid DNA molecules. Mol. Cell. Biol. 1982, 2, 1372–1387. [Google Scholar] [CrossRef]
- Folger, K.R.; Thomas, K.; Capecchi, M.R. Nonreciprocal exchanges of information between DNA duplexes coinjected into mammalian cell nuclei. Mol. Cell. Biol. 1985, 5, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Brouillette, S.; Chartrand, P. Intermolecular recombination assay for mammalian cells that produces recombinants carrying both homologous and nonhomologous junctions. Mol. Cell. Biol. 1987, 7, 2248–2255. [Google Scholar] [CrossRef]
- Gening, L.; Takeshita, M.; Levine, R.L.; Peden, K.W.; Grollman, A.P. Extrachromosomal unequal homologous recombination and gene conversion in simian kidney cells: Effects of UV damage. Mutat. Res.-DNA Repair 1998, 407, 11–24. [Google Scholar] [CrossRef]
- Rohan, R.M.; King, D.; Frels, W.I. Direct sequencing of PCR-amplified junction fragments from tandemly repeated transgenes. Nucleic Acids Res. 1990, 18, 6089–6095. [Google Scholar] [CrossRef]
- Brinster, R.L.; Chen, H.Y.; Trumbauer, M.E.; Yagle, M.K.; Palmiter, R.D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc. Natl. Acad. Sci. USA 1985, 82, 4438–4442. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Sasaki, H.; Seki, R.; Sakaki, Y. Mechanism of chromosomal integration of transgenes in microinjected mouse eggs: Sequence analysis of genome-transgene and transgene-transgene junctions at two loci. Gene 1993, 128, 197–202. [Google Scholar] [CrossRef]
- Würtele, H.; Gusew, N.; Lussier, R.; Chartrand, P. Characterization of in vivo recombination activities in the mouse embryo. Mol. Genet. Genomics 2005, 273, 252–263. [Google Scholar] [CrossRef]
- Pieper, F.R.; Wit, I.C.M.D.; Pronk, A.C.J.; Kooiman, P.M.; Strijker, R.; Krimpenfort, P.J.A.; Nuyens, J.H.; Boer, H.A.D. Efficient generation of functional transgenes by homologous recombination in murine zygotes. Nucleic Acids Res. 1992, 20, 1259–1264. [Google Scholar] [CrossRef][Green Version]
- Migchielsen, A.A.J.; Breuer, M.L.; Hershfield, M.S.; Valerio, D. Full genetic rescue of adenosine deaminase-deficient mice through introduction of the human gene. Hum. Mol. Genet. 1996, 5, 1523–1532. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tacken, P.J.; Zee, A.V.D.; Beumer, T.L.; Florijn, R.J.; Gijpels, M.J.J.; Havekes, L.M.; Frants, R.R.; Dijk, K.W.V.; Hofker, M.H. Effective generation of very low density lipoprotein receptor transgenic mice by overlapping genomic DNA fragments: High testis expression and disturbed spermatogenesis. Transgenic Res. 2001, 10, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Ramsden, D.A.; Carvajal-Garcia, J.; Gupta, G.P. Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nat. Rev. Mol. Cell Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Truong, L.N.; Li, Y.; Shi, L.Z.; Hwang, P.Y.-H.; He, J.; Wang, H.; Razavian, N.; Berns, M.W.; Wu, X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7720–7725. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Fishman, V.; Yunusova, A.; Korablev, A.; Serova, I.; Skryabin, B.V.; Rozhdestvensky, T.S.; Battulin, N. DNA barcoding reveals that injected transgenes are predominantly processed by homologous recombination in mouse zygote. Nucleic Acids Res. 2019, 48, 719–735. [Google Scholar] [CrossRef]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef]
- Schimmel, J.; Kool, H.; van Schendel, R.; Tijsterman, M. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 2017, 36, 3634–3649. [Google Scholar] [CrossRef]
- Dai, J.; Cui, X.; Zhu, Z.; Hu, W. Non-Homologous End Joining Plays a Key Role in Transgene Concatemer Formation in Transgenic Zebrafish Embryos. Int. J. Biol. Sci. 2010, 6, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, K.M.; Sun, C.-W.; Higgins, N.P.; Townes, T.M. End joining of genomic DNA and transgene DNA in fertilized mouse eggs. Gene 1995, 165, 173–181. [Google Scholar] [CrossRef]
- Suemizu, H.; Muguruma, K.; Maruyama, C.; Tomisawa, M.; Kimura, M.; Hioki, K.; Shimozawa, N.; Ohnishi, Y.; Tamaoki, N.; Nomura, T. Transgene stability and features of rasH2 mice as an animal model for short-term carcinogenicity testing. Mol. Carcinog. 2002, 34, 1–9. [Google Scholar] [CrossRef]
- Zelensky, A.N.; Schimmel, J.; Kool, H.; Kanaar, R.; Tijsterman, M. Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 2017, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Li, D.; Gou, K. Homologous illegitimate random integration of foreign DNA into the X chromosome of a transgenic mouse line. BMC Mol. Biol. 2010, 11, 58. [Google Scholar] [CrossRef]
- Nakade, S.; Tsubota, T.; Sakane, Y.; Kume, S.; Sakamoto, N.; Obara, M.; Daimon, T.; Sezutsu, H.; Yamamoto, T.; Sakuma, T.; et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014, 5, 5560. [Google Scholar] [CrossRef]
- Masumura, K.; Sakamoto, Y.; Kumita, W.; Honma, M.; Nishikawa, A.; Nohmi, T. Genomic integration of lambda EG10 transgene in gpt delta transgenic rodents. Genes Environ. 2015, 37, 24. [Google Scholar] [CrossRef]
- Mark, W.H.; Signorelli, K.; Blum, M.; Kwee, L.; Lacy, E. Genomic structure of the locus associated with an insertional mutation in line 4 transgenic mice. Genomics 1992, 13, 159–166. [Google Scholar] [CrossRef]
- Murnane, J.P.; Yu, L.C. Acquisition of telomere repeat sequences by transfected DNA integrated at the site of a chromosome break. Mol. Cell. Biol. 1993, 13, 977–983. [Google Scholar] [CrossRef]
- Nicholls, P.K.; Bellott, D.W.; Cho, T.-J.; Pyntikova, T.; Page, D.C. Locating and Characterizing a Transgene Integration Site by Nanopore Sequencing. G3 Genes|Genomes|Genet. 2019, 9, 1481–1486. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.K.; Park, J.S.; Lee, C.S.; Yeom, Y.I.; Chung, A.S.; Lee, K.K. Efficient integration of short interspersed element-flanked foreign DNA via homologous recombination. J. Biol. Chem. 1999, 274, 36585–36591. [Google Scholar] [CrossRef]
- Ganapathiraju, M.K.; Subramanian, S.; Chaparala, S.; Karunakaran, K.B. A reference catalog of DNA palindromes in the human genome and their variations in 1000 Genomes. Hum. Genome Var. 2020, 7, 40. [Google Scholar] [CrossRef]
- Ditch, S.; Sammarco, M.C.; Banerjee, A.; Grabczyk, E. Progressive GAA·TTC repeat expansion in human cell lines. PLoS Genet. 2009, 5, e1000704. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Kockler, Z.; Evans, R.; Downing, B.D.; Malkova, A. Single-strand annealing between inverted DNA repeats: Pathway choice, participating proteins, and genome destabilizing consequences. PLoS Genet. 2018, 14, e1007543. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.S.; Mitsuda, S.H.; Shimizu, N. How a replication origin and matrix attachment region accelerate gene amplification under replication stress in mammalian cells. PLoS ONE 2014, 9, e103439. [Google Scholar] [CrossRef] [PubMed]
- Hasty, P.; Montagna, C. Chromosomal rearrangements in cancer. Mol. Cell. Oncol. 2014, 1, e29904. [Google Scholar] [CrossRef]
- Trombetta, B.; Cruciani, F. Y chromosome palindromes and gene conversion. Hum. Genet. 2017, 136, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.K.; Bellott, D.W.; Cho, T.J.; Skaletsky, H.; Hughes, J.F.; Pyntikova, T.; Page, D.C. Large palindromes on the primate X Chromosome are preserved by natural selection. Genome Res. 2021, 31, 1337–1352. [Google Scholar] [CrossRef]
- Inagaki, H.; Ohye, T.; Kogo, H.; Tsutsumi, M.; Kato, T.; Tong, M.; Emanuel, B.S.; Kurahashi, H. Two sequential cleavage reactions on cruciform DNA structures cause palindrome-mediated chromosomal translocations. Nat. Commun. 2013, 4, 1592. [Google Scholar] [CrossRef]
- Inagaki, H.; Kato, T.; Tsutsumi, M.; Ouchi, Y.; Ohye, T.; Kurahashi, H. Palindrome-mediated translocations in humans: A new mechanistic model for gross chromosomal rearrangements. Front. Genet. 2016, 7, 125. [Google Scholar] [CrossRef]
- Voineagu, I.; Narayanan, V.; Lobachev, K.S.; Mirkin, S.M. Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 9936–9941. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vasquez, K.M. Effects of replication and transcription on DNA Structure-Related genetic instability. Genes 2017, 8, 17. [Google Scholar] [CrossRef]
- Holkers, M.; De Vries, A.A.F.; Gonçalves, M.A.F.V. Nonspaced inverted DNA repeats are preferential targets for homology-directed gene repair in mammalian cells. Nucleic Acids Res. 2012, 40, 1984–1999. [Google Scholar] [CrossRef]
- Chen, H.; Lisby, M.; Symington, L. RPA Coordinates DNA End Resection and Prevents Formation of DNA Hairpins. Mol. Cell 2013, 50, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Hároníková, L.; Liao, J.C.C.; Fridrichová, H.; Jagelská, E.B. Strong preference of BRCA1 protein to topologically constrained non-B DNA structures. BMC Mol. Biol. 2016, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Coufal, J. Recognition of local DNA structures by p53 protein. Int. J. Mol. Sci. 2017, 18, 375. [Google Scholar] [CrossRef]
- Lobachev, K.S.; Gordenin, D.A.; Resnick, M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 2002, 108, 183–193. [Google Scholar] [CrossRef]
- Ait Saada, A.; Costa, A.B.; Sheng, Z.; Guo, W.; Haber, J.E.; Lobachev, K.S. Structural parameters of palindromic repeats determine the specificity of nuclease attack of secondary structures. Nucleic Acids Res. 2021, 49, 3932–3947. [Google Scholar] [CrossRef]
- Rass, U.; Compton, S.A.; Matos, J.; Singleton, M.R.; Ip, S.C.Y.; Blanco, M.G.; Griffith, J.D.; West, S.C. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 2010, 24, 1559–1569. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kirschner, K.; Melton, D.W. Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Res. 2010, 30, 3223–3232. [Google Scholar]
- Halabi, A.; Fuselier, K.T.B.; Grabczyk, E. GAA•TTC repeat expansion in human cells is mediated by mismatch repair complex MutL and depends upon the endonuclease domain in MLH3 isoform one. Nucleic Acids Res. 2018, 46, 4022–4032. [Google Scholar] [CrossRef]
- Svetec Miklenić, M.; Gatalica, N.; Matanović, A.; Žunar, B.; Štafa, A.; Lisnić, B.; Svetec, I.K. Size-dependent antirecombinogenic effect of short spacers on palindrome recombinogenicity. DNA Repair 2020, 90, 102848. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Akgün, E.; Jasin, M. Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc. Natl. Acad. Sci. USA 2001, 98, 8326–8333. [Google Scholar] [CrossRef]
- Akgün, E.; Zahn, J.; Baumes, S.; Brown, G.; Liang, F.; Romanienko, P.J.; Lewis, S.; Jasin, M. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 1997, 17, 5559–5570. [Google Scholar] [CrossRef][Green Version]
- Cunningham, L.A.; Coté, A.G.; Cam-Ozdemir, C.; Lewis, S.M. Rapid, Stabilizing Palindrome Rearrangements in Somatic Cells by the Center-Break Mechanism. Mol. Cell. Biol. 2003, 23, 8740–8750. [Google Scholar] [CrossRef]
- Mikhailov, K.V.; Efeykin, B.D.; Panchin, A.Y.; Knorre, D.A.; Logacheva, M.D.; Penin, A.A.; Muntyan, M.S.; Nikitin, M.A.; Popova, O.V.; Zanegina, O.N.; et al. Coding palindromes in mitochondrial genes of Nematomorpha. Nucleic Acids Res. 2019, 47, 6858–6870. [Google Scholar] [CrossRef]
- Daley, J.M.; Gaines, W.A.; Kwon, Y.; Sung, P. Regulation of DNA Pairing in Homologous Recombination. Cold Spring Harb. Perspect. Biol. 2014, 6, a017954. [Google Scholar] [CrossRef]
- Yong, C.S.M.; Sharkey, J.; Duscio, B.; Venville, B.; Wei, W.-Z.; Jones, R.F.; Slaney, C.Y.; Mir Arnau, G.; Papenfuss, A.T.; Schröder, J.; et al. Embryonic Lethality in Homozygous Human Her-2 Transgenic Mice Due to Disruption of the Pds5b Gene. PLoS ONE 2015, 10, e0136817. [Google Scholar] [CrossRef]
- Chiang, C.; Jacobsen, J.C.; Ernst, C.; Hanscom, C.; Heilbut, A.; Blumenthal, I.; Mills, R.E.; Kirby, A.; Lindgren, A.M.; Rudiger, S.R.; et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 2012, 44, 390–397. [Google Scholar] [CrossRef]
- Ranjha, L.; Howard, S.M.; Cejka, P. Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 2018, 127, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Bizard, A.H.; Hickson, I.D. The dissolution of double Holliday junctions. Cold Spring Harb. Perspect. Biol. 2014, 6, a016477. [Google Scholar] [CrossRef]
- Gutierrez-Triana, J.A.; Tavhelidse, T.; Thumberger, T.; Thomas, I.; Wittbrodt, B.; Kellner, T.; Anlas, K.; Tsingos, E.; Wittbrodt, J. Efficient single-copy HDR by 5′ modified long dsDNA donors. Elife 2018, 7, e39468. [Google Scholar] [CrossRef]
- Canaj, H.; Hussmann, J.A.; Li, H.; Beckman, K.A.; Goodrich, L.; Cho, N.H.; Li, Y.J.; Santos, D.A.; McGeever, A.; Stewart, E.M.; et al. Deep profiling reveals substantial heterogeneity of integration outcomes in CRISPR knock-in experiments. bioRxiv 2019. [Google Scholar] [CrossRef]
- Hartlerode, A.J.; Willis, N.A.; Rajendran, A.; Manis, J.P.; Scully, R. Complex Breakpoints and Template Switching Associated with Non-canonical Termination of Homologous Recombination in Mammalian Cells. PLOS Genet. 2016, 12, e1006410. [Google Scholar] [CrossRef]
- Wyatt, D.W.; Feng, W.; Conlin, M.P.; Yousefzadeh, M.J.; Roberts, S.A.; Mieczkowski, P.; Wood, R.D.; Gupta, G.P.; Ramsden, D.A. Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks. Mol. Cell 2016, 63, 662–673. [Google Scholar] [CrossRef]
- Shibata, A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat. Res. Mol. Mech. Mutagen. 2017, 803–805, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Paix, A.; Folkmann, A.; Goldman, D.H.; Kulaga, H.; Grzelak, M.J.; Rasoloson, D.; Paidemarry, S.; Green, R.; Reed, R.R.; Seydoux, G. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl. Acad. Sci. USA 2017, 114, E10745–E10754. [Google Scholar] [CrossRef]
- Verma, P.; Greenberg, R.A. Noncanonical views of homology-directed DNA repair. Genes Dev. 2016, 30, 1138–1154. [Google Scholar] [CrossRef]
- Piazza, A.; Heyer, W.-D. Homologous Recombination and the Formation of Complex Genomic Rearrangements. Trends Cell Biol. 2019, 29, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Wright, W.D.; Heyer, W.-D. Multi-invasions Are Recombination Byproducts that Induce Chromosomal Rearrangements. Cell 2017, 170, 760–773.e15. [Google Scholar] [CrossRef]
- Zhou, Y.; Caron, P.; Legube, G.; Paull, T.T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014, 42, e19. [Google Scholar] [CrossRef] [PubMed]
- Fus-Kujawa, A.; Prus, P.; Bajdak-Rusinek, K.; Teper, P.; Gawron, K.; Kowalczuk, A.; Sieron, A.L. An Overview of Methods and Tools for Transfection of Eukaryotic Cells in vitro. Front. Bioeng. Biotechnol. 2021, 9, 634. [Google Scholar] [CrossRef]
- Dorer, D.R.; Henikoff, S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 1994, 77, 993–1002. [Google Scholar] [CrossRef]
- Catalanotto, C.; Nolan, T.; Cogoni, C. Homology effects in Neurospora crassa. FEMS Microbiol. Lett. 2006, 254, 182–189. [Google Scholar] [CrossRef][Green Version]
- Ye, F.; Signer, E.R. RIGS (repeat-induced gene silencing) in Arabidopsis is transcriptional and alters chromatin configuration. Proc. Natl. Acad. Sci. USA 1996, 93, 10881–10886. [Google Scholar] [CrossRef]
- Henikoff, S. Conspiracy of silence among repeated transgenes. BioEssays 1998, 20, 532–535. [Google Scholar] [CrossRef]
- Garrick, D.; Fiering, S.; Martin, D.I.K.; Whitelaw, E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998, 8, 56–59. [Google Scholar] [CrossRef]
- Girton, J.R.; Johansen, K.M. Chapter 1 Chromatin Structure and the Regulation of Gene Expression: The Lessons of PEV in Drosophila. Adv. Genet. 2008, 61, 1–43. [Google Scholar]
- Sidorenko, L.V.; Lee, T.F.; Woosley, A.; Moskal, W.A.; Bevan, S.A.; Merlo, P.A.O.; Walsh, T.A.; Wang, X.; Weaver, S.; Glancy, T.P.; et al. GC-rich coding sequences reduce transposon-like, small RNA-mediated transgene silencing. Nat. Plants 2017, 3, 875–884. [Google Scholar] [CrossRef]
- Fu, X.; Kohli, A.; Twyman, R.M.; Christou, P. Alternative silencing effects involve distinct types of non-spreading cytosine methylation at a three-gene, single-copy transgenic locus in rice. Mol. Gen. Genet. 2000, 263, 106–118. [Google Scholar] [CrossRef]
- Kostyrko, K.; Neuenschwander, S.; Junier, T.; Regamey, A.; Iseli, C.; Schmid-Siegert, E.; Bosshard, S.; Majocchi, S.; Le Fourn, V.; Girod, P.-A.; et al. MAR-Mediated transgene integration into permissive chromatin and increased expression by recombination pathway engineering. Biotechnol. Bioeng. 2017, 114, 384–396. [Google Scholar] [CrossRef]
- Eszterhas, S.K.; Bouhassira, E.E.; Martin, D.I.K.; Fiering, S. Transcriptional Interference by Independently Regulated Genes Occurs in Any Relative Arrangement of the Genes and Is Influenced by Chromosomal Integration Position. Mol. Cell. Biol. 2002, 22, 469–479. [Google Scholar] [CrossRef]
- Carver, J.; Ng, D.; Zhou, M.; Ko, P.; Zhan, D.; Yim, M.; Shaw, D.; Snedecor, B.; Laird, M.W.; Lang, S.; et al. Maximizing antibody production in a targeted integration host by optimization of subunit gene dosage and position. Biotechnol. Prog. 2020, 36, e2967. [Google Scholar] [CrossRef]
- Zimak, J.; Wagoner, Z.W.; Nelson, N.; Waechtler, B.; Schlosser, H.; Kopecky, M.; Wu, J.; Zhao, W. Epigenetic silencing directs expression heterogeneity of stably integrated multi-transcript unit genetic circuits. Sci. Rep. 2021, 11, 2424. [Google Scholar] [CrossRef]
- Rosser, J.M.; An, W. Repeat-induced gene silencing of L1 transgenes is correlated with differential promoter methylation. Gene 2010, 456, 15–23. [Google Scholar] [CrossRef][Green Version]
- Williams, A.; Harker, N.; Ktistaki, E.; Veiga-fernandes, H.; Roderick, K.; Tolaini, M.; Norton, T.; Williams, K.; Kioussis, D. Position effect variegation and imprinting of transgenes in lymphocytes. Nucleic Acids Res. 2008, 36, 2320–2329. [Google Scholar] [CrossRef]
- Alonso-González, L.; Couldrey, C.; Meinhardt, M.W.; Cole, S.A.; Wells, D.N.; Laible, G. Primary transgenic bovine cells and their rejuvenated cloned equivalents show transgene-specific epigenetic differences. PLoS ONE 2012, 7, e35619. [Google Scholar] [CrossRef]
- Ordovás, L.; Boon, R.; Pistoni, M.; Chen, Y.; Wolfs, E.; Guo, W.; Sambathkumar, R.; Bobis-Wozowicz, S.; Helsen, N.; Vanhove, J.; et al. Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Rep. 2015, 5, 918–931. [Google Scholar] [CrossRef]
- Gödecke, N.; Zha, L.; Spencer, S.; Behme, S.; Riemer, P.; Rehli, M.; Hauser, H.; Wirth, D. Controlled re-activation of epigenetically silenced Tet promoter-driven transgene expression by targeted demethylation. Nucleic Acids Res. 2017, 45, e147. [Google Scholar] [CrossRef]
- Klatt, D.; Cheng, E.; Hoffmann, D.; Santilli, G.; Thrasher, A.J.; Brendel, C.; Schambach, A. Differential Transgene Silencing of Myeloid-Specific Promoters in the AAVS1 Safe Harbor Locus of Induced Pluripotent Stem Cell-Derived Myeloid Cells. Hum. Gene Ther. 2020, 31, 199–210. [Google Scholar] [CrossRef]
- Luo, Z.; Chen, Z. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 2007, 19, 943–958. [Google Scholar] [CrossRef]
- Pérez-González, A.; Caro, E. Effect of transcription terminator usage on the establishment of transgene transcriptional gene silencing. BMC Res. Notes 2018, 11, 511. [Google Scholar] [CrossRef]
- Klattenhoff, C.; Xi, H.; Li, C.; Lee, S.; Xu, J.; Khurana, J.S.; Zhang, F.; Schultz, N.; Koppetsch, B.S.; Nowosielska, A.; et al. The Drosophila HP1 Homolog Rhino Is Required for Transposon Silencing and piRNA Production by Dual-Strand Clusters. Cell 2009, 138, 1137–1149. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Hashiguchi, N.; Janicki, S.M.; Tumbar, T.; Belmont, A.S.; Spector, D.L. Visualization of gene activity in living cells. Nat. Cell Biol. 2000, 2, 871–878. [Google Scholar] [CrossRef]
- Wang, F.; Koyama, N.; Nishida, H.; Haraguchi, T.; Reith, W.; Tsukamoto, T. The Assembly and Maintenance of Heterochromatin Initiated by Transgene Repeats Are Independent of the RNA Interference Pathway in Mammalian Cells. Mol. Cell. Biol. 2006, 26, 4028–4040. [Google Scholar] [CrossRef]
- Mazur, A.K.; Gladyshev, E. Partition of Repeat-Induced Point Mutations Reveals Structural Aspects of Homologous DNA-DNA Pairing. Biophys. J. 2018, 115, 605–615. [Google Scholar] [CrossRef]
- McBurney, M.W.; Mai, T.; Yang, X.; Jardine, K. Evidence for Repeat-Induced Gene Silencing in Cultured Mammalian Cells: Inactivation of Tandem Repeats of Transfected Genes. Exp. Cell Res. 2002, 274, 1–8. [Google Scholar] [CrossRef]
- Calero-Garcia, M.; Gaspar, H.B. Gene-ectomy: Gene ablation with CRISPR/Cas9 in human hematopoietic cells. Cell Stem Cell 2014, 15, 529–530. [Google Scholar] [CrossRef][Green Version]
- Osterlehner, A.; Simmeth, S.; Göpfert, U. Promoter methylation and transgene copy numbers predict unstable protein production in recombinant chinese hamster ovary cell lines. Biotechnol. Bioeng. 2011, 108, 2670–2681. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Luque, F.J.; Kempen, M.J.H.C.; Gerdes, P.; Vargas-Landin, D.B.; Richardson, S.R.; Troskie, R.L.; Jesuadian, J.S.; Cheetham, S.W.; Carreira, P.E.; Salvador-Palomeque, C.; et al. LINE-1 Evasion of Epigenetic Repression in Humans. Mol. Cell 2019, 75, 590–604.e12. [Google Scholar] [CrossRef] [PubMed]
- Fukuma, M.; Ganmyo, Y.; Miura, O.; Ohyama, T.; Shimizu, N. Cloning and characterization of a human genomic sequence that alleviates repeat-induced gene silencing. PLoS ONE 2016, 11, e0153338. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Xiang, X.; Chen, Q.; Pan, X.; Cheng, H.; Wang, F. HP1 cooperates with CAF-1 to compact heterochromatic transgene repeats in mammalian cells. Sci. Rep. 2018, 8, 14141. [Google Scholar] [CrossRef]
- Janicki, S.M.; Tsukamoto, T.; Salghetti, S.E.; Tansey, W.P.; Sachidanandam, R.; Prasanth, K.V.; Ried, T.; Shav-Tal, Y.; Bertrand, E.; Singer, R.H.; et al. From silencing to gene expression: Real-time analysis in single cells. Cell 2004, 116, 683–698. [Google Scholar] [CrossRef]
- Taniguchi, R.; Utani, K.; Thakur, B.; Ishine, K.; Aladjem, M.I.; Shimizu, N. SIRT1 stabilizes extrachromosomal gene amplification and contributes to repeat-induced gene silencing. J. Biol. Chem. 2021, 296, 100356. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Wu, J.; Cao, T.; Zhi, S.; Chen, Y.; Aagaard, L.; Zhen, P.; Huang, Y.; Zhong, J.; Huang, J. Methylation silencing and reactivation of exogenous genes in lentivirus-mediated transgenic mice. Transgenic Res. 2021, 30, 63–76. [Google Scholar] [CrossRef]
- Cain-Hom, C.; Splinter, E.; van Min, M.; Simonis, M.; van de Heijning, M.; Martinez, M.; Asghari, V.; Cox, J.C.; Warming, S. Efficient mapping of transgene integration sites and local structural changes in Cre transgenic mice using targeted locus amplification. Nucleic Acids Res. 2017, 45, gkw1329. [Google Scholar] [CrossRef][Green Version]
- Goodwin, L.O.; Splinter, E.; Davis, T.L.; Urban, R.; He, H.; Braun, R.E.; Chesler, E.J.; Kumar, V.; van Min, M.; Ndukum, J.; et al. Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res. 2019, 29, 494–505. [Google Scholar] [CrossRef]
- Brueckner, L.; Zhao, P.A.; Schaik, T.; Leemans, C.; Sima, J.; Peric-Hupkes, D.; Gilbert, D.M.; Steensel, B. Local rewiring of genome–nuclear lamina interactions by transcription. EMBO J. 2020, 39, e103159. [Google Scholar] [CrossRef]
- Maksimenko, O.; Gasanov, N.B.; Georgiev, P. Regulatory Elements in Vectors for Efficient Generation of Cell Lines Producing Target Proteins. Acta Nat. 2015, 7, 15–26. [Google Scholar] [CrossRef]
- Lau, S.; Jardine, K.; McBurney, M.W. DNA methylation pattern of a tandemly repeated lacZ transgene indicates that most copies are silent. Dev. Dyn. 1999, 215, 126–138. [Google Scholar] [CrossRef]
- Xu, L.; Seki, M. Recent advances in the detection of base modifications using the Nanopore sequencer. J. Hum. Genet. 2020, 65, 25–33. [Google Scholar] [CrossRef]
- Herzog, V.A.; Reichholf, B.; Neumann, T.; Rescheneder, P.; Bhat, P.; Burkard, T.R.; Wlotzka, W.; von Haeseler, A.; Zuber, J.; Ameres, S.L. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 2017, 14, 1198–1204. [Google Scholar] [CrossRef]
- Hafford-Tear, N.J.; Tsai, Y.C.; Sadan, A.N.; Sanchez-Pintado, B.; Zarouchlioti, C.; Maher, G.J.; Liskova, P.; Tuft, S.J.; Hardcastle, A.J.; Clark, T.A.; et al. CRISPR/Cas9-targeted enrichment and long-read sequencing of the Fuchs endothelial corneal dystrophy–associated TCF4 triplet repeat. Genet. Med. 2019, 21, 2092–2102. [Google Scholar] [CrossRef]
- Watson, C.M.; Crinnion, L.A.; Hewitt, S.; Bates, J.; Robinson, R.; Carr, I.M.; Sheridan, E.; Adlard, J.; Bonthron, D.T. Cas9-based enrichment and single-molecule sequencing for precise characterization of genomic duplications. Lab. Investig. 2020, 100, 135–146. [Google Scholar] [CrossRef]
- McDonald, T.L.; Zhou, W.; Castro, C.P.; Mumm, C.; Switzenberg, J.A.; Mills, R.E.; Boyle, A.P. Cas9 targeted enrichment of mobile elements using nanopore sequencing. Nat. Commun. 2021, 12, 3586. [Google Scholar] [CrossRef]
- Wilkie, T.M.; Palmiter, R.D. Analysis of the integrant in MyK-103 transgenic mice in which males fail to transmit the integrant. Mol. Cell. Biol. 1987, 7, 1646–1655. [Google Scholar] [CrossRef] [PubMed]
- Sailer, S.; Coassin, S.; Lackner, K.; Fischer, C.; McNeill, E.; Streiter, G.; Kremser, C.; Maglione, M.; Green, C.M.; Moralli, D.; et al. When the genome bluffs: A tandem duplication event during generation of a novel Agmo knockout mouse model fools routine genotyping. Cell Biosci. 2021, 11, 54. [Google Scholar] [CrossRef]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef]
- Payne, A.; Holmes, N.; Rakyan, V.; Loose, M. Bulkvis: A graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 2019, 35, 2193–2198. [Google Scholar] [CrossRef] [PubMed]
- Van Haasteren, J.; Munis, A.M.; Gill, D.R.; Hyde, S.C. Genome-wide integration site detection using Cas9 enriched amplification-free long-range sequencing. Nucleic Acids Res. 2021, 49, e16. [Google Scholar] [CrossRef]
- Li, S.; Jia, S.; Hou, L.; Nguyen, H.; Sato, S.; Holding, D.; Cahoon, E.; Zhang, C.; Clemente, T.; Yu, B. Mapping of transgenic alleles in soybean using a nanopore-based sequencing strategy. J. Exp. Bot. 2019, 70, 3825–3833. [Google Scholar] [CrossRef] [PubMed]
- Slesarev, A.; Viswanathan, L.; Tang, Y.; Borgschulte, T.; Achtien, K.; Razafsky, D.; Onions, D.; Chang, A.; Cote, C. CRISPR/Cas9 targeted CAPTURE of mammalian genomic regions for characterization by NGS. Sci. Rep. 2019, 9, 3587. [Google Scholar] [CrossRef]
- Gilpatrick, T.; Lee, I.; Graham, J.E.; Raimondeau, E.; Bowen, R.; Heron, A.; Downs, B.; Sukumar, S.; Sedlazeck, F.J.; Timp, W. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020, 38, 433–438. [Google Scholar] [CrossRef]
- Buck, D.; Weirather, J.L.; de Cesare, M.; Wang, Y.; Piazza, P.; Sebastiano, V.; Wang, X.J.; Au, K.F. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Research 2017, 6, 100. [Google Scholar] [CrossRef]
- Delahaye, C.; Nicolas, J. Sequencing DNA with nanopores: Troubles and biases. PLoS ONE 2021, 16, e0257521. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef]
- Spealman, P.; Burrell, J.; Gresham, D. Inverted duplicate DNA sequences increase translocation rates through sequencing nanopores resulting in reduced base calling accuracy. Nucleic Acids Res. 2020, 48, 4940–4945. [Google Scholar] [CrossRef] [PubMed]
- Khokhlova, E.V.; Fesenko, Z.S.; Sopova, J.V.; Leonova, E.I. Features of dna repair in the early stages of mammalian embryonic development. Genes 2020, 1, 1138. [Google Scholar] [CrossRef]
- Wilde, J.J.; Aida, T.; del Rosario, R.C.H.; Kaiser, T.; Qi, P.; Wienisch, M.; Zhang, Q.; Colvin, S.; Feng, G. Efficient embryonic homozygous gene conversion via RAD51-enhanced interhomolog repair. Cell 2021, 184, 3267–3280.e18. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Inoue, K.-I.; Furuta, Y.; Kiyonari, H. Pronuclear Microinjection during S-Phase Increases the Efficiency of CRISPR-Cas9-Assisted Knockin of Large DNA Donors in Mouse Zygotes. Cell Rep. 2020, 31, 107653. [Google Scholar] [CrossRef]
- Schep, R.; Brinkman, E.K.; Leemans, C.; Vergara, X.; van der Weide, R.H.; Morris, B.; van Schaik, T.; Manzo, S.G.; Peric-Hupkes, D.; van den Berg, J.; et al. Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance. Mol. Cell 2021, 81, 2216–2230.e10. [Google Scholar] [CrossRef]
- Roidos, P.; Sungalee, S.; Benfatto, S.; Serçin, Ö.; Stütz, A.M.; Abdollahi, A.; Mauer, J.; Zenke, F.T.; Korbel, J.O.; Mardin, B.R. A scalable CRISPR/Cas9-based fluorescent reporter assay to study DNA double-strand break repair choice. Nat. Commun. 2020, 1, 1–15. [Google Scholar] [CrossRef]
- Miyaoka, Y.; Mayerl, S.J.; Chan, A.H.; Conklin, B.R. Detection and Quantification of HDR and NHEJ Induced by Genome Editing at Endogenous Gene Loci Using Droplet Digital PCR. Methods Mol. Biol. 2018, 1768, 349–362. [Google Scholar] [CrossRef]
- Watry, H.L.; Feliciano, C.M.; Gjoni, K.; Takahashi, G.; Miyaoka, Y.; Conklin, B.R.; Judge, L.M. Rapid, precise quantification of large DNA excisions and inversions by ddPCR. Sci. Rep. 2020, 10, 14896. [Google Scholar] [CrossRef]
- Gu, B.; Posfai, E.; Rossant, J. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol. 2018, 36, 632–637. [Google Scholar] [CrossRef]
- Yesbolatova, A.; Saito, Y.; Kitamoto, N.; Makino-Itou, H.; Ajima, R.; Nakano, R.; Nakaoka, H.; Fukui, K.; Gamo, K.; Tominari, Y.; et al. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 2020, 11, 5701. [Google Scholar] [CrossRef]
- Morozov, V.; Wawrousek, E.F. Single-strand DNA-mediated targeted mutagenesis of genomic DNA in early mouse embryos is stimulated by Rad51/54 and by Ku70/86 inhibition. Gene Ther. 2008, 15, 468–472. [Google Scholar] [CrossRef][Green Version]
- Canny, M.D.; Moatti, N.; Wan, L.C.K.; Fradet-Turcotte, A.; Krasner, D.; Mateos-Gomez, P.A.; Zimmermann, M.; Orthwein, A.; Juang, Y.C.; Zhang, W.; et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat. Biotechnol. 2018, 36, 95–102. [Google Scholar] [CrossRef]
- Nambiar, T.S.; Billon, P.; Diedenhofen, G.; Hayward, S.B.; Taglialatela, A.; Cai, K.; Huang, J.-W.; Leuzzi, G.; Cuella-Martin, R.; Palacios, A.; et al. Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat. Commun. 2019, 10, 3395. [Google Scholar] [CrossRef]
- Cao, X.; Kouyama-Suzuki, E.; Pang, B.; Kurihara, T.; Mori, T.; Yanagawa, T.; Shirai, Y.; Tabuchi, K. Inhibition of DNA ligase IV enhances the CRISPR/Cas9-mediated knock-in efficiency in mouse brain neurons. Biochem. Biophys. Res. Commun. 2020, 533, 449–457. [Google Scholar] [CrossRef]
- Yu, C.; Liu, Y.; Ma, T.; Liu, K.; Xu, S.; Zhang, Y.; Liu, H.; La Russa, M.; Xie, M.; Ding, S.; et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 2015, 16, 142–147. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Minev, D.; Guerra, R.; Kishi, J.Y.; Smith, C.; Krieg, E.; Said, K.; Hornick, A.; Sasaki, H.M.; Filsinger, G.; Beliveau, B.J.; et al. Rapid in vitro production of single-stranded DNA. Nucleic Acids Res. 2019, 47, 11956–11962. [Google Scholar] [CrossRef]
- Inoue, Y.U.; Morimoto, Y.; Yamada, M.; Kaneko, R.; Shimaoka, K.; Oki, S.; Hotta, M.; Asami, J.; Koike, E.; Hori, K.; et al. An optimized preparation method for long ssDNA donors to facilitate quick knock-in mouse generation. Cells 2021, 10, 1076. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smirnov, A.; Battulin, N. Concatenation of Transgenic DNA: Random or Orchestrated? Genes 2021, 12, 1969. https://doi.org/10.3390/genes12121969
Smirnov A, Battulin N. Concatenation of Transgenic DNA: Random or Orchestrated? Genes. 2021; 12(12):1969. https://doi.org/10.3390/genes12121969
Chicago/Turabian StyleSmirnov, Alexander, and Nariman Battulin. 2021. "Concatenation of Transgenic DNA: Random or Orchestrated?" Genes 12, no. 12: 1969. https://doi.org/10.3390/genes12121969
APA StyleSmirnov, A., & Battulin, N. (2021). Concatenation of Transgenic DNA: Random or Orchestrated? Genes, 12(12), 1969. https://doi.org/10.3390/genes12121969




