Genome-Wide Analyses for Osteosarcoma in Leonberger Dogs Reveal the CDKN2A/B Gene Locus as a Major Risk Locus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Clinical Phenotypes
2.2. SNP Array Genotyping and Genome-Wide Association Study
2.3. Estimation of Heritability
2.4. Determination of Genomic Architecture
3. Results
3.1. Animals
3.2. Genome-Wide Association Study
3.3. Allele Frequencies
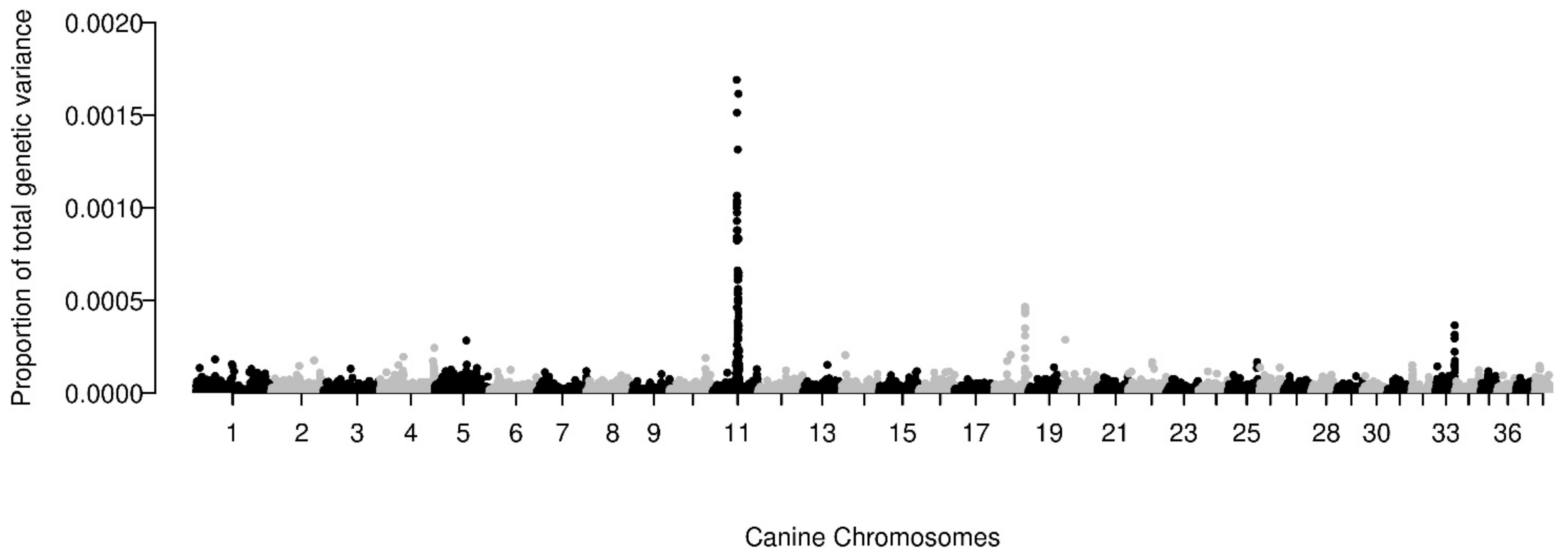
3.4. Heritability and Genetic Architecture
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franceschini, N.; Lam, S.W.; Cleton-Jansen, A.-M.; Bovée, J.V.M.G. What’s new in bone forming tumours of the skeleton? Virchows Arch. 2020, 476, 147–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Bahrami, A.; Pappo, A.; Easton, J.; Dalton, J.; Hedlund, E.; Ellison, D.; Shurtleff, S.; Wu, G.; Wei, L.; et al. Recurrent Somatic Structural Variations Contribute to Tumorigenesis in Pediatric Osteosarcoma. Cell Rep. 2014, 7, 104–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Liu, Z. Systematic meta-analysis of genetic variants associated with osteosarcoma susceptibility. Medicine 2018, 97, e12525. [Google Scholar] [CrossRef]
- Morello, E.; Martano, M.; Buracco, P. Biology, diagnosis and treatment of canine appendicular osteosarcoma: Similarities and differences with human osteosarcoma. Vet. J. 2011, 189, 268–277. [Google Scholar] [CrossRef]
- Simpson, S.; Dunning, M.D.; de Brot, S.; Grau-Roma, L.; Mongan, N.P.; Rutland, C.S. Comparative review of human and canine osteosarcoma: Morphology, epidemiology, prognosis, treatment and genetics. Acta Vet. Scand. 2017, 59, 71. [Google Scholar] [CrossRef]
- Ostrander, E.A.; Dreger, D.L.; Evans, J.M. Canine Cancer Genomics: Lessons for Canine and Human Health. Annu. Rev. Anim. Biosci. 2019, 7, 449–472. [Google Scholar] [CrossRef]
- Mueller, F.; Fuchs, B.; Kaser-Hotz, B. Comparative biology of human and canine osteosarcoma. Anticancer Res. 2007, 27, 155–164. [Google Scholar]
- Fan, T.; Khanna, C. Comparative Aspects of Osteosarcoma Pathogenesis in Humans and Dogs. Vet. Sci. 2015, 2, 210–230. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.C.; Stephenson, B.; Hauck, M.; Dillberger, J. Heritability and segregation analysis of osteosarcoma in the Scottish deerhound. Genomics 2007, 90, 354–363. [Google Scholar] [CrossRef] [Green Version]
- Momen, M.; Kohler, N.L.; Binversie, E.E.; Dentino, M.; Sample, S.J. Heritability and genetic variance estimation of Osteosarcoma (OSA) in Irish Wolfhound, using deep pedigree information. Canine Med. Genet. 2021, 8, 9. [Google Scholar] [CrossRef]
- Egenvall, A.; Nødtvedt, A.; von Euler, H. Bone tumors in a population of 400 000 insured Swedish dogs up to 10 y of age: Incidence and survival. Can. J. Vet. Res. 2007, 71, 292–299. [Google Scholar]
- Edmunds, G.L.; Smalley, M.J.; Beck, S.; Errington, R.J.; Gould, S.; Winter, H.; Brodbelt, D.C.; O’Neill, D.G. Dog breeds and body conformations with predisposition to osteosarcoma in the UK: A case-control study. Canine Med. Genet. 2021, 8, 2. [Google Scholar] [CrossRef]
- Vonholdt, B.M.; Pollinger, J.P.; Lohmueller, K.E.; Han, E.; Parker, H.G.; Quignon, P.; Degenhardt, J.D.; Boyko, A.R.; Earl, D.A.; Auton, A.; et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 2010, 464, 898–902. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.C.; Lembcke, L.; Chamberlin, T. A novel locus for canine osteosarcoma (OSA1) maps to CFA34, the canine orthologue of human 3q26. Genomics 2010, 96, 220–227. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, E.K.; Sigurdsson, S.; Ivansson, E.; Thomas, R.; Elvers, I.; Wright, J.; Howald, C.; Tonomura, N.; Perloski, M.; Swofford, R.; et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 2013, 14, R132. [Google Scholar] [CrossRef]
- Sakthikumar, S.; Elvers, I.; Kim, J.; Arendt, M.L.; Thomas, R.; Turner-Maier, J.; Swofford, R.; Johnson, J.; Schumacher, S.E.; Alföldi, J.; et al. SETD2 Is Recurrently Mutated in Whole-Exome Sequenced Canine Osteosarcoma. Cancer Res. 2018, 78, 3421–3431. [Google Scholar] [CrossRef] [Green Version]
- Shearin, A.L.; Hedan, B.; Cadieu, E.; Erich, S.A.; Schmidt, E.V.; Faden, D.L.; Cullen, J.; Abadie, J.; Kwon, E.M.; Gröne, A.; et al. The MTAP-CDKN2A Locus Confers Susceptibility to a Naturally Occurring Canine Cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1019–1027. [Google Scholar] [CrossRef] [Green Version]
- Hédan, B.; Cadieu, É.; Rimbault, M.; Vaysse, A.; Dufaure de Citres, C.; Devauchelle, P.; Botherel, N.; Abadie, J.; Quignon, P.; Derrien, T.; et al. Identification of common predisposing loci to hematopoietic cancers in four dog breeds. PLoS Genet. 2021, 17, e1009395. [Google Scholar] [CrossRef]
- Letko, A.; Minor, K.M.; Jagannathan, V.; Seefried, F.R.; Mickelson, J.R.; Oliehoek, P.; Drögemüller, C. Genomic diversity and population structure of the Leonberger dog breed. Genet. Sel. Evol. 2020, 52, 61. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies By Use of Localized Haplotype Clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letko, A.; Minor, K.M.; Friedenberg, S.G.; Shelton, G.D.; Salvador, J.P.; Mandigers, P.J.J.; Leegwater, P.A.J.; Winkler, P.A.; Petersen-Jones, S.M.; Stanley, B.J.; et al. A CNTNAP1 Missense Variant Is Associated with Canine Laryngeal Paralysis and Polyneuropathy. Genes 2020, 11, 1426. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv 2014, 81, 559–575. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Speed, D.; Hemani, G.; Johnson, M.R.; Balding, D.J. Improved Heritability Estimation from Genome-wide SNPs. Am. J. Hum. Genet. 2012, 91, 1011–1021. [Google Scholar] [CrossRef] [Green Version]
- Moser, G.; Lee, S.H.; Hayes, B.J.; Goddard, M.E.; Wray, N.R.; Visscher, P.M. Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model. PLoS Genet. 2015, 11, e1004969. [Google Scholar] [CrossRef]
- Mollandin, F.; Rau, A.; Croiseau, P. An evaluation of the predictive performance and mapping power of the BayesR model for genomic prediction. G3 Genes Genomes Genet. 2021, 11, jkab225. [Google Scholar] [CrossRef]
- Zapata, I.; Moraes, L.E.; Fiala, E.M.; Zaldivar-Lopez, S.; Couto, C.G.; Rowell, J.L.; Alvarez, C.E. Risk-modeling of dog osteosarcoma genome scans shows individuals with Mendelian-level polygenic risk are common. BMC Genom. 2019, 20, 226. [Google Scholar] [CrossRef]
- Megquier, K.; Turner-Maier, J.; Swofford, R.; Kim, J.-H.; Sarver, A.L.; Wang, C.; Sakthikumar, S.; Johnson, J.; Koltookian, M.; Lewellen, M.; et al. Comparative Genomics Reveals Shared Mutational Landscape in Canine Hemangiosarcoma and Human Angiosarcoma. Mol. Cancer Res. 2019, 17, 2410–2421. [Google Scholar] [CrossRef] [Green Version]
- Sándor, S.; Tátrai, K.; Czeibert, K.; Egyed, B.; Kubinyi, E. CDKN2A Gene Expression as a Potential Aging Biomarker in Dogs. Front. Vet. Sci. 2021, 8, 660435. [Google Scholar] [CrossRef]
- Guim, T.N.; Bianchi, M.V.; De Lorenzo, C.; Gouvêa, A.S.; Gerardi, D.G.; Driemeier, D.; Pavarini, S.P.; Sonne, L. Relationship Between Clinicopathological Features and Prognosis in Appendicular Osteosarcoma in Dogs. J. Comp. Pathol. 2020, 180, 91–99. [Google Scholar] [CrossRef]
- Ehrhart, N.P.; Ryan, S.D.; Fan, T.M. Tumors of the Skeletal System. In Withrow and MacEwen’s Small Animal Clinical Oncology; Elsevier: St. Louis, MI, USA, 2013; pp. 463–503. [Google Scholar]
- Tuohy, J.L.; Shaevitz, M.H.; Garrett, L.D.; Ruple, A.; Selmic, L.E. Demographic characteristics, site and phylogenetic distribution of dogs with appendicular osteosarcoma: 744 dogs (2000-2015). PLoS ONE 2019, 14, e0223243. [Google Scholar] [CrossRef] [Green Version]
- Boerman, I.; Selvarajah, G.T.; Nielen, M.; Kirpensteijn, J. Prognostic factors in canine appendicular osteosarcoma—A meta-analysis. BMC Vet. Res. 2012, 8, 56. [Google Scholar] [CrossRef] [Green Version]

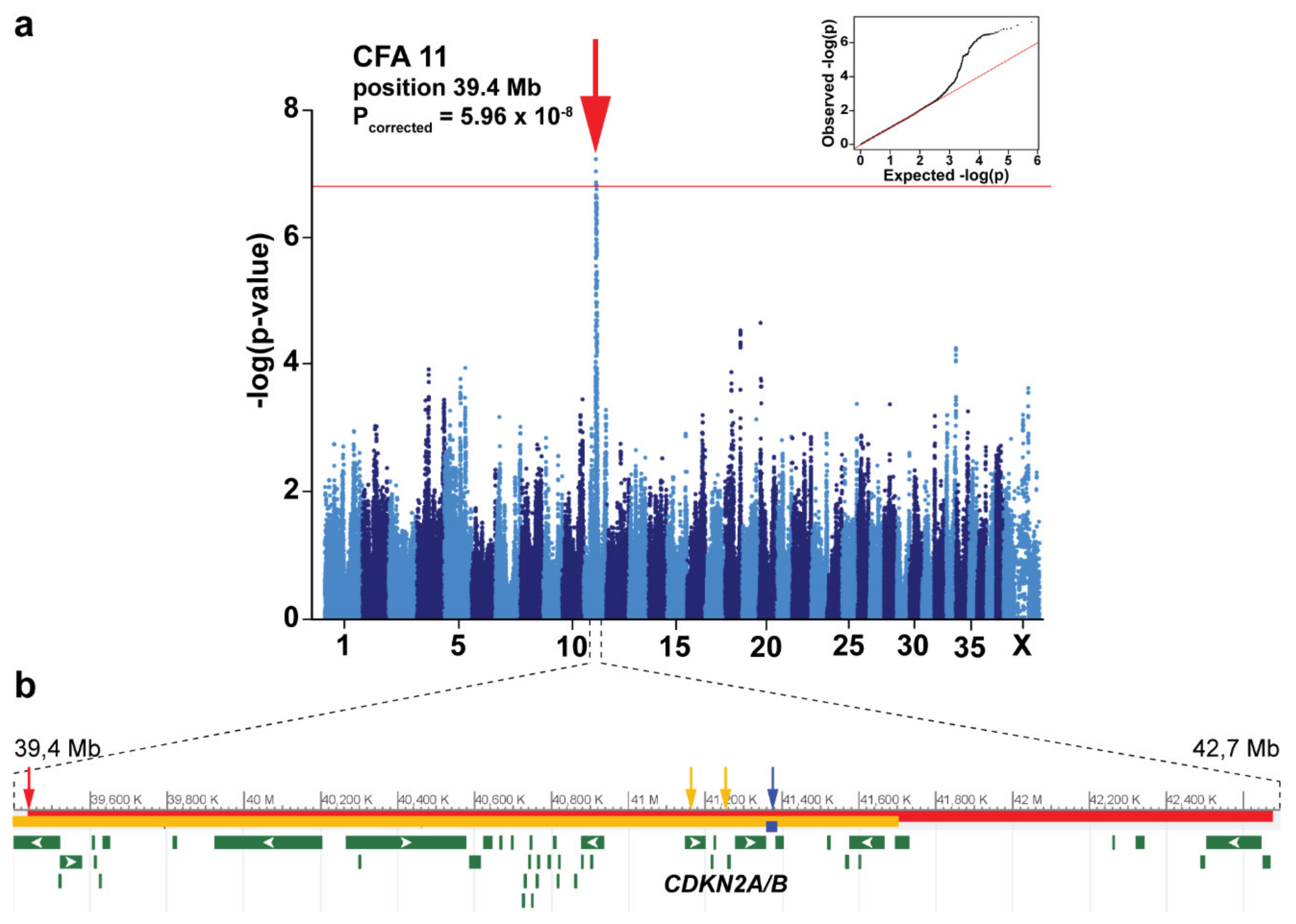
| Position | Alleles | Discovery Breed | AF in Leonbergers | Leonberger p-Value 1 | Reference | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| 39,434,964 | G/A | Leonberger | 0.64 | 0.47 | 1.64 × 10−9 | current study |
| 41,375,800 | C/A | Greyhound | 0.73 | 0.63 | 1.16 × 10−4 | [16] |
| 41,161,441 | T/C | Bernese mountain dog | 0.93 | 0.95 | 0.16 | [19] |
| 41,252,822 | T/C | Flat-coated retriever, Rottweiler | 0.71 | 0.61 | 1.15 × 10−4 | [19] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Letko, A.; Minor, K.M.; Norton, E.M.; Marinescu, V.D.; Drögemüller, M.; Ivansson, E.; Megquier, K.; Noh, H.J.; Starkey, M.; Friedenberg, S.G.; et al. Genome-Wide Analyses for Osteosarcoma in Leonberger Dogs Reveal the CDKN2A/B Gene Locus as a Major Risk Locus. Genes 2021, 12, 1964. https://doi.org/10.3390/genes12121964
Letko A, Minor KM, Norton EM, Marinescu VD, Drögemüller M, Ivansson E, Megquier K, Noh HJ, Starkey M, Friedenberg SG, et al. Genome-Wide Analyses for Osteosarcoma in Leonberger Dogs Reveal the CDKN2A/B Gene Locus as a Major Risk Locus. Genes. 2021; 12(12):1964. https://doi.org/10.3390/genes12121964
Chicago/Turabian StyleLetko, Anna, Katie M. Minor, Elaine M. Norton, Voichita D. Marinescu, Michaela Drögemüller, Emma Ivansson, Kate Megquier, Hyun Ji Noh, Mike Starkey, Steven G. Friedenberg, and et al. 2021. "Genome-Wide Analyses for Osteosarcoma in Leonberger Dogs Reveal the CDKN2A/B Gene Locus as a Major Risk Locus" Genes 12, no. 12: 1964. https://doi.org/10.3390/genes12121964







