Chromosome Number, Ploidy Level, and Nuclear DNA Content in 23 Species of Echeveria (Crassulaceae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Mitotic Chromosome Counts
2.3. Estimation of Nuclear DNA Content
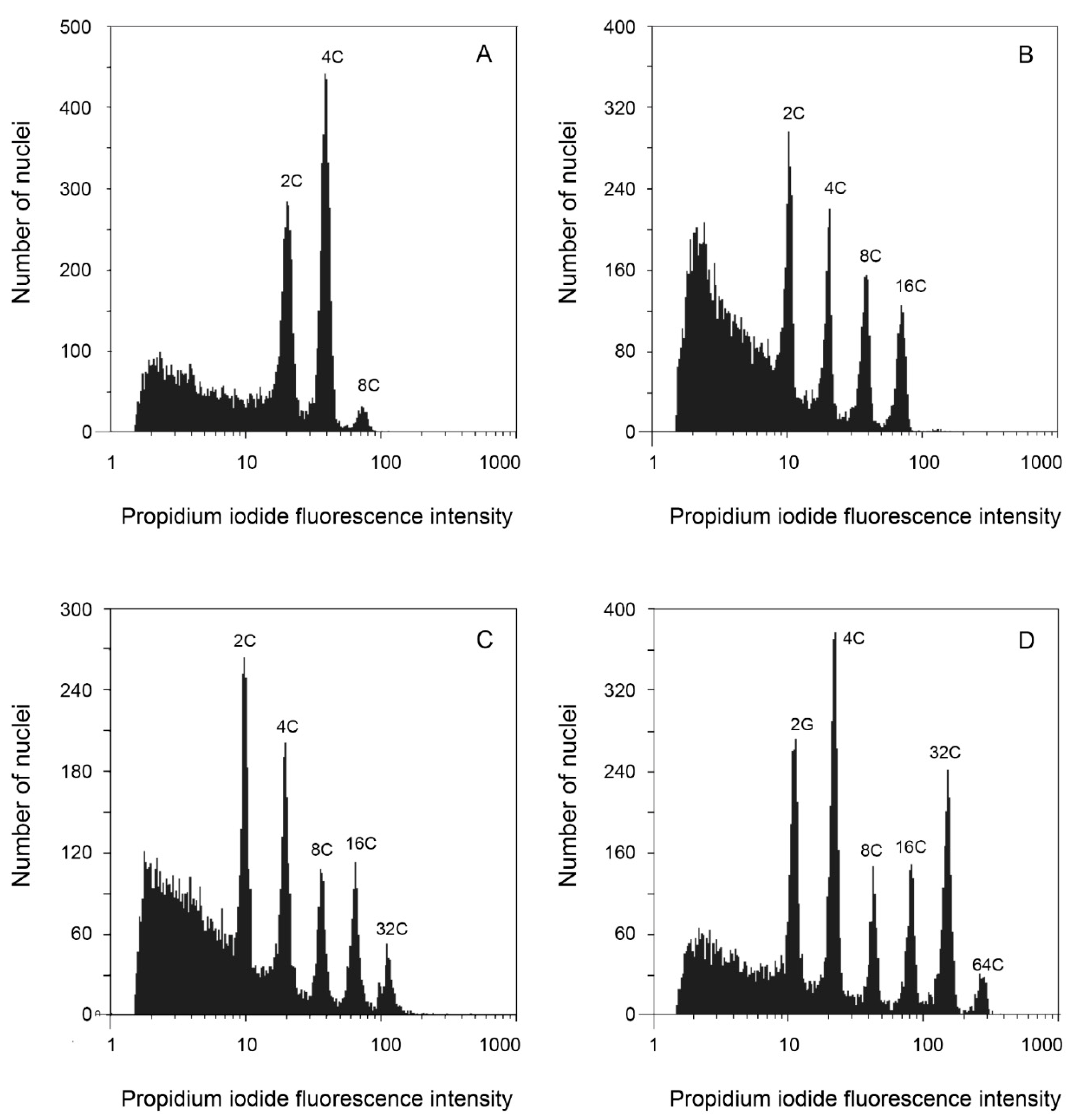
2.4. Endopolyploidy Determination
2.5. Statistical Analyses
3. Results
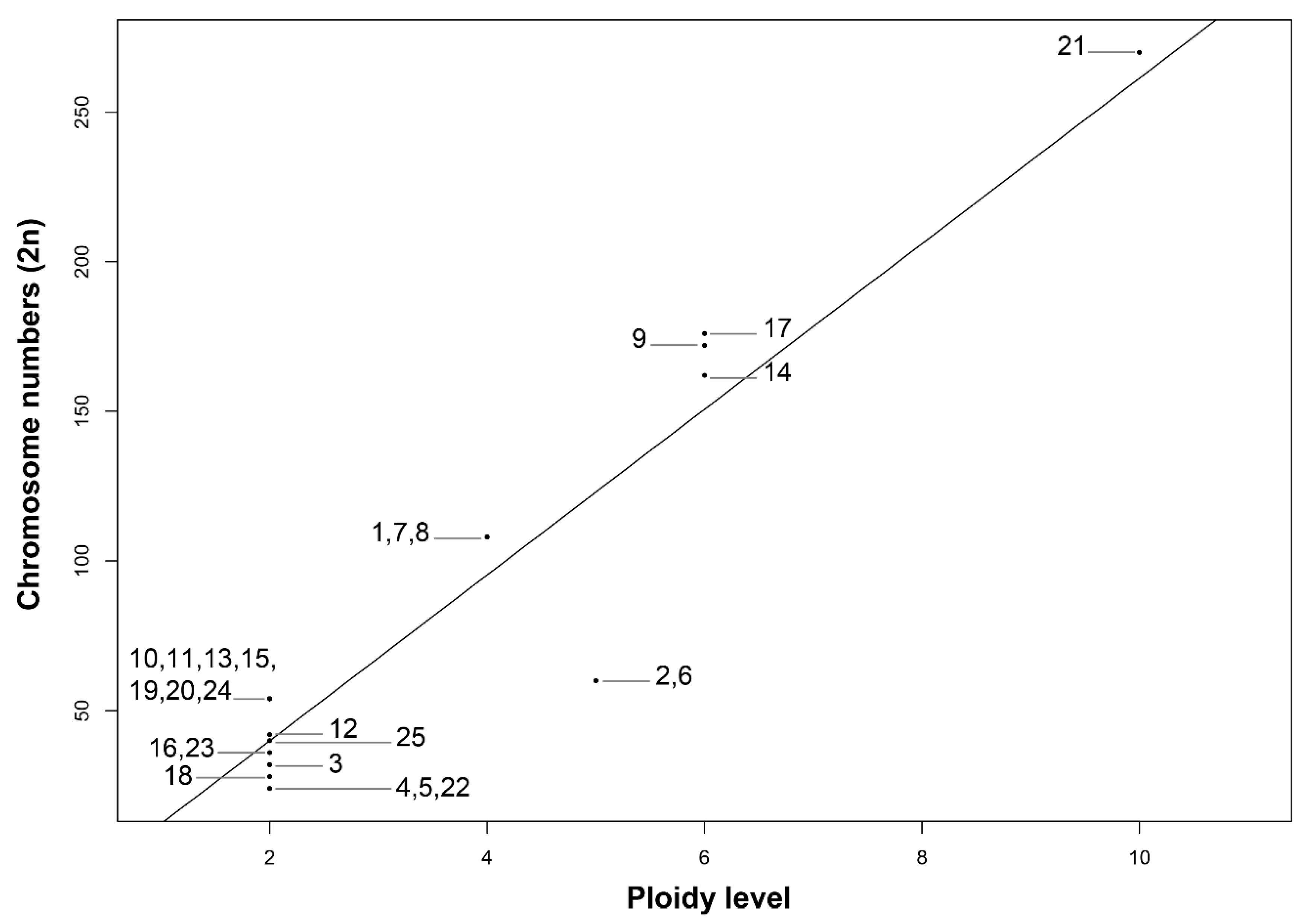
3.1. Endemism, Chromosome Numbers, and Ploidy Level
3.2. Nuclear DNA Content
3.3. Endopolyploidy
3.4. Correlation Polyploidy, Chromosome Number, and 2C DNA Content
4. Discussion
4.1. Endemism, Chromosome Numbers, and Polyploidy in Echeveria
4.2. Nuclear DNA Content and Ploidy Levels in Echeveria
4.3. Endopolyploidy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Low, J.E. The Echeverias, some facts about this genus, flowers, culture. Int. Crassulaceae Netw. 2007, 1–2. Available online: https://www.crassulaceae.ch/de/artikel?akID=48 (accessed on 9 March 2021).
- Reyes, S.J.; Islas-Luna, M.A.; González-Zorzano, O. Guía Práctica de Propagación y Cultivo de las Especies del Género Echeveria; Editorial Universidad Nacional Autónoma de México: Mexico City, Mexico, 2014; p. 111. [Google Scholar]
- Reyes, S.J.; Brachet, I.C.; González, Z.O.; Islas-Luna, M.A.; López, C.L. Four new taxa of the genus Echeveria from the state of Oaxaca, México. Haseltonia 2015, 21, 80–91. [Google Scholar]
- Uhl, C.H. Some cytotaxonomic problems in the Crassulaceae. Evolution 1961, 15, 375–383. [Google Scholar] [CrossRef] [Green Version]
- Uhl, C.H. Polyploidy, dysploidy, and chromosome pairing in Echeveria (Crassulaceae) and its hybrids. Am. J. Bot. 1992, 5, 556–566. [Google Scholar] [CrossRef]
- Kimnach, M. Echeveria. In Illustrated Handbook of Succulent Plants: Crassulaceae; Eggli, U., Ed.; Springer: Berlin, Germany, 2003; pp. 103–128. [Google Scholar]
- Reyes, S.J.; Islas, A.; González-Zorzano, O.; Cerrillo-Reyes, P.; Vergara-Silva, F.; Brachet, C. Echeveria: Manual del Perfil Diagnóstico del Género Echeveria en México; Universidad Autónoma Chapingo: Texcoco, Mexico, 2011; p. 139. [Google Scholar]
- Etter, J.; Kristen, M. Data Base, Agavaceae and Crassulaceae. Available online: https://www.crassulaceae.com/crassulaceae/craseinfuehrung_en.asp (accessed on 9 March 2021).
- Reyes, S.J.; González, O.; Gutiérrez, A. Pachyphytum brachetii, una nueva especie del estado de Hidalgo, México. Cactáceas Suculentas Mexicanas 2007, 2, 53–63. [Google Scholar]
- Vazquez-Cotero, C.; Sosa, V.; Carrillo-Reyes, P. Phylogenetic position of Echeveria heterosepala (Crasulaceae) a rare species with diagnostic characters of Pachyphytum. Bot. Sci. 2017, 95, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Uhl, C.H. Chromosomes, hybrids and ploidy of Sedum cremnophila and Echeveria linguifolia (Crassulaceae). Am. J. Bot. 1976, 6, 806–820. [Google Scholar] [CrossRef]
- Uhl, C.H. Chromosomes and hybrids of Echeveria X. South American species of Series Nudae. Haseltonia 2006, 12, 31–40. [Google Scholar] [CrossRef]
- Uhl, C.H. Chromosomes and hybrids of Echeveria (Crassulaceae). VII. Series Gibbiflorae (Baker) Berger. Haseltonia 2002, 9, 121–145. [Google Scholar]
- Uhl, C.H. Chromosomes and hybrids of Echeveria VIII. Central American species. Haseltonia 2004, 10, 71–82. [Google Scholar]
- Uhl, C.H. Chromosomes and hybrids of Echeveria XI. South American species of Series Racemosae. Haseltonia 2007, 13, 3–22. [Google Scholar] [CrossRef]
- Uhl, C.H. The problem of ploidy in Echeveria (Crassulaceae) I. Diploidy in E. ciliata. Am. J. Bot. 1982, 5, 843–854. [Google Scholar] [CrossRef]
- Uhl, C.H. The problem of ploidy in Echeveria (Crassulaceae) II. Tetraploidy in E. secunda. Am. J. Bot. 1982, 9, 1497–1511. [Google Scholar] [CrossRef]
- Uhl, C.H. Chromosomes and hybrids of Echeveria II. Series Occidentales Moran (Crassulaceae). Haseltonia 1995, 3, 25–33. [Google Scholar]
- Uhl, C.H. Chromosomes and hybrids of Echeveria III. Series Secundae (Baker) Berger (Crassulaceae). Haseltonia 1995, 3, 34–48. [Google Scholar]
- Uhl, C.H. Chromosomes and hybrids of Echeveria. 4. Series Urceolatae E. Walther. Haseltonia 1996, 4, 66–88. [Google Scholar]
- Uhl, C.H. Chromosomes of Mexican Sedum II. Section Pachysedum. Rhodora 1978, 80, 491–512. [Google Scholar]
- Leitch, I.J.; Leitch, A.R. Genome size diversity and evolution in land plants. In Plant Genome Diversity. Physical Structure, Behaviour and Evolution of Plant Genomes; Leitch, I.J., Greilhulber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2, pp. 307–322. [Google Scholar]
- Soltis, D.E.; Soltis, P.S.; Bennett, M.D.; Leitch, I.J. Evolution of genome size in the angiosperms. Am. J. Bot. 2003, 90, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Palomino, G.; Martínez, J.; Méndez, I.; Muñoz-Urias, A.; Cepeda-Cornejo, V.; Pimienta-Barrios, E. Nuclear genome size, ploidy level and endopolyploidy pattern in six species of Opuntia (Cactaceae). Cytologia 2016, 1, 82–89. [Google Scholar]
- Hart, H.T. Evolutionary and systematic significance of hybridization in Mediterranean Sedoideae (Crassulaceae). Lagascalia 1997, 19, 57–58. [Google Scholar]
- Hart, H.T.; Tomlik, A.; Alpinar, K. Biosystematic studies in Sedum (Crassulaceae) from Turkey. 4. The cytology of Sedum subsect. Spathulata Boriss. Acta Botanica Neerlandica 1993, 3, 289–298. [Google Scholar] [CrossRef]
- Poggio, L.; Realini, M.F.; Fourastié, M.F.; García, A.M.; González, G.E. Genome downsizing and karyotype constancy in diploid and polyploid congeners: A model of genome size variation. AoB Plants 2014, 6, plu029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelli, M.; Miller, C.H.; Schmidt-lebuhn, A.N. Polyploidization and genome size evolution in Australian billy buttons (Craspidia, Asteraceae: Gnaphakieae). Int. J. Plant. Sci. 2017, 5, 352–361. [Google Scholar] [CrossRef]
- Levin, D.A. The Role of Chromosomal Change in Plan Evolution; Oxford Series in Ecology and Evolution; Oxford University Press: Oxford, UK, 2002; p. 230. [Google Scholar]
- Soltis, D.E.; Alvart, V.A.; Leebens-Mack, J.; Bell, C.D.; Paterson, A.H.; Zheng, C.; Sankoff, D.; de Pamphilis, C.W.; Wall, P.K.; Soltis, P.S. Polyploidy and angiosperm diversification. Am. J. Bot. 2009, 1, 336–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltis, S.P.; Soltis, D.E. Angiosperm phylogeny: A framework for studies of genome evolution. In Plant Genome Diversity. Physical Structure, Behaviour and Evolution of Plant Genomes; Leitch, I.J., Greilhulber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2, pp. 1–11. [Google Scholar]
- Soto-Trejo, F.; Palomino, G.; Villaseñor, J.L.; Crawford, D.J. Polyploidy in Asteraceae of the xerophytic scrub of the ecological reserve of the Pedregal of San Angel. México City. Bot. J. Linn. Soc. 2013, 2, 211–229. [Google Scholar] [CrossRef] [Green Version]
- Maluszynska, J.; Kolano, B.; Sas-Nowosielska, H. Endopolyploidy in plants. In Plant Genome Diversity. Physical Structure, Behaviour and Evolution of Plant Genomes; Leitch, I.J., Greilhulber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2, pp. 99–119. [Google Scholar]
- Palomino, G.; Dolezel, J.; Cid, R.; Brunner, I.; Mendez, I.; Rubluo, A. Nuclear genome stability of Mammillaria San-Angelesis (Cactaceae) regenerants induced by auxins in long-term in vitro culture. Plant. Sci. 1999, 141, 191–200. [Google Scholar] [CrossRef]
- Palomino, G. Genome analysis of Mexican flora. Genet. Mol. Biol. 2000, 23, 921–924. [Google Scholar] [CrossRef]
- Del Angel, C.; Palomino, G.; García, A.; Méndez, I. Nuclear genome size and karyotype analysis in Mammillaria species (Cactaceae). Caryologia 2006, 59, 177–186. [Google Scholar]
- Dolezel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 9, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.D.; Leitch, I.J. Nuclear DNA amounts in angiosperms: Target, trends and tomorrow. Ann. Bot. 2011, 107, 467–590. [Google Scholar] [CrossRef]
- Greilhuber, J.; Leitch, I.J. Genome size and the phenotype. In Plant Genome Diversity. Physical Structure, Behaviour and Evolution of Plant Genomes, Leitch, I.J., Greilhulber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2, pp. 323–344. [Google Scholar]
- Leitch, I.J.; Johnston, E.; Pellicer, J.; Hidalgo, O.; Bennett, M.D. Plant DNA C-Values Database. Release 7.1. April 2019. Available online: https://cvalues.science.kew.org/ (accessed on 9 March 2021).
- Greilhuber, J.; Borsch, T.; Müller, K.; Worberg, A.; Porembski, S.; Barthlott, W. Smallest angiosperm genomes found in Lentibulariaceae with chromosomes of bacterial size. Plant. Biol. 2006, 8, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, A.; Michael, T.P.; Rivadavia, F.; Sousa, A.; Wang, W.; Temsch, E.M.; Greilhuber, J.; Muller, K.F.; Heubl, G. Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperm. Ann. Bot. 2014, 114, 1651–1663. [Google Scholar] [CrossRef] [Green Version]
- Husband, B.C.; Balwin, S.J.; Suda, J. The incidence of polyploidy in natural plant population: Major patterns and evolutionary processes. In Plant Genome Diversity. Physical Structure, Behaviour and Evolution of Plant Genomes; Leitch, I.J., Greilhulber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2, pp. 255–276. [Google Scholar]
- Clark, J.; Hidalgo, O.; Pellicer, J.; Liu, H.; Marquardt, J.; Robert, Y.; Christenhusz, M.; Zhang, S.; Gibby, M.; Leith, I.J.; et al. Genome evolution of ferns: Evidence for relative stasis of genome size across the fern phylogeny. New Phytol. 2016, 3, 1072–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetter, M.G.; Schmid, K.J. Analysis of phylogenetic relationships and genome size evolution of the Amaranthus genus using GBS indicates the ancestors of an ancient crop. Mol. Phylogenet. Evol. 2017, 109, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Palomino, G.; Martínez, J.; Romero, P.; Barba-González, R.; Rodríguez-Garay, B. Nuclear genome size and karyotype analysis of Agave angustifolia Haw. ‘Cimarron’ and ‘Lineño’ (Asparagales, Asparagaceae). Caryologia 2017, 70, 93–101. [Google Scholar] [CrossRef]
- Palomino, G.; Soto-Trejo, F.; Correa, H.; Méndez, I.; Villaseñor, J.L. Nuclear genome size and chromosome number in the Mexican genus Pittocaulon (Asteraceae). Caryologia 2018, 2, 113–119. [Google Scholar] [CrossRef]
- Palomino, G.; Martínez, J.; Cepeda-Cornejo, V.; Pimienta-Barrios, E. Nuclear genome size and cytotype analysis in Agave cupreata Trel. & Berger (Agavaceae). Caryologia 2013, 65, 281–294. [Google Scholar] [CrossRef] [Green Version]
- D’Amato, F. Endopolyploidy as a factor in plant tissue development. Caryologia 1964, 17, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Leitch, I.; Dodsworth, S. Endopolyploidy in plants. In Encyclopedia of Life Sciences, Hetherington; John Wiley & Sons, Ltd: Hoboken, NJ, USA, 2017; pp. 1–10. [Google Scholar] [CrossRef]
- Joubès, J.; Chevalier, C. Endoreduplication in higher plants. Plant. Mol. Biol. 2000, 43, 735–745. [Google Scholar] [CrossRef]
- Edgar, B.A.; Orr-Weaver, T.L. Endoreplication Cell Cycles: More for Less. Cell 2001, 105, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Scholes, D.R.; Paige, K.N. Plasticity in ploidy: A generalized stress response. Trends Plant. Sci. 2015, 20, 165–175. [Google Scholar] [CrossRef]
- Bennett, M.D.; Leitch, I.J.; Price, J.; Johnston, J.S. Comparisons with Caenorhabditis (∼100 Mb) and Drosophila (∼175 Mb) using flow cytometry show genome size in Arabidopsis to be ∼157 Mb and thus ∼25 % larger than the Arabidopsis genome initiative estimate of ∼125 Mb. Ann. Bot. 2003, 91, 547–557. [Google Scholar] [CrossRef]
- Nodal-Moreno, S.A.; Palomino, G.; Almaguer-Sierra, P.; Blanco-Macías, F.; Barrientos-Lozano, L.; Flores-Gracia, J. Nuclear genome size, polyploidy and endopolyploidy pattern in populations of Nopalea cochenillifera (L). Salm-Dyc (Cactaceae) in Tamaulipas, México. Acta Univ. Multidiscip. Sci. J. 2019, 29, e2238. [Google Scholar]
- Pilbeam, J. The Genus Echeveria; The British Cactus & Succulent Society: Great Malvern, UK, 2008. [Google Scholar]
- García, V.A. Técnicas y Procedimientos de Citogenética Vegetal; Colegio de Posgraduados, Talleres Gráficos de la Nación: Mexico, Mexico, 1990; p. 144. [Google Scholar]
- Conger, D.D.; Fairchield, L.M. A quick freeze method for making smear slides permanent. Stain Technol. 1953, 28, 281–283. [Google Scholar] [CrossRef]
- Temsch, E.M.; Koutecký, P.; Urfus, T.; Šmarda, P.; Doležel, J. Reference standards for flow cytometric estimation of absolute nuclear DNA content in plants. Cytom. Part A 2021, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Barow, M.; Meister, A. Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant. Cell Environ. 2003, 4, 571–584. [Google Scholar] [CrossRef]
- Barow, M. Endopolyploidy in seed plants. BioEssays 2006, 3, 271–281. [Google Scholar] [CrossRef]
- R. Core Team. A Language and Environment for Statistical Computing; R. Foundation for Statistical Computing: Vienna, Austria, 2015; Available online: https://www.R-project.org/ (accessed on 2 October 2018).
- Reyes, S.J.; Pérez, C.J.; Brachet, I.C. Echeveria cuicatecana, una nueva especie para el estado de Oaxaca, México. Cactáceas Suculentas Mexicanas 2004, 3, 80–90. [Google Scholar]
- Barow, M.; Jovtchev, G. Endopolyploidy in plants and its analysis by flow cytometry. In Flow Cytometry with Plant Cells, Analysis of Genes, Chromosomes and Genomes; Dolezel, J., Greilhulber, J., Suda, J., Eds.; Wiley-VCH: Hoboken, NJ, USA, 2007; pp. 349–372. [Google Scholar]
- Leitch, A.R.; Leitch, I.J. Genomic plasticity and the diversity of polyploid plants. Science 2008, 320, 481–483. [Google Scholar] [CrossRef]
- Dorone, Y. Diploidización of Meiosis in Autotetraploids; Harvard University: Cambridge, MA, USA, 2012; pp. 1–6. [Google Scholar]
- Wendel, J.F. The wondrous cycles of polyploidy in plants. Am. J. Bot. 2015, 102, 1753–1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandáková, T.; Lysak, M.A. Post-polyploid diploidization and diversification through dysploid changes. Curr. Opin. Plant. Biol. 2018, 42, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Gilles, A.; Randolph, L.F. Reduction in quadrivalent frequency in autotraploid maize during a period of 10 years. Amer. J. Bot. 1951, 38, 12–16. [Google Scholar] [CrossRef]
- Kato, Y.T.A. Mecanismo de diploidización en Zea perennis (Hitchcock) Reeves y Mangelsdorf. Agrociencia 1984, 58, 113–126. [Google Scholar]
- Jauhar, P.P. Genetics regulation of diploid-like chromosome pairing in the hexaploid species, Festuca arundinacea Schreb and F. rubra (Graminae). Chromosoma 1993, 52, 363–382. [Google Scholar] [CrossRef]
- Martínez, J.; Palomino, G. Evidence of heterozygous chromosome interchange and chromatid Exchange in autotetraploid cytotype of Gibasis schiedeana (Tradescantiae-Commelinacea). Cytologia 1997, 62, 275–281. [Google Scholar] [CrossRef] [Green Version]
- De la Cruz-López, L.E.; Vergara-Silva, F.; Reyes, S.J.; Espino-Ortega, G.; Carrillo-Reyes, P.; Kuzmina, M. Phylogenetic relationships of Echeveria (Crassulaceae) and related genera from Mexico, based on three DNA barcoding loci. Phytotaxa 2019, 1, 33–57. [Google Scholar] [CrossRef]
- Loureiro, J.; Rodriguez, E.; Dolezel, J.; Santos, C. Comparison of four nuclear isolation buffers for plant DNA flow cytometry. Ann. Bot. 2006, 98, 679–689. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M.; Leitch, I.; Bennet, M.D. First nuclear DNA amounts in more than 300 angiosperms. Ann. Bot. 2005, 96, 229–244. [Google Scholar] [CrossRef] [Green Version]
- Hart, H. Evolution and classification of European Sedum species Crassulaceae. Flora Mediterr. 1991, 1, 31–61. [Google Scholar]
- Wang, X.; Morton, J.A.; Pellicer, J.; Leitch, I.J.; Leitch, A.R. Genome downsizing after polyploidy: Mechanisms, rates and selection pressures. Plant J. 2021, 107, 1003–1015. [Google Scholar] [CrossRef]
- Fawcett, J.A.; van de Peer, Y.; Meare, S. Significance and Biological consecuences of poliploidization in land plant evolution. In Plant Genome Diversity. Physical Structure, Behaviour and Evolution of Plant Genomes; Leitch, I.J., Greilhulber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2, pp. 277–293. ISBN 978-37091-1151-8. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M. The DNA weights per nucleus (genome size) of more than 2350 species of the Flora of The Netherlands, of which 1370 are new to science, including the pattern of their DNA peaks. Forum Geobot. 2019, 8, 24–78. [Google Scholar]
- De Rocher, E.J.; Harkins, K.; Galbraith, D.; Bohnert, H. Developmentally regulated systemic endopolyploidy in succulents with small genomes. Science 1990, 4977, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Cushman, J.C. Crassulacean acid metabolism. A plastic photosynthetic adaptation to arid environments. Plant. Physiol. 2001, 4, 1439–1448. [Google Scholar] [CrossRef]
- Nagl, W. DNA endoreduplication and polyteny understood as evolutionary strategies. Nature 1976, 261, 614–615. [Google Scholar] [CrossRef] [PubMed]
- Escudero, M.; Wendel, J.F. The grand sweep of chromosomal evolution in angiosperms. New Phytol. 2020, 228, 805–808. [Google Scholar] [CrossRef]
- Sakhanokho, H.F.; Rinehart, T.A.; Stringer, S.J.; Islam-Faridi, M.N.; Pounders, C.T. Variation in nuclear DNA content and chromosome numbers in blueberry. Scientia Horticulturae 2018, 233, 108–113. [Google Scholar] [CrossRef]
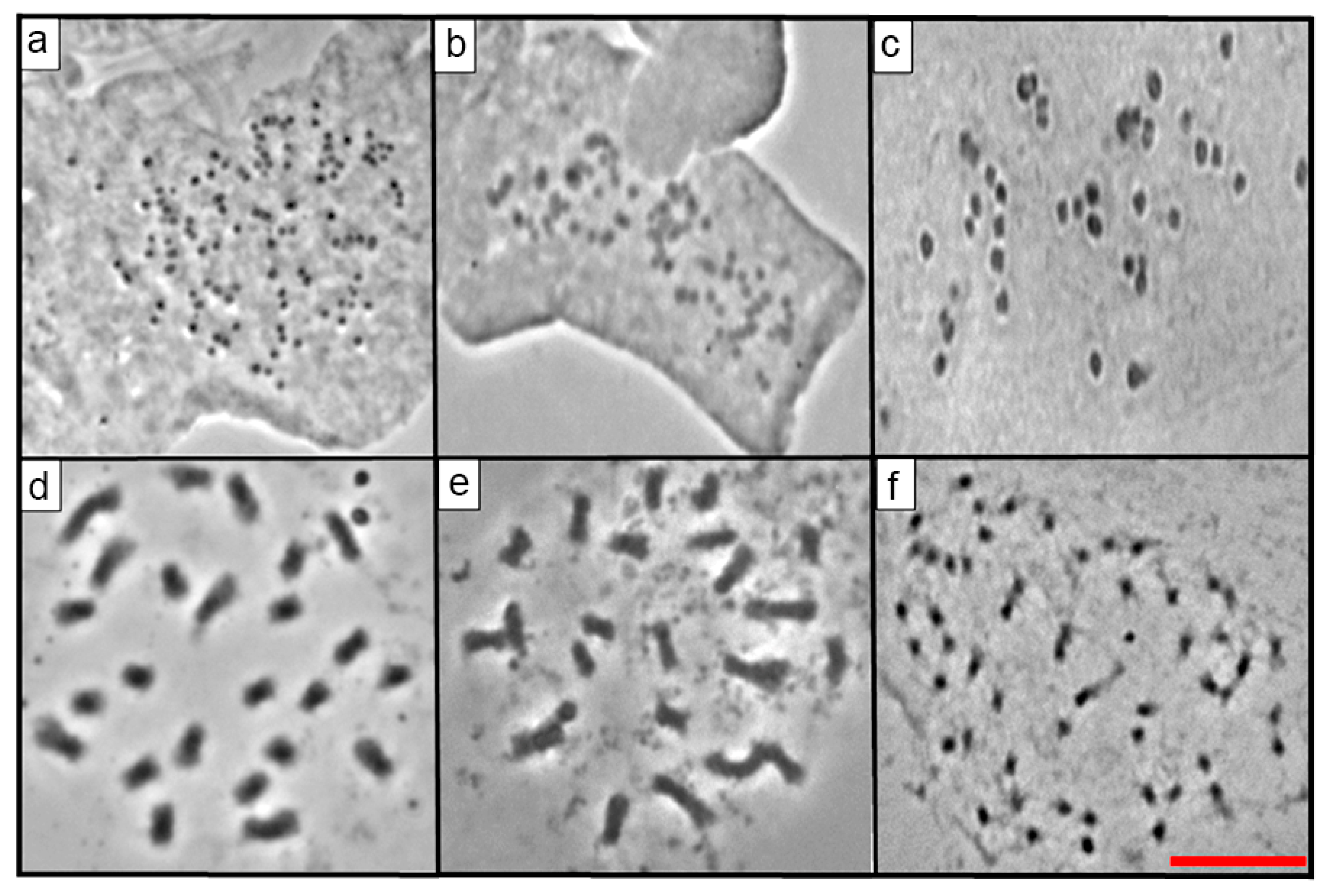
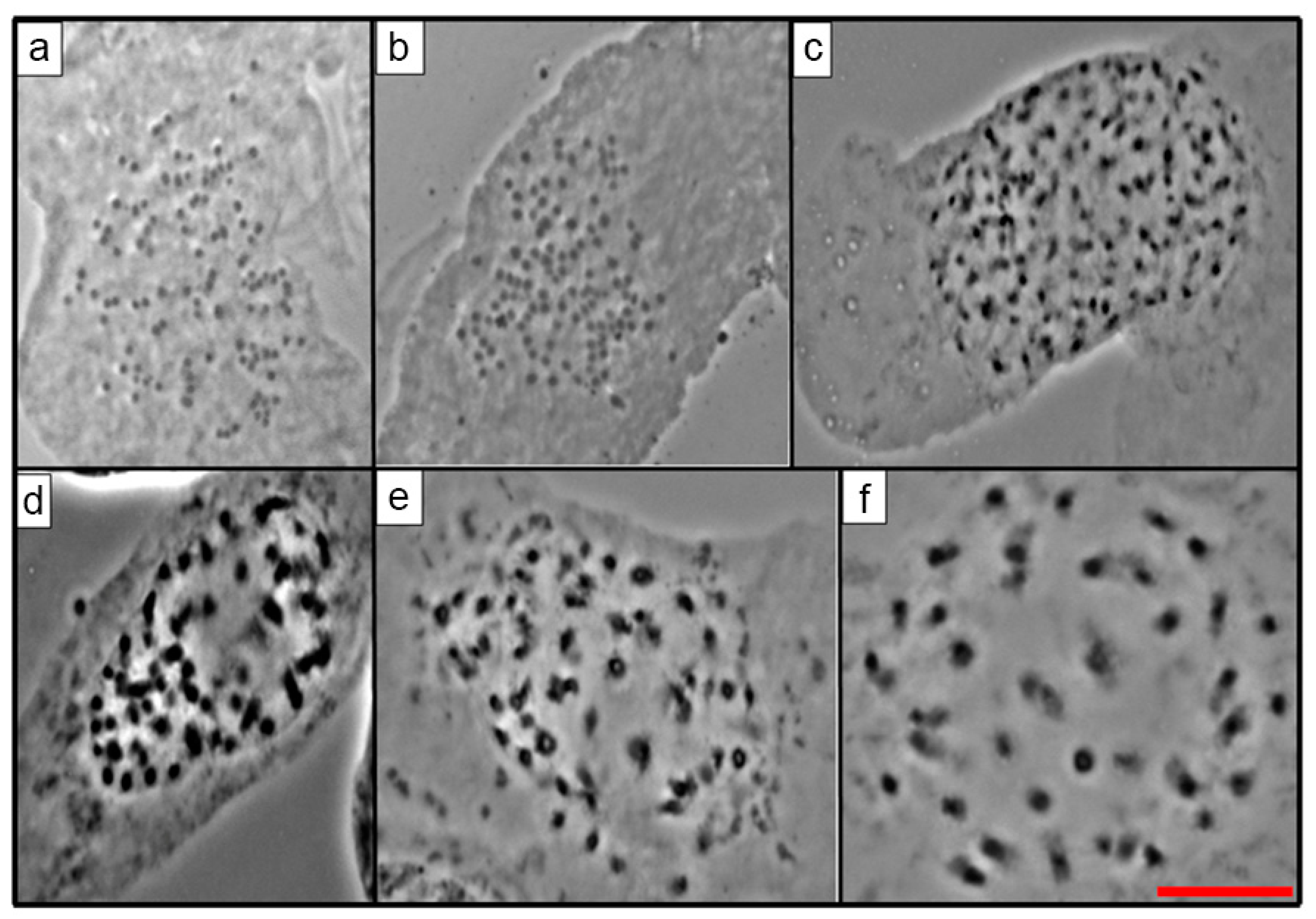
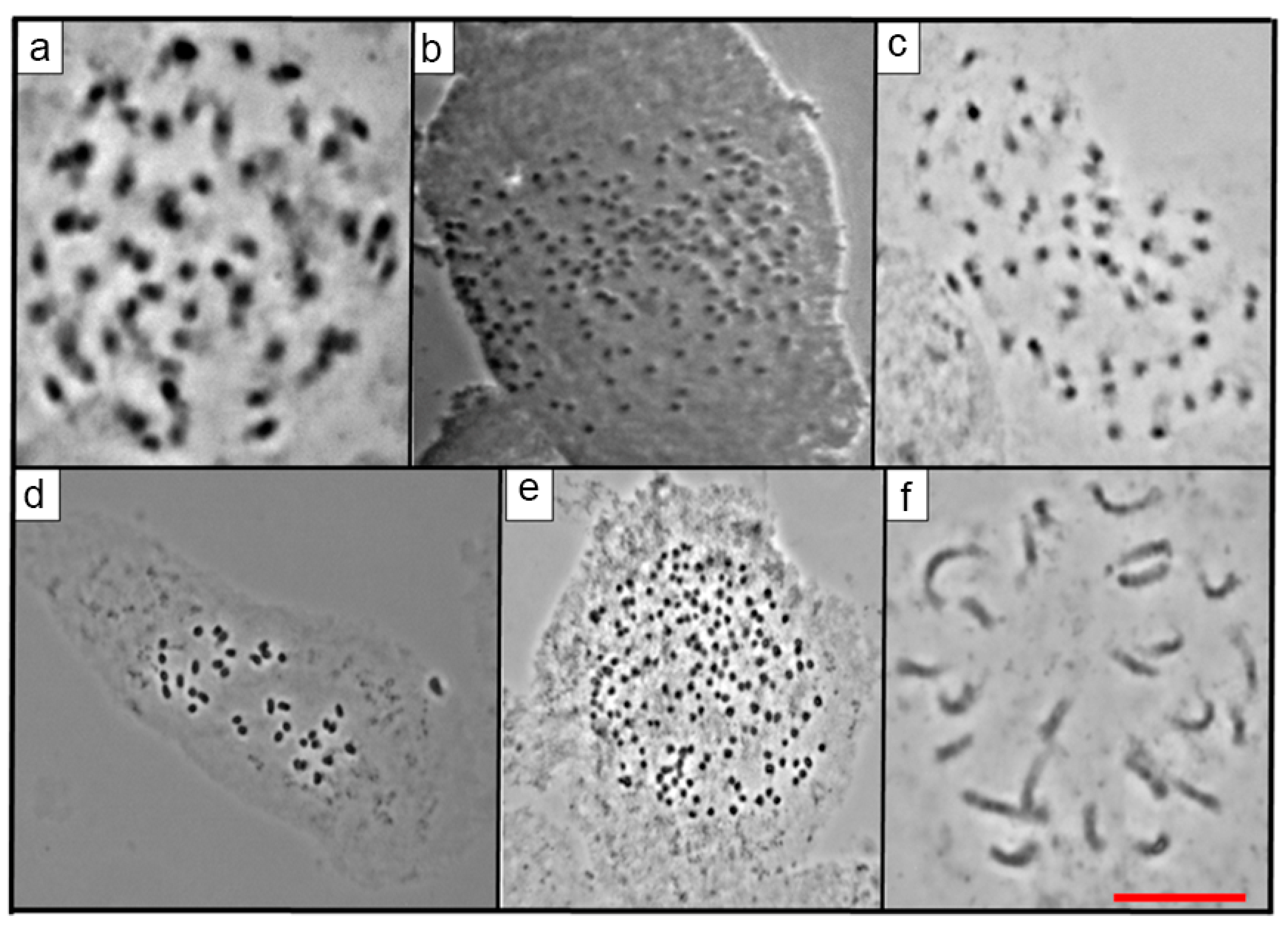



| No. | Taxon | Serie | Accession Number | Locality |
|---|---|---|---|---|
| 1 | E. altamirae | Gibbiflorae | JE-7548 | San Vicente Muñú, Oaxaca, Mexico. Near Anama. 17°22′10″ N 97°26′42″ W. 1828 m asl |
| 2 | E. caamanoi | Urbinae | JE-8311 | Ixtacamaxtitlan, Puebla, Mexico. Course path from Ixtacamaxtitlan–Texocoixpan. 19°36′52.5″ N 97°48′47.7″ W. 2347 m asl |
| 3 | E. carnicolor | Racemosae | JE-8374 | Tenampa, Veracruz, Mexico. Barranca de tenampa. 19°15′27.7″ N 96°52′59″ W. 762 m asl |
| 4 | E. catorce | Angulatae | JE-5469 | Catorce, San Luis Potosí, Mexico. 0.5 Km Dirt Road Catorce-Vanegas. 23°41′40.67″ N 100°53′18.88″ W. 2690 m asl |
| 5 | E. catorce | Angulatae | EK-3223 | Catorce, San Luis Potosí, Mexico. Matehuala-Real de Catorce. 23°41′30.9″ N 100°53′15.2″ W. 2713 m asl |
| 6 | E. cuicatecana | Pruinosae | JP-584 | San Juan Bautista Cuicatlán, Oaxaca, Mexico. Between Santo Dominguito and San Juan Tonaltepec. 17°41′21.5″ N 96°53′28.9″ W. 800 m asl |
| 7 | E. cupreata | Gibbiflorae | JE-6807 | San Vicente Lachixio, Oaxaca, Mexico. Km. 20 between the caves of San Sebastian and Vicente Guerrero. 16°41′6.2″ N 96°56.5″ W. 2093 m asl |
| 8 | E. dactylifera | Gibbiflorae | EK-2603 | San Dimas, Durango, Mexico. Mexico to Durango Road, Km. 172.5. 23°39′ N 105°47′9.9 W. 2493 m asl |
| 9 | E. gibbiflora | Gibbiflorae | EK-3427 | Tlaxiaco City, Oaxaca, Mexico. Putla-Nundaco and Atatlahuca. 17°12′0.7″ N 97°43 39.4″ W. 2125 m asl |
| 10 | E. guerrerensis | Gibbiflorae | JE-7521 | San Miguel Totolapan, Guerrero México. 3 Km NW from Station Toro Muerto. 17°34′42 N 100°17′0″ W. 2731 m asl |
| 11 | E. guerrerensis | Gibbiflorae | JE-7526 | Zihuatanejo de Azueta, Guerrero, Mexico Zihuatanejo to Ciudad Altamirano road. 17°56′2″ N 101°17′1″ W. 1446 m asl |
| 12 | E. helmutiana | Racemosae | JE-7075 | Santiago Juxtlahuaca, Oaxaca, Mexico. Dirt Road Juxtlahuaca-Yucunicoco. 17°17′44″ N 97°59′9.8″ W. 2476 m asl |
| 13 | E. juarezensis | Gibbiflorae | JE-7538 | Santa Catarina Ixtepeji, Oaxaca, Mexico. 10.5 Km South from “la Cumbre”. Km. 191.5 of the Oaxaca-Tuxtepec Road. 17°10′157″ N 96°36′15″ W. 2726 m asl |
| 14 | E. longiflora | Gibbiflorae | JE-6770 | Taxco, Guerrero México. Km. 21, of the Taxco-Ixcateopan Road. 18°31′54.5 N 99°42′54.9″ W. 2293 m asl |
| E. longiflora | Gibbiflorae | JE-6923 | Ixcateopan de Cuahtemoc, Guerrero, Mexico. Near “Los Naranjos” 3 Km. far from Ixcateopan. 18°31′32.4″ N 99°44′42″ W. 2256 m asl | |
| E. longiflora | Gibbiflorae | JE-6013 | Taxco, Guerrero, Mexico. “Cruz Verde, El Puerto” between Taxco and Ixcateopan, 17 Km. far from Taxco. | |
| 15 | E. magnifica | Gibbiflorae | JE-6270 | San Juan Ozolotepec, Oaxaca, Mexico. “Rio Grande” 2 Km North-Northeast from San Juan Ozolotepec. 16°7′37″ N 96°15′18″ W. 1942 m asl |
| 16 | E. multicaulis | Nudae | JE-7501 | Leonardo Bravo, Guerrero, Mexico. “Filo de Caballo” in the way to “Cruz de Ocote”. 17°37′0″ N 99°50′30″ W. 2455 m asl |
| 17 | E. novogaliciana | Gibbiflorae | JE-6580 | Calvillo Municipality, Aguascalientes, Mexico. “Barranca el Montoro” between “Potrero de Lopez” and Milpillas. 21°59′48.87″ N 35′30.8″ W. 2343 m asl |
| E. novogaliciana | Gibbiflorae | JE-6823 | Zapopan, Jalisco, Mexico. Collin Hill, SE from football “las Chivas” stadium. 20°39′45.7″ N 103°27′39.3″ W. 1872 m asl | |
| 18 | E. olivacea | Racemosae | JE-6402 | San Miguel Tenango Municipality, Oaxaca, Mexico. San Pedro Hill. 16°16′38″ N 95°31′44″ W. 1282 m asl |
| E. olivacea | Racemosae | EK-3899 | San Miguel Tenango Municipality, Oaxaca, Mexico. Tehuantepec-Jalapa de Marquez to San Miguel Tenango. 16°16′41.3″ N 95°31′44.5″ W. 1302 m asl | |
| 19 | E. pallida | Gibbiflorae | JE-6475 | San Juan Guichicovi, Oaxaca, Mexico. 4 Km West from “Hierba Santa”. Near to the train track and Malatenco riverbank. 16°17′30″ N 95°1′30″ W. 100 m asl |
| 20 | E. perezcalixii | Gibbiflorae | PCR-6322 | Teul de Gonzalez Ortega, Zacatecas, Mexico. Conejo-Milpillas Road. 21°21′4″ N 103°33′59″ W. 1700 m asl |
| 21 | E. roseiflora | Gibbiflorae | JE-6744 | Morelia, Michoacan, Mexico. “Cañada del Cerro Azul”, 3 Km South from “San Miguel del Monte”. 19°34′57.6″ N 101°7′38.7″ W. 2253 m asl |
| E. roseiflora | Gibbiflorae | JE-6821 | “La Mascota” Municipality, Jalisco, Mexico. at 1.5 Km al oeste de Juanacatlan. 20°35′42.9″ N 104°42′29″ W. 2235 m asl | |
| 22 | E. schaffneri | Angulatae | OZ-54 | Guadalcazar, San Luis Potosi, Mexico. Path to Santa Rita del Rocio to “El Jaujal”, next to the path. 23°3′35″ N 100°17′47″ W. 1641 m asl |
| 23 | E. triquiana | Gibbiflorae | JE-6396 | San Sebastian Tecomaxtlahuaca, Oaxaca, Mexico. “Laguna Encantada”, 5 Km North from Santiago Juxtlahuaca. 17°22′1″ N 98°1′26″ W. 1703 m asl |
| 24 | E. uhlii | Racemosae | JE-6437 | San Pedro Nopala, Oaxaca, Mexico. River Elite, at North from San Pedro Nopala. 17°55′13″ N 97°26′27″ W. 2220 m asl |
| E. uhlii | Racemosae | JE-8553 | San Pedro Nopala, Oaxaca, Mexico. “Cañada del Cerro Pericón”, Nopala. 17°50′7.2″ N 97°33′4.1″ W. 2512 m asl | |
| 25 | E. zorzaniana | Echeveria | JE-7237 | Villa Diaz Ordaz, Oaxaca, Mexico. 3 Km. North from San Miguel del Valle, dirty road from “El Carrizal”-Diaz Ordaz. 17°4′39″ N 96°24′9″ W. 2758 m asl |
| Taxon | Chromosome Number | Internal Standard | 2C DNA Content (pg) ( SE) | 1Cx-Value Mpb | Tukey’s Grouping | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2n | x | Ploidy Level | |||||||||||||
| E. roseifloraδ* | 270 | 27 | 10x | P. sativum | 7.70 ± 0.10 | 753 | a | ||||||||
| E. novogaliciana * | 176 | 27 | 6x + 14 | P. sativum | 5.81 ± 0.06 | 947 | b | ||||||||
| E. gibbiflora | 172 | 27 | 6x + 10 | Z. mays | 3.68 ± 0.05 | 599 | c | ||||||||
| E. altamiraeδ* | 108 | 27 | 4x | Z. mays | 3.54 ± 0.10 | 865 | c | d | |||||||
| E. perezcalixii * | 54 | 27 | 2x | S. lycopersicum | 2.96 ± 0.05 | 1447 | d | e | |||||||
| E. dactylifera | 108 | 27 | 4x | Z. mays | 2.90 ± 0.10 | 709 | d | e | f | ||||||
| E. guerrerensisδ* | 54 | 27 | 2x | P. sativum | 2.88 ± 0.10 | 1408 | d | e | f | ||||||
| E. helmutianaδ* | 42 | 12 | 2x | P. sativum | 2.81 ± 0.10 | 1374 | d | e | f | ||||||
| E. pallida δ | 54 | 27 | 2x | S. lycopersicum | 2.79 ± 0.07 | 1364 | d | e | f | ||||||
| E. guerrerensisδ* | 54 | 27 | 2x | P. sativum | 2.73 ± 0.10 | 1335 | d | e | f | g | |||||
| E. longifloraδ* | 162 | 27 | 6x | P. sativum | 2.54 ± 0.06 | 414 | e | f | g | ||||||
| E. cupreataδ* | 108 | 27 | 4x | P. sativum | 2.50 ± 0.07 | 611 | e | f | g | ||||||
| E. cuicatecanaδ° | 60 | 12 | 5x | Z. mays | 2.44 ± 0.05 | 477 | f | g | |||||||
| E. uhliiδ* | 54 | 27 | 2x | Z. mays | 2.36 ± 0.04 | 1154 | f | g | h | ||||||
| E. carnicolor δ | 36 | 18 | 2x | Z. mays | 2.31 ± 0.10 | 1130 | f | g | h | ||||||
| E. triquianaδ* | 32 | 16 | 2x | Z. mays | 2.07 ± 0.10 | 1012 | g | h | |||||||
| E. olivaceaδ* | 28 | 14 | 2x | Z. mays | 1.96 ± 0.02 | 958 | h | ||||||||
| E. caamanoiδ* | 60 | 12 | 5x | Z. mays | 1.95 ± 0.06 | 381 | h | ||||||||
| E. schaffneri * | 24 | 12 | 2x | Z. mays | 1.50 ± 0.09 | 733 | i | ||||||||
| E. zorzanianaδ* | 40 | 20 | 2x | Z. mays | 1.49 ± 0.00 | 729 | i | ||||||||
| E. multicaulis | 32 | 16 | 2x | S. lycopersicum | 1.40 ± 0.07 | 685 | i | ||||||||
| E. magnificaδ* | 54 | 27 | 2x | S. lycopersicum | 1.36 ± 0.05 | 665 | i | ||||||||
| E. juarezensis δ | 54 | 27 | 2x | S. lycopersicum | 1.31 ± 0.06 | 641 | i | ||||||||
| E. catorce * | 24 | 12 | 2x | S. lycopersicum | 1.29 ± 0.02 | 631 | i | ||||||||
| E. catorce * | 24 | 12 | 2x | S. lycopersicum | 1.26 ± 0.02 | 616 | i | ||||||||
| Series | Mean of Chromosome Number | Standard Error | Tukey–Kramer Test | ||
|---|---|---|---|---|---|
| Gibbiflorae | 104.3 | 15.6 | A | ||
| Racemosae | 40 | 29.2 | B | ||
| Angulatae | 24 | 33.7 | C | ||
| Taxon | Accession Number | Percentage of Nuclei Populations | Number of Endocycles | Cycle Value | Tukey’s Grouping | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2C | 4C | 8C | 16C | 32C | 64C | ||||||||||
| E. schaffneri | 54 | 8.80 | 25.71 | 10.55 | 18.23 | 28.84 | 7.87 | 4 | 2.562 | a | |||||
| E. catorce | 3223 | 12.49 | 19.00 | 11.25 | 22.68 | 27.18 | 7.39 | 4 | 2.552 | a | |||||
| E. cuicatecana | 584 | 20.23 | 11.89 | 10.83 | 22.83 | 28.38 | 5.84 | 4 | 2.448 | a | |||||
| E. olivacea | 6402 | 22.34 | 13.91 | 14.54 | 40.13 | 9.08 | 3 or 4 * | 2.207 | a | b | |||||
| E. catorce | 5469 | 17.99 | 23.72 | 13.82 | 29.15 | 15.31 | 3 or 4 * | 2.240 | a | b | c | ||||
| E. juarezensis | 7538 | 27.15 | 35.07 | 19.16 | 12.37 | 6.26 | 3 or 4 * | 2.155 | a | b | c | d | |||
| E. helmutiana | 7075 | 12.34 | 21.65 | 13.61 | 31.97 | 20.44 | 3 | 2.265 | a | b | c | d | e | ||
| E. caamanoi | 8311 | 22.61 | 25.59 | 10.96 | 12.79 | 23.68 | 4.37 | 4 | 2.024 | a | b | c | d | e | |
| E. uhlii | 8553 | 16.77 | 13.81 | 30.48 | 31.62 | 7.33 | 3 | 1.989 | a | b | c | d | e | ||
| E. magnifica | 6270 | 19.36 | 18.96 | 16.42 | 35.54 | 9.71 | 3 | 1.973 | a | b | c | d | e | ||
| E. guerrerensis | 7526 | 19.50 | 27.19 | 18.70 | 13.42 | 16.90 | 4.30 | 4 | 1.939 | a | b | c | d | e | f |
| E. carnicolor | 8374 | 26.29 | 15.11 | 22.60 | 27.03 | 8.97 | 3 | 1.773 | a | b | c | d | e | f | |
| E. zorzaniana | 7237 | 26.34 | 16.28 | 17.49 | 34.34 | 5.56 | 3 | 1.765 | a | b | c | d | e | f | |
| E. perezcalixii | 6322 | 25.67 | 24.13 | 14.29 | 16.62 | 19.28 | 3 or 4 * | 1.688 | a | b | c | d | e | f | |
| E. novogaliciana | 6823 | 16.31 | 28.60 | 36.25 | 18.85 | 2 | 1.576 | a | b | c | d | e | f | ||
| E. gibiflora | 3427 | 35.77 | 27.16 | 16.38 | 14.20 | 6.48 | 3 | 1.285 | a | b | c | d | e | f | |
| E. multicaulis | 7501 | 26.42 | 32.98 | 36.26 | 4.34 | 2 | 1.185 | a | b | c | d | e | f | ||
| E. dactylifera | 2603 | 41.23 | 22.07 | 18.03 | 16.91 | 1.75 | 3 | 1.159 | a | b | c | d | e | f | |
| E. pallida | 6475 | 38.75 | 18.93 | 14.12 | 17.18 | 11.02 | 3 | 1.428 | b | c | d | e | f | ||
| E. guerrerensis | 7521 | 40.24 | 21.71 | 16.74 | 13.97 | 7.33 | 3 | 1.264 | b | c | d | e | f | ||
| E. triquiana | 6396 | 29.81 | 30.11 | 31.93 | 8.15 | 2 | 1.184 | b | c | d | e | f | |||
| E. altamirae | 7548 | 39.49 | 22.33 | 20.97 | 17.21 | 2 | 1.159 | c | d | e | f | ||||
| E. longiflora | 6923 | 42.54 | 19.67 | 21.98 | 15.80 | 2 or 3 * | 1.105 | d | e | f | |||||
| E. cupreata | 6807 | 65.65 | 16.91 | 6.08 | 5.55 | 5.81 | 3 | 0.690 | e | f | |||||
| E. roseiflora | 6744 | 37.57 | 48.36 | 14.07 | 1 or 2 * | 0.815 | f | ||||||||
| Taxon | Accession Number | Percentage of Nuclei Populations | Number of Endocycles | Cycle Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| 2C | 4C | 8C | 16C | 32C | 64C | ||||
| E. catorce | 5469 | 17.99 | 23.72 | 13.82 | 29.15 | 15.31 | 3 | 2.001 | |
| E. catorce | 5469 | 13.22 | 21.31 | 9.25 | 21.00 | 30.99 | 4.23 | 4 | 2.479 |
| E. olivacea | 6402 | 22.34 | 13.91 | 14.54 | 40.13 | 9.08 | 3 | 1.997 | |
| E. olivacea | 6402 | 17.54 | 19.40 | 10.71 | 14.71 | 31.55 | 6.10 | 4 | 2.416 |
| E. juarezensis | 7538 | 27.15 | 35.07 | 19.16 | 12.37 | 6.26 | 3 | 1.355 | |
| E. juarezensis | 7538 | 15.66 | 16.68 | 12.86 | 16.48 | 27.96 | 10.36 | 4 | 2.555 |
| E. perezcalixii | 6322 | 25.67 | 24.13 | 14.29 | 16.62 | 19.28 | 3 | 1.797 | |
| E. perezcalixii | 6322 | 40.96 | 19.81 | 11.73 | 11.15 | 11.35 | 5.00 | 4 | 1.471 |
| E. longiflora | 6923 | 42.54 | 19.67 | 21.98 | 15.80 | 2 | 1.110 | ||
| E. longiflora | 6013 | 50.84 | 15.29 | 9.49 | 21.91 | 2.46 | 3 | 1.099 | |
| E. roseiflora | 6744 | 37.57 | 48.36 | 14.07 | 1 | 0.765 | |||
| E. roseiflora | 6744 | 37.50 | 33.51 | 23.96 | 5.03 | 2 | 0.965 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palomino, G.; Martínez-Ramón, J.; Cepeda-Cornejo, V.; Ladd-Otero, M.; Romero, P.; Reyes-Santiago, J. Chromosome Number, Ploidy Level, and Nuclear DNA Content in 23 Species of Echeveria (Crassulaceae). Genes 2021, 12, 1950. https://doi.org/10.3390/genes12121950
Palomino G, Martínez-Ramón J, Cepeda-Cornejo V, Ladd-Otero M, Romero P, Reyes-Santiago J. Chromosome Number, Ploidy Level, and Nuclear DNA Content in 23 Species of Echeveria (Crassulaceae). Genes. 2021; 12(12):1950. https://doi.org/10.3390/genes12121950
Chicago/Turabian StylePalomino, Guadalupe, Javier Martínez-Ramón, Verónica Cepeda-Cornejo, Miriam Ladd-Otero, Patricia Romero, and Jerónimo Reyes-Santiago. 2021. "Chromosome Number, Ploidy Level, and Nuclear DNA Content in 23 Species of Echeveria (Crassulaceae)" Genes 12, no. 12: 1950. https://doi.org/10.3390/genes12121950
APA StylePalomino, G., Martínez-Ramón, J., Cepeda-Cornejo, V., Ladd-Otero, M., Romero, P., & Reyes-Santiago, J. (2021). Chromosome Number, Ploidy Level, and Nuclear DNA Content in 23 Species of Echeveria (Crassulaceae). Genes, 12(12), 1950. https://doi.org/10.3390/genes12121950






