Genetic Polymorphism and Natural Selection of Apical Membrane Antigen-1 in Plasmodium falciparum Isolates from Vietnam
Abstract
:1. Introduction
2. Materials and Methods
2.1. Blood Samples and Ethics
2.2. Amplification and Sequence Analysis of pfama-1
2.3. Nucleotide Sequence Polymorphism and Neutrality Test
2.4. Population Diversity of pfama-1 among Global P. falciparum Isolates
3. Results
3.1. Sequence Polymorphism of Vietnamese pfama-1
3.2. Amino Acid Polymorphisms in Vietnamese pfama-1 Compared with Global pfama-1
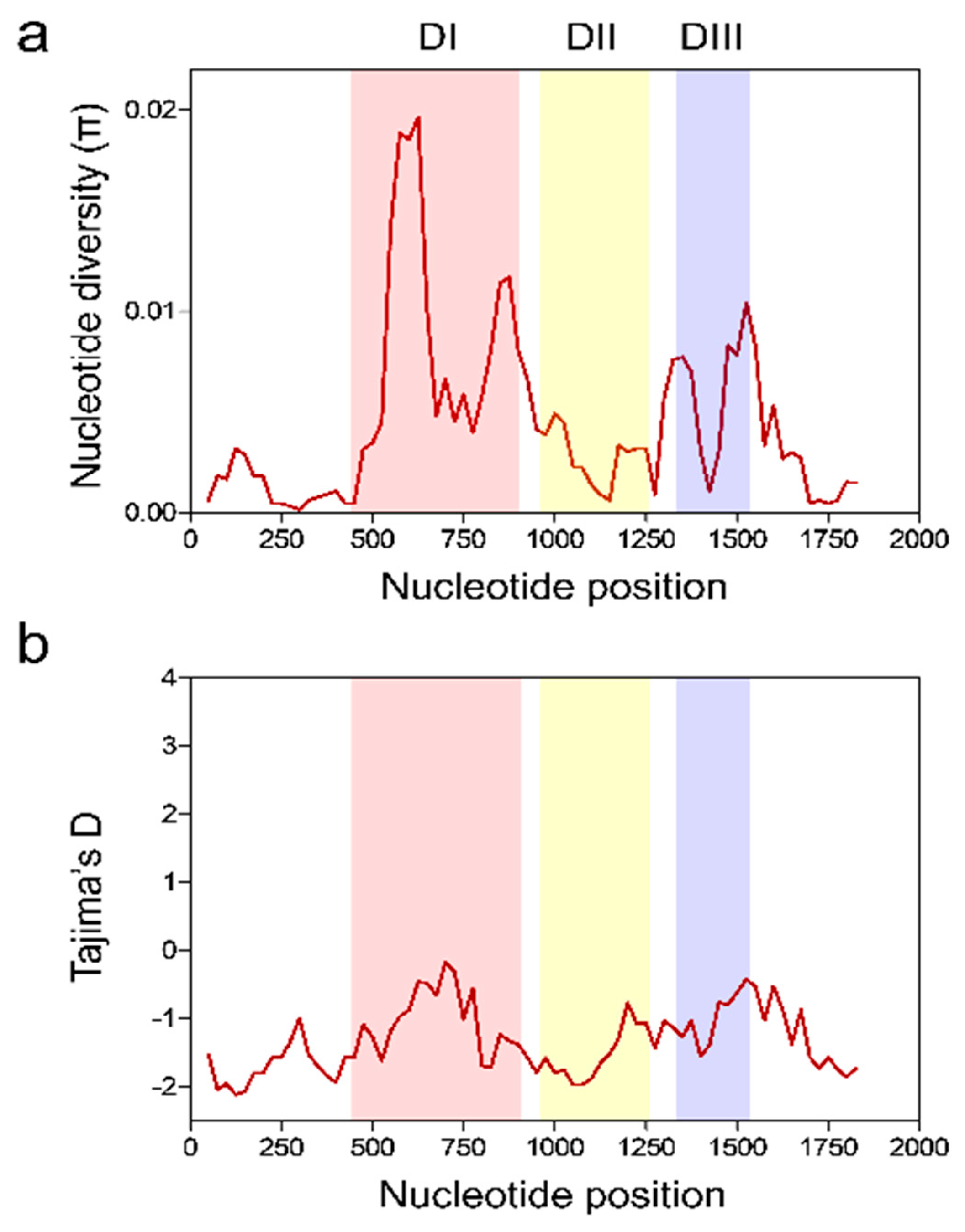
3.3. Nucleotide Diversity and Natural Selection of Vietnamese pfama-1
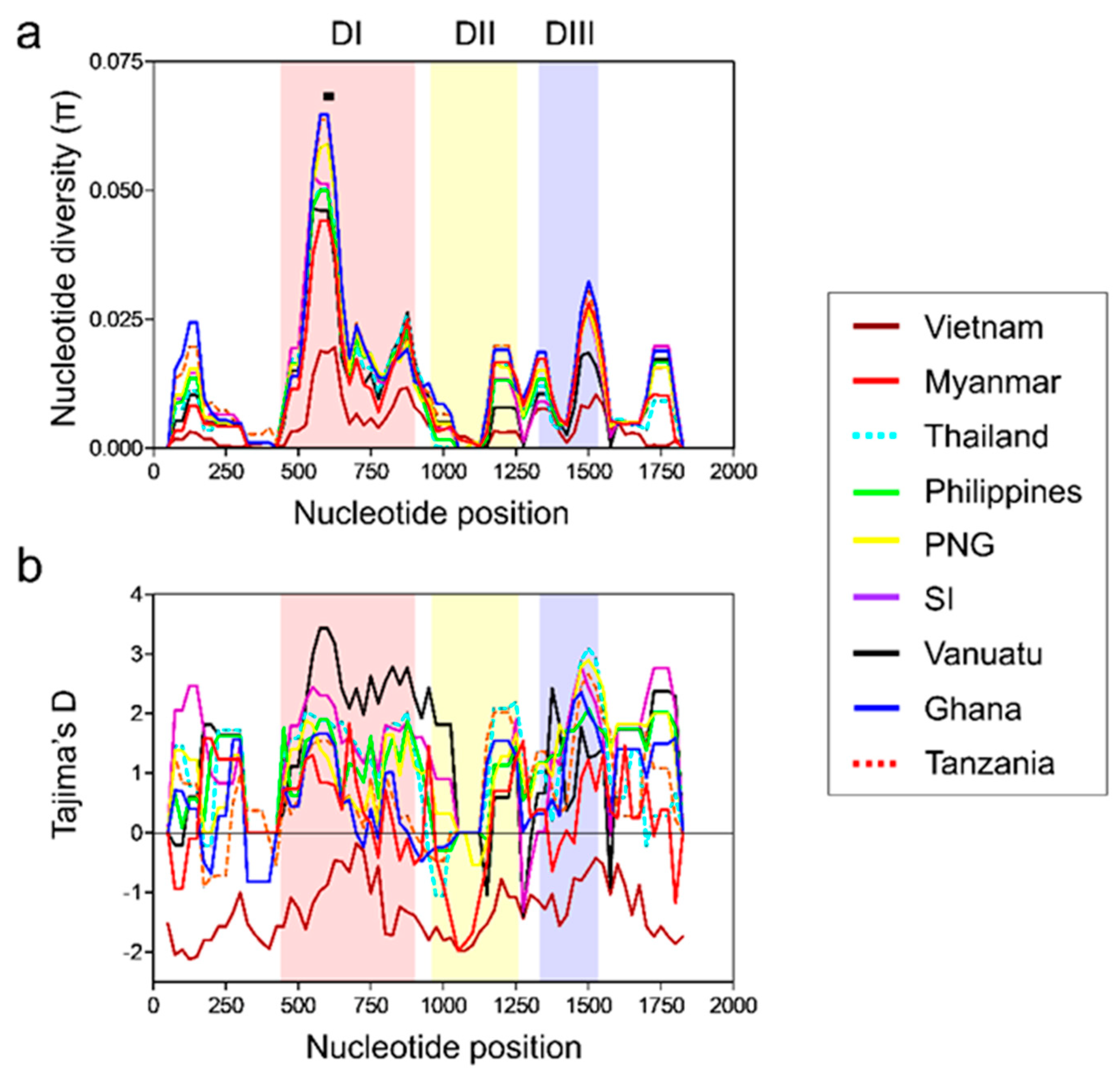
3.4. Nucleotide Diversity and Natural Selection of Global pfama-1
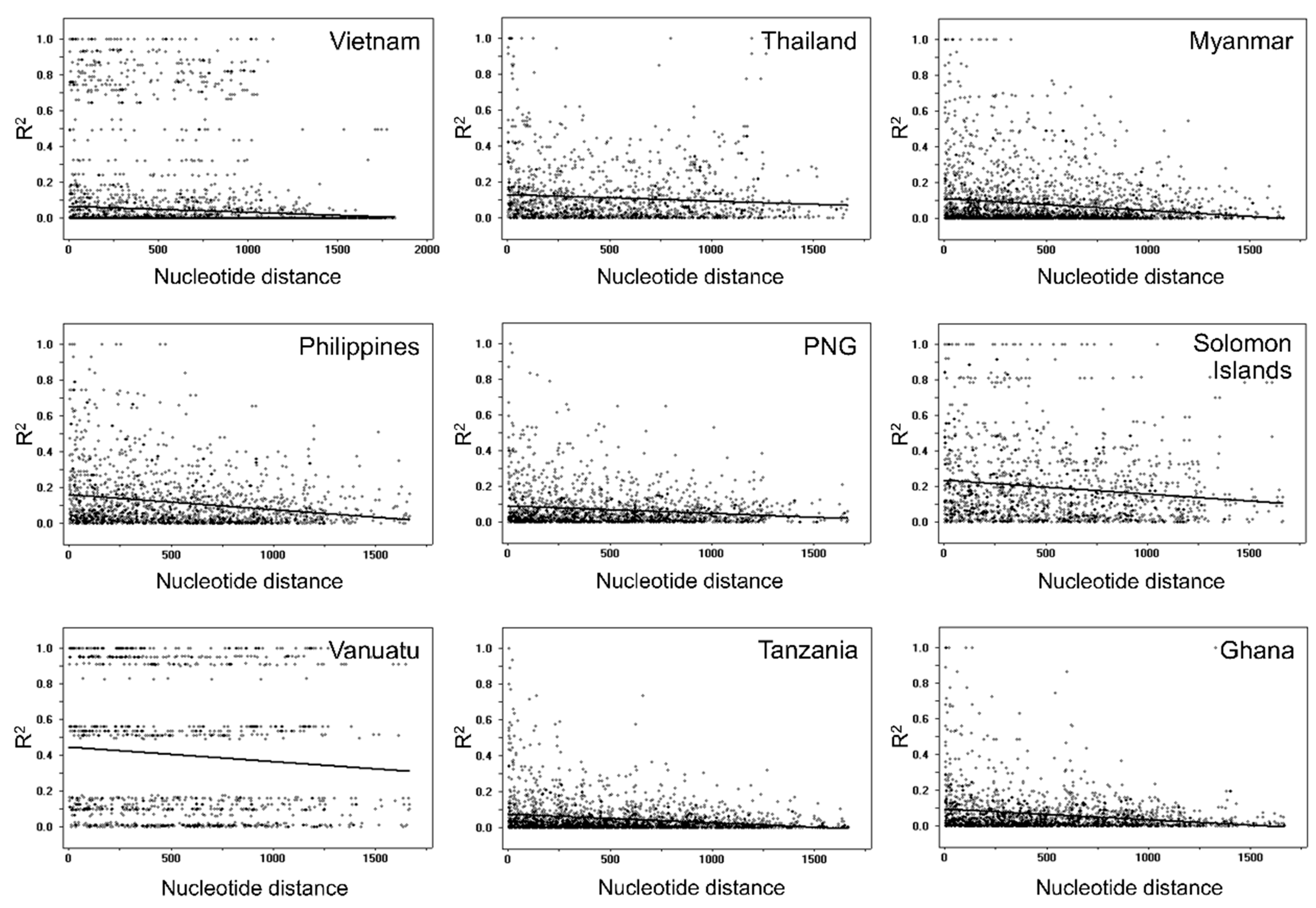
3.5. Recombination and Linkage Disequilibrium
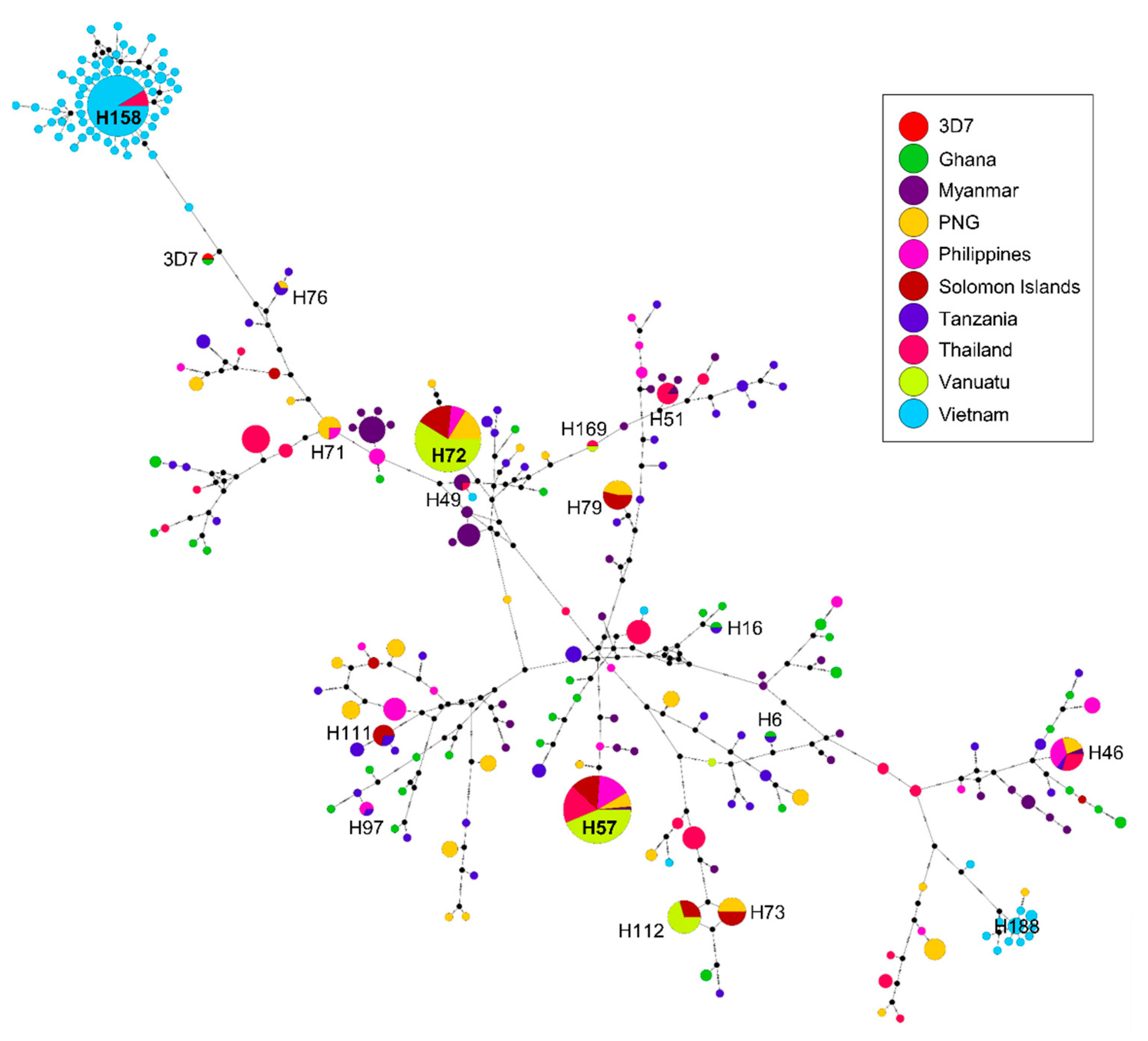
3.6. Haplotype Network Analysis
3.7. Nucleotide Differentiation among Global pfama-1
3.8. Association between Natural Selection and Host Immune Pressure
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Malaria Report: 20 Years of Global Progress and Challenges; World Health Organization: Geneva, Switzerland, 2020.
- Menard, D.; Dondorp, A. Antimalarial drug resistance: A threat to malaria elimination. Cold Spring Harb. Perspect. Med. 2017, 7, a025619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodder, A.N.; Crewther, P.E.; Matthew, M.L.S.M.; Reid, G.E.; Moritz, R.L.; Simpson, R.J.; Anders, R.F. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 1996, 271, 29446–29452. [Google Scholar] [CrossRef] [Green Version]
- Peterson, M.G.; Marshall, V.M.; Smythe, J.A.; Crewther, P.E.; Lew, A.; Silva, A.; Anders, R.F.; Kemp, D.J. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol. Cell. Biol. 1989, 9, 3151–3154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Healer, J.; Crawford, S.; Ralph, S.; McFadden, G.; Cowman, A.F. Independent translocation of two micronemal proteins in developing Plasmodium falciparum merozoites. Infect. Immun. 2002, 70, 5751–5758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, G.H.; Thomas, A.W.; Margos, G.; Dluzewski, A.R.; Bannister, L.H. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect. Immun. 2004, 72, 154–158. [Google Scholar] [CrossRef] [Green Version]
- Silvie, O.; Franetich, J.-F.; Charrin, S.; Mueller, M.S.; Siau, A.; Bodescot, M.; Rubinstein, E.; Hannoun, L.; Charoenvit, Y.; Kocken, C.; et al. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J. Biol. Chem. 2004, 279, 9490–9496. [Google Scholar] [CrossRef] [Green Version]
- Yap, A.; Azevedo, M.; Gilson, P.R.; Weiss, G.E.; O’Neill, M.T.; Wilson, D.W.; Crabb, B.S.; Cowman, A.F. Conditional expression of apical membrane antigen 1 in Plasmodium falciparum shows it is required for erythrocyte invasion by merozoites. Cell. Microbiol. 2014, 16, 642–656. [Google Scholar] [CrossRef] [Green Version]
- Udhayakumar, V.; Kariuki, S.; Nahlen, B.L.; Kolczack, M.; A Lal, A.; Roberts, J.M.; Oloo, A.J.; Girma, M. Longitudinal study of natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in a holoendemic region of malaria in western Kenya: Asembo Bay Cohort Project VIII. Am. J. Trop. Med. Hyg. 2001, 65, 100–107. [Google Scholar] [CrossRef] [Green Version]
- Cortés, A.; Mellombo, M.; Masciantonio, R.; Murphy, V.J.; Reeder, J.C.; Anders, R.F. Allele specificity of naturally acquired antibody responses against Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 2005, 73, 422–430. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, M.H.C.; Rodrigues, K.M.; Oliveira, T.R.; Cômodo, A.N.; Rodrigues, M.M.; Kocken, C.H.M.; Thomas, A.W.; Soares, I.S. Antibody response of naturally infected individuals to recombinant Plasmodium vivax apical membrane antigen-1. Int. J. Parasitol. 2005, 35, 185–192. [Google Scholar] [CrossRef]
- Moncunill, G.; Mayor, A.; Jiménez, A.; Nhabomba, A.; Vila, N.C.; Puyol, L.; Campo, J.J.; Manaca, M.N.; Aguilar, R.; Pinazo, M.-J.; et al. High antibody responses against Plasmodium falciparum in immigrants after extended periods of interrupted exposure to malaria. PLoS ONE 2013, 8, e73624. [Google Scholar] [CrossRef] [Green Version]
- Remarque, E.J.; Faber, B.W.; Kocken, C.H.M.; Thomas, A.W. Apical membrane antigen 1: A malaria vaccine candidate in review. Trends Parasitol. 2008, 24, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Thera, M.A.; Coulibaly, D.; Kone, A.K.; Guindo, A.B.; Traoré, K.; Sall, A.H.; Diarra, I.; Daou, M.; Traore, I.M.; Tolo, Y.; et al. Phase 1 randomized controlled trial to evaluate the safety and immunogenicity of recombinant Pichia pastoris-expressed Plasmodium falciparum apical membrane antigen 1 (PfAMA1-FVO [25-545]) in healthy Malian adults in Bandiagara. Malar. J. 2016, 15, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polley, S.D.; Mwangi, T.; Kocken, C.H.M.; Thomas, A.W.; Dutta, S.; Lanar, D.E.; Remarque, E.; Ross, A.; Williams, T.N.; Mwambingu, G.; et al. Human antibodies to recombinant protein constructs of Plasmodium falciparum apical membrane antigen 1 (AMA1) and their associations with protection from malaria. Vaccine 2004, 23, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Takala, S.L.; Coulibaly, D.; Thera, M.A.; Batchelor, A.H.; Cummings, M.P.; Escalante, A.A.; Ouattara, A.; Traoré, K.; Niangaly, A.; Djimdé, A.A.; et al. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: Implications for vaccine development. Sci. Transl. Med. 2009, 1, 2ra5. [Google Scholar] [CrossRef] [Green Version]
- Mugyenyi, C.K.; Elliott, S.R.; McCallum, F.J.; Anders, R.F.; Marsh, K.; Beeson, J.G. Antibodies to polymorphic invasion-inhibitory and non-inhibitory epitopes of Plasmodium falciparum apical membrane antigen 1 in human malaria. PLoS ONE 2013, 8, e68304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escalante, A.A.; Grebert, H.M.; Chaiyaroj, S.C.; Magris, M.; Biswas, S.; Nahlen, B.L.; Lal, A.A. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol. Biochem. Parasitol. 2001, 113, 279–287. [Google Scholar] [CrossRef]
- Polley, S.D.; Conway, D.J. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 2001, 158, 1505–1512. [Google Scholar] [CrossRef]
- Moon, S.-U.; Na, B.-K.; Kang, J.-M.; Kim, J.-Y.; Cho, S.-H.; Park, Y.-K.; Sohn, W.-M.; Lin, K.; Kim, T.-S. Genetic polymorphism and effect of natural selection at domain I of apical membrane antigen-1 (AMA-1) in Plasmodium vivax isolates from Myanmar. Acta Trop. 2010, 114, 71–75. [Google Scholar] [CrossRef]
- Arnott, A.; Mueller, I.; Ramsland, P.A.; Siba, P.M.; Reeder, J.C.; Barry, A.E. Global population structure of the genes encoding the malaria vaccine candidate, Plasmodium vivax apical membrane antigen 1 (PvAMA1). PLoS Negl. Trop. Dis. 2013, 7, e2506. [Google Scholar] [CrossRef] [Green Version]
- Terheggen, U.; Drew, D.R.; Hodder, A.N.; Cross, N.J.; Mugyenyi, C.K.; Barry, A.E.; Anders, R.F.; Dutta, S.; Osier, F.H.A.; Elliott, S.R.; et al. Limited antigenic diversity of Plasmodium falciparumapical membrane antigen 1 supports the development of effective multi-allele vaccines. BMC Med. 2014, 12, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.-M.; Lee, J.; Cho, P.-Y.; Moon, S.-U.; Ju, H.-L.; Ahn, S.K.; Sohn, W.-M.; Lee, H.-W.; Kim, T.-S.; Na, B.-K. Population genetic structure and natural selection of apical membrane antigen-1 in Plasmodium vivax Korean isolates. Malar. J. 2015, 14, 455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Zhao, P.; Wang, S.; Liu, F.; Liu, J.; Wang, J.; Yang, Z.; Yan, G.; Fan, Q.; Cao, Y.; et al. Analysis of Pvama1 genes from China-Myanmar border reveals little regional genetic differentiation of Plasmodium vivax populations. Parasites Vectors 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.-M.; Lee, J.-Y.; Moe, M.; Jun, H.; Lê, H.G.; Kim, T.I.; Thái, T.L.; Sohn, W.-M.; Myint, M.K.; Lin, K.; et al. Population genetic structure and natural selection of Plasmodium falciparum apical membrane antigen-1 in Myanmar isolates. Malar. J. 2018, 17, 1–17. [Google Scholar] [CrossRef]
- Afridi, S.G.; Irfan, M.; Ahmad, H.; Aslam, M.; Nawaz, M.; Ilyas, M.; Khan, A. Population genetic structure of domain I of apical membrane antigen-1 in Plasmodium falciparum isolates from Hazara division of Pakistan. Malar. J. 2018, 17, 389. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-N.; Lin, M.; Liang, X.-Y.; Chen, J.-T.; Xie, D.-D.; Wang, Y.-L.; Ehapo, C.S.; Eyi, U.M.; Huang, H.-Y.; Wu, J.-L.; et al. Natural selection and genetic diversity of domain I of Plasmodium falciparum apical membrane antigen-1 on Bioko Island. Malar. J. 2019, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Health; National Malaria Control and Elimination Programme. National Strategy for Malaria Control and Elimination for the Period of 2012–2015; National Institute of Malariology Parasitology and Entomology: Hanoi, Vietnam, 2012. [Google Scholar]
- Goldlust, S.M.; Thuan, P.D.; Giang, D.D.H.; Thang, N.D.; Thwaites, G.E.; Farrar, J.; Thanh, N.V.; Nguyen, T.D.; Grenfell, B.T.; Boni, M.F.; et al. The decline of malaria in Vietnam, 1991–2014. Malar. J. 2018, 17, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Long, B.; Allen, G.; Brauny, M.; Linh, L.T.K.; Pallerla, S.R.; Huyen, T.T.T.; van Tong, H.; Toan, N.L.; Quyet, D.; Son, H.A.; et al. Molecular surveillance and temporal monitoring of malaria parasites in focal Vietnamese provinces. Malar. J. 2020, 19, 1–7. [Google Scholar] [CrossRef]
- Võ, T.C.; Lê, H.G.; Kang, J.-M.; Naw, H.; Fan, C.-K.; Trinh, N.T.M.; Quang, H.H.; Na, B.-K. Molecular surveillance of malaria in the Central Highlands, Vietnam. Parasitol. Int. 2021, 83, 102374. [Google Scholar] [CrossRef]
- Phan, G.T.; de Vries, P.J.; Tran, B.Q.; Le, H.Q.; Nguyen, N.V.; Nguyen, T.V.; Heisterkamp, S.H.; Kager, P.A. Artemisinin or chloroquine for blood stage Plasmodium vivax malaria in Vietnam. Trop. Med. Int. Health 2002, 7, 858–864. [Google Scholar] [CrossRef]
- Phuc, B.Q.; Rasmussen, C.; Duong, T.T.; Dong, L.T.; Loi, M.A.; Menard, D.; Tarning, J.; Bustos, D.; Ringwald, P.; Galappaththy, G.L.; et al. Treatment failure of dihydroartemisinin/piperaquine for Plasmodium falciparum malaria, Vietnam. Emerg. Infect. Dis. 2017, 23, 715–717. [Google Scholar] [CrossRef] [Green Version]
- Ménard, D.; Fidock, D. Accelerated evolution and spread of multidrug-resistant Plasmodium falciparum takes down the latest first-line antimalarial drug in southeast Asia. Lancet Infect. Dis. 2019, 19, 916–917. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, W.; Amato, R.; van der Pluijm, R.W.; Jacob, C.G.; Quang, H.H.; Thuy-Nhien, N.T.; Hien, T.T.; Hongvanthong, B.; Chindavongsa, K.; Mayxay, M.; et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: A genomic epidemiology study. Lancet Infect. Dis. 2019, 19, 943–951. [Google Scholar] [CrossRef]
- Snounou, G.; Singh, B. Nested PCR analysis of Plasmodium parasites. Malar. Methods Protoc. 2002, 72, 189–204. [Google Scholar] [CrossRef]
- Kang, J.-M.; Cho, P.-Y.; Moe, M.; Lee, J.; Jun, H.; Lee, H.-W.; Ahn, S.K.; Kim, T.I.; Pak, J.H.; Myint, M.K.; et al. Comparison of the diagnostic performance of microscopic examination with nested polymerase chain reaction for optimum malaria diagnosis in Upper Myanmar. Malar. J. 2017, 16, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X.; Li, W.-H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Urquiza, M.; Suarez, J.E.; Cardenas, C.; Lopez, R.; Puentes, A.; Chavez, F.; Calvo, J.C.; Patarroyo, M.E. Plasmodium falciparum AMA-1 erythrocyte binding peptides implicate AMA-1 as erythrocyte binding protein. Vaccine 2000, 19, 508–513. [Google Scholar] [CrossRef]
- Mehrizi, A.A.; Sepehri, M.; Karimi, F.; Djadid, N.D.; Zakeri, S. Population genetics, sequence diversity and selection in the gene encoding the Plasmodium falciparum apical membrane antigen 1 in clinical isolates from the south-east of Iran. Infect. Genet. Evol. 2013, 17, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Bai, T.; Becker, M.; Gupta, A.; Strike, P.; Murphy, V.J.; Anders, R.F.; Batchelor, A.H. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. USA 2005, 102, 12736–12741. [Google Scholar] [CrossRef] [Green Version]
- Cortés, A.; Mellombo, M.; Mueller, I.; Benet, A.; Reeder, J.C.; Anders, R.F. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 2003, 71, 1416–1426. [Google Scholar] [CrossRef] [Green Version]
- Basu, M.; Maji, A.K.; Mitra, M.; Sengupta, S. Natural selection and population genetic structure of domain-I of Plasmodium falciparum apical membrane antigen-1 in India. Infect. Genet. Evol. 2013, 18, 247–256. [Google Scholar] [CrossRef]
- Al-Qahtani, A.A.; Abdel-Muhsin, A.-M.A.; Dajem, S.M.B.; AlSheikh, A.A.H.; Bohol, M.F.F.; Al-Ahdal, M.N.; Putaporntip, C.; Jongwutiwes, S. Comparative sequence analysis of domain I of Plasmodium falciparum apical membrane antigen 1 from Saudi Arabia and worldwide isolates. Infect. Genet. Evol. 2016, 39, 381–388. [Google Scholar] [CrossRef]
- Zhu, X.; Zhao, Z.; Feng, Y.; Li, P.; Liu, F.; Liu, J.; Yang, Z.; Yan, G.; Fan, Q.; Cao, Y.; et al. Genetic diversity of the Plasmodium falciparum apical membrane antigen I gene in parasite population from the China–Myanmar border area. Infect. Genet. Evol. 2016, 39, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Balloux, F.; Lugon-Moulin, N. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 2002, 11, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.-M.; Lee, J.-Y.; Cho, P.-Y.; Kim, T.I.; Sohn, W.-M.; Park, J.-W.; Kim, T.-S.; Na, B.-K. Dynamic changes of Plasmodium vivax population structure in South Korea. Infect. Genet. Evol. 2016, 45, 90–94. [Google Scholar] [CrossRef]
- Maneerattanasak, S.; Gosi, P.; Krudsood, S.; Tongshoob, J.; Lanteri, C.A.; Snounou, G.; Khusmith, S. Genetic diversity among Plasmodium vivax isolates along the Thai–Myanmar border of Thailand. Malar. J. 2016, 15, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Tuo, F.; Liang, Y.; Wu, W.; Wu, G.; Huang, S.; Zhong, Q.; Su, X.Z.; Zhang, H.; Li, M.; et al. Temporal changes in genetic diversity of msp-1, msp-2, and msp-3 in Plasmodium falciparum isolates from Grande Comore Island after introduction of ACT. Malar. J. 2018, 17, 1–14. [Google Scholar] [CrossRef]
- Ngomo, J.N.; M’Bondoukwe, N.P.; Yavo, W.; Mavoungou, L.B.; Bouyou-Akotet, M.K.; Mawili-Mboumba, D.P. Spatial and temporal distribution of Pfmsp1 and Pfmsp2 alleles and genetic profile change of Plasmodium falciparum populations in Gabon. Acta Trop. 2018, 178, 27–33. [Google Scholar] [CrossRef]
- Lê, H.G.; Kang, J.-M.; Jun, H.; Lee, J.-Y.; Thái, T.L.; Myint, M.K.; Aye, K.S.; Sohn, W.-M.; Shin, H.-J.; Kim, T.-S.; et al. Changing pattern of the genetic diversities of Plasmodium falciparum merozoite surface protein-1 and merozoite surface protein-2 in Myanmar isolates. Malar. J. 2019, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Eisen, D.P.; Marshall, V.M.; Billman-Jacobe, H.; Coppel, R.L. A Plasmodium falciparum apical membrane antigen-1 (AMA-1) gene apparently generated by intragenic recombination. Mol. Biochem. Parasitol. 1999, 100, 243–246. [Google Scholar] [CrossRef]
- Garg, S.; Alam, M.T.; Das, M.K.; Dev, V.; Kumar, A.; Dash, A.P.; Sharma, Y.D. Sequence diversity and natural selection at domain I of the apical membrane antigen 1 among Indian Plasmodium falciparum populations. Malar. J. 2007, 6, 154. [Google Scholar] [CrossRef] [Green Version]
- Coley, A.M.; Parisi, K.; Masciantonio, R.; Hoeck, J.; Casey, J.L.; Murphy, V.J.; Harris, K.S.; Batchelor, A.H.; Anders, R.F.; Foley, M. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect. Immun. 2006, 74, 2628–2636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coley, A.M.; Gupta, A.; Murphy, V.J.; Bai, T.; Kim, H.; Anders, R.F.; Foley, M.; Batchelor, A.H. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog. 2007, 3, e138-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, S.; Lee, S.Y.; Batchelor, A.H.; Lanar, D.E. Structural basis of antigenic escape of a malaria vaccine candidate. Proc. Natl. Acad. Sci. USA 2007, 104, 12488–12493. [Google Scholar] [CrossRef] [Green Version]
- Harris, K.S.; Adda, C.G.; Khore, M.; Drew, D.R.; Valentini-Gatt, A.; Fowkes, F.J.I.; Beeson, J.G.; Dutta, S.; Anders, R.F.; Foley, M. Use of immunodampening to overcome diversity in the malarial vaccine candidate apical membrane antigen 1. Infect. Immun. 2014, 82, 4707–4717. [Google Scholar] [CrossRef] [Green Version]
- Collins, C.R.; Withers-Martinez, C.; Bentley, G.A.; Batchelor, A.H.; Thomas, A.W.; Blackman, M.J. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. J. Biol. Chem. 2007, 282, 7431–7441. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Fragment | Segregating Sites (S) | Singleton Variable Sites | Parsimony Informative Sites | Total No. of Mutations | K | H | Hd ± SD | π ± SD | dN−dS | Tajima’s D |
|---|---|---|---|---|---|---|---|---|---|---|
| 5′-Terminal | 21 | 12 | 9 | 21 | 0.485 | 20 | 0.358 ± 0.055 | 0.0011 ± 0.0002 | −0.0018 | −2.4673 (p < 0.01) |
| Domain I | 41 | 12 | 29 | 42 | 4.198 | 30 | 0.575 ± 0.052 | 0.0091 ± 0.0014 | 0.0084 | −1.3889 (p > 0.1) |
| Domain II | 17 | 4 | 13 | 18 | 0.915 | 21 | 0.465 ± 0.053 | 0.0031 ± 0.0004 | 0.0020 | −1.9940 (p < 0.05) |
| Domain III | 14 | 5 | 9 | 14 | 1.526 | 16 | 0.455 ± 0.053 | 0.0076 ± 0.0012 | −0.0003 | −1.0724 (p > 0.1) |
| 3′-Terminal | 14 | 9 | 5 | 14 | 0.772 | 17 | 0.432 ± 0.053 | 0.0023 ± 0.0003 | −0.0052 | −1.8470 (p < 0.05) |
| Full | 114 | 47 | 67 | 116 | 8.062 | 73 | 0.837 ± 0.034 | 0.0043 ± 0.0006 | 0.0009 | −2.0046 (p < 0.05) |
| Isolates | Segregating Sites (S) | Singleton Variable Sites | Parsimony Informative Sites | Total No. of Mutations | K | H | Hd ± SD | π ± SD | dN/dS | Tajima’s D | Fu and Li’s D | Fu and Li’s F |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vietnam (n = 131) | 114 | 47 | 67 | 116 | 8.062 | 73 | 0.837 ± 0.034 | 0.0043 ± 0.0006 | 1.250 | −2.0046 (p < 0.05) | −3.1913 (p < 0.05) | −3.1959 (p < 0.02) |
| Myanmar # (n = 58) | 78 | 16 | 62 | 79 | 19.886 | 37 | 0.948 ± 0.019 | 0.0106 ± 0.0005 | 6.524 | 0.5719 (p > 0.1) | 0.1677 (p > 0.1) | 0.3851 (p > 0.1) |
| Thailand # (n = 80) | 58 | 5 | 53 | 61 | 20.621 | 21 | 0.920 ± 0.013 | 0.0110 ± 0.0002 | 6.698 | 2.2222 (p < 0.05) | 0.9572 (p > 0.1) | 1.7516 (p < 0.05) |
| Philippines # (n = 55) | 61 | 3 | 58 | 62 | 21.232 | 19 | 0.916 ± 0.019 | 0.0114 ± 0.0003 | 7.200 | 1.9545 (0.1 > p > 0.05) | 1.6123 (p < 0.05) | 2.07960 (p < 0.02) |
| PNG # (n = 90) | 68 | 2 | 66 | 75 | 22.923 | 28 | 0.954 ± 0.007 | 0.0123 ± 0.0002 | 7.497 | 1.8082 (0.1 > p > 0.05) | 1.6759 (p < 0.05) | 2.0760 (p < 0.02) |
| Solomon Islands # (n = 50) | 56 | 3 | 53 | 57 | 22.044 | 9 | 0.862 ± 0.021 | 0.0118 ± 0.0003 | 7.023 | 2.5455 (p < 0.05) | 1.5447 (p < 0.05) | 2.2804 (p < 0.02) |
| Vanuatu # (n = 85) | 50 | 6 | 44 | 50 | 18.905 | 5 | 0.633 ± 0.028 | 0.0101 ± 0.0003 | 6.471 | 2.8973 (p < 0.01) | 0.8642 (p > 0.1) | 2.0039 (p < 0.05) |
| Ghana # (n = 37) | 75 | 7 | 68 | 85 | 26.261 | 32 | 0.992 ± 0.008 | 0.0141 ± 0.0003 | 7.423 | 1.0674 (p > 0.1) | 1.2179 (p > 0.1) | 1.38500 (p > 0.1) |
| Tanzania # (n = 62) | 81 | 8 | 73 | 89 | 25.444 | 47 | 0.989 ± 0.005 | 0.0136 ± 0.0002 | 8.135 | 1.1819 (p > 0.1) | 1.1635 (p > 0.1) | 1.4020 (p > 0.1) |
| n | Ra | Rb | Rm | |
|---|---|---|---|---|
| Vietnam | 131 | 0.0000 | 0.001 | 20 |
| Myanmar # | 78 | 0.0141 | 26.3 | 24 |
| Thailand # | 38 | 0.0243 | 45.3 | 20 |
| Philippines # | 61 | 0.0218 | 40.8 | 17 |
| PNG # | 68 | 0.0500 | 93.4 | 28 |
| Solomon Islands # | 56 | 0.0120 | 22.5 | 14 |
| Vanuatu # | 50 | 0.0010 | 1.8 | 9 |
| Ghana # | 75 | 0.0921 | 172 | 27 |
| Tanzania # | 81 | 0.0915 | 171 | 28 |
| Vietnam | Myanmar # | Thailand # | Philippines # | PNG # | SI # | Vanuatu # | Ghana # | Tanzania # | |
|---|---|---|---|---|---|---|---|---|---|
| Vietnam | 11.56767 | 12.82408 | 11.95679 | 14.11411 | 11.92480 | 12.32934 | 12.07054 | 13.64608 | |
| Myanmar # | 0.40780 | 20.15798 | 20.35295 | 21.58878 | 20.68805 | 19.15425 | 22.14497 | 20.15798 | |
| Thailand # | 0.37881 | 0.06073 | 20.87018 | 21.83959 | 21.16846 | 19.73726 | 22.40481 | 22.72679 | |
| Philippines # | 0.41382 | 0.08390 | 0.03818 | 22.28146 | 21.61888 | 19.81950 | 23.25483 | 23.46398 | |
| PNG # | 0.37343 | 0.05281 | 0.03818 | 0.02319 | 22.60884 | 20.97135 | 23.89528 | 25.95093 | |
| SI # | 0.42009 | 0.09605 | 0.06636 | 0.03529 | 0.03529 | 20.06783 | 23.83759 | 23.92600 | |
| Vanuatu # | 0.40551 | 0.09740 | 0.10535 | 0.06689 | 0.06689 | 0.04933 | 21.13622 | 21.66300 | |
| Ghana # | 0.37955 | 0.07802 | 0.04140 | 0.04167 | 0.03640 | 0.04953 | 0.12425 | 25.74924 | |
| Tanzania # | 0.39934 | 0.08139 | 0.05865 | 0.05787 | 0.03662 | 0.05782 | 0.14141 | 0.00019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, J.-M.; Lê, H.G.; Võ, T.C.; Naw, H.; Yoo, W.G.; Sohn, W.-M.; Trinh, N.T.M.; Quang, H.-H.; Na, B.-K. Genetic Polymorphism and Natural Selection of Apical Membrane Antigen-1 in Plasmodium falciparum Isolates from Vietnam. Genes 2021, 12, 1903. https://doi.org/10.3390/genes12121903
Kang J-M, Lê HG, Võ TC, Naw H, Yoo WG, Sohn W-M, Trinh NTM, Quang H-H, Na B-K. Genetic Polymorphism and Natural Selection of Apical Membrane Antigen-1 in Plasmodium falciparum Isolates from Vietnam. Genes. 2021; 12(12):1903. https://doi.org/10.3390/genes12121903
Chicago/Turabian StyleKang, Jung-Mi, Hương Giang Lê, Tuấn Cường Võ, Haung Naw, Won Gi Yoo, Woon-Mok Sohn, Nguyen Thi Minh Trinh, Huynh-Hong Quang, and Byoung-Kuk Na. 2021. "Genetic Polymorphism and Natural Selection of Apical Membrane Antigen-1 in Plasmodium falciparum Isolates from Vietnam" Genes 12, no. 12: 1903. https://doi.org/10.3390/genes12121903
APA StyleKang, J.-M., Lê, H. G., Võ, T. C., Naw, H., Yoo, W. G., Sohn, W.-M., Trinh, N. T. M., Quang, H.-H., & Na, B.-K. (2021). Genetic Polymorphism and Natural Selection of Apical Membrane Antigen-1 in Plasmodium falciparum Isolates from Vietnam. Genes, 12(12), 1903. https://doi.org/10.3390/genes12121903








