Transcriptomics Analysis of Lens from Patients with Posterior Subcapsular Congenital Cataract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. RNA Extracting, Library Construction and RNA-seq
2.3. Gene Expression Data Analysis
2.4. Real-time Quantitative PCR (RT-qPCR)
2.5. Immunohistochemical Staining
2.6. Statistical Analysis
3. Results
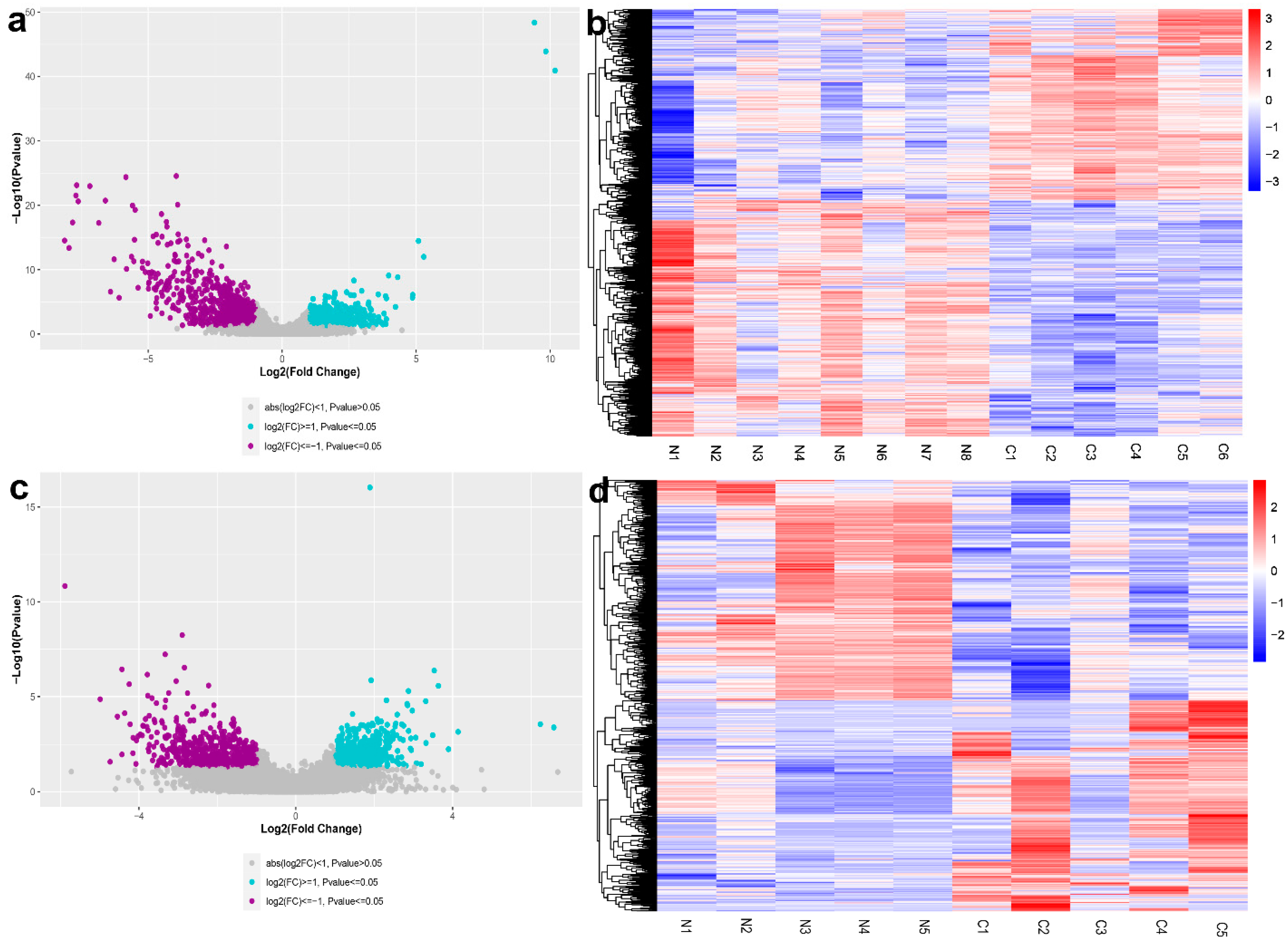
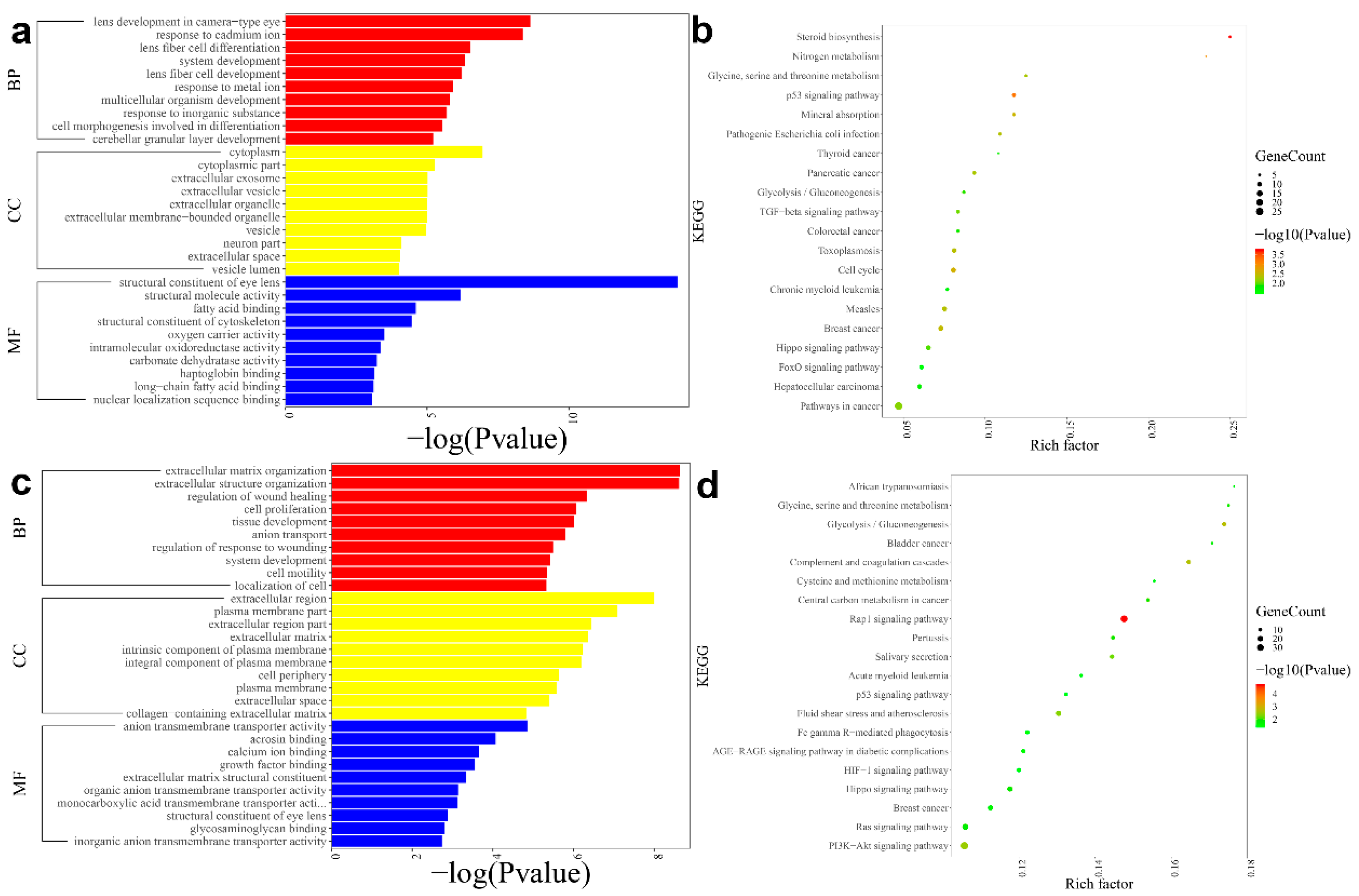
3.1. Differential Expression and Functional Enrichment Analysis in the Anterior Lens Epithelial Cells
3.2. Differential Expression and Functional Enrichment Analysis in the Fibre Cells
3.3. PPI Network/Strings Network
3.4. RT-qPCR Validates the Results of RNA-seq
3.5. Lens-Associated Differential Gene Expression to Identify High-Priority Candidate Genes
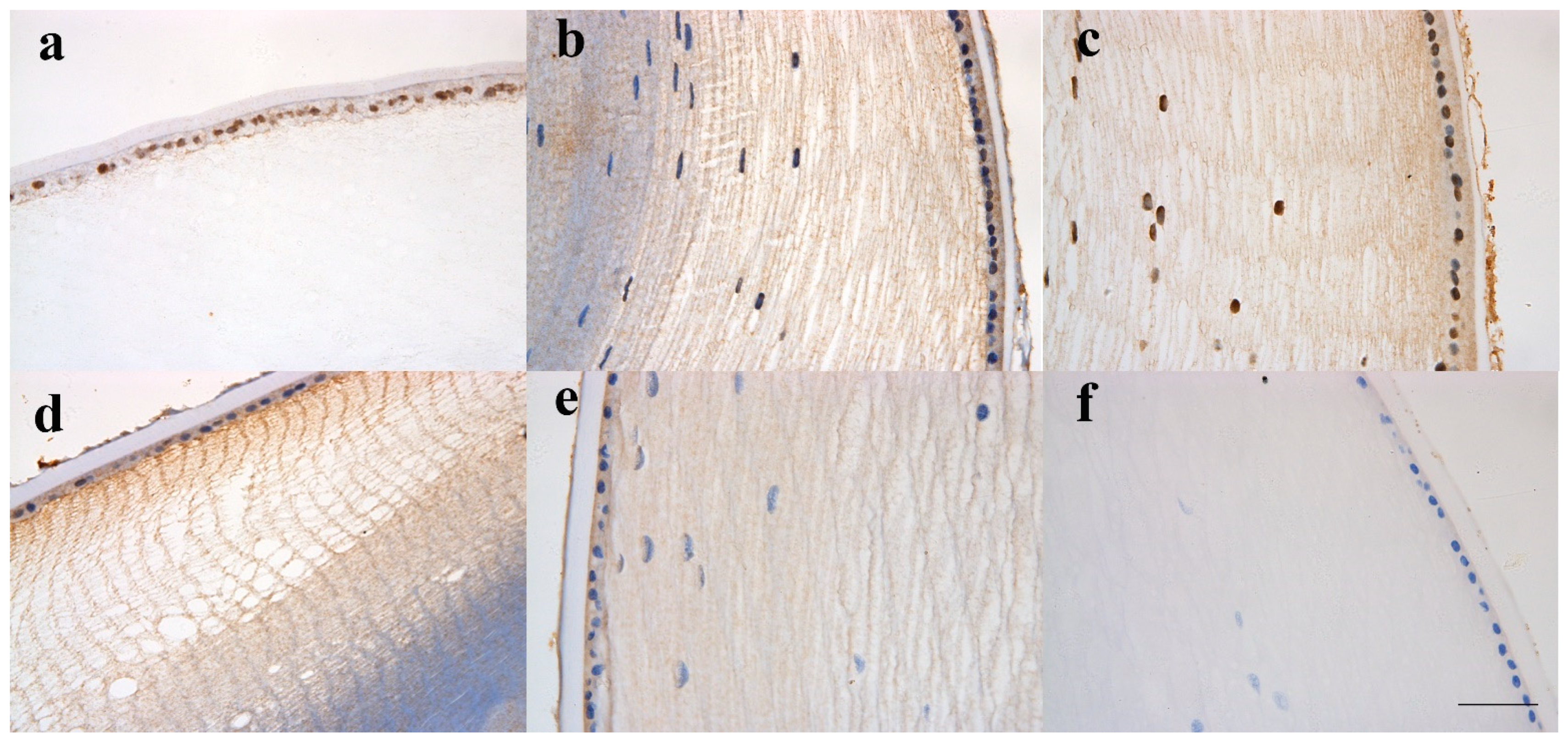
3.6. A Protein Marker of Cell Growth and Epithelial Cell Proliferation Confirms RNA-seq Transcriptome Findings
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| cDNA | complementary DNA |
| GO | gene ontology |
| KEGG | Kyoto Encyclopaedia of Genes and Genomes |
| FPKM | per million fragments mapped |
| DEG | differentially expressed genes |
| PPI | protein–protein interaction |
| RT-qPCR | real-time quantitative PCR |
| RIPA | radio-immunoprecipitation assay |
| SDS-PAGE | sodium dodecylsulphate-polyacrylamide gel electrophoresis |
| PVDF | transferred to a polyvinylidene difluoride |
| GRIFIN | galectin-related inter-fibre protein |
| TGF-β | transforming growth factor-β |
| ECM | extracellular matrix |
| α-SMA | deposition of α-smooth muscle actin |
| FDR | false discovery rate |
| PBS | phosphate-buffered saline |
| RNA-seq | RNA sequencing |
References
- Mohammadpour, M.; Shaabani, A.; Sahraian, A.; Momenaei, B.; Tayebi, F.; Bayat, R.; Mirshahi, R. Updates on managements of pediatric cataract. J. Curr. Ophthalmol. 2019, 31, 118–126. [Google Scholar] [CrossRef]
- Churchill, A.; Graw, J. Clinical and experimental advances in congenital and paediatric cataracts. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 1234–1249. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Long, E.; Lin, H.; Liu, Y. Prevalence and epidemiological characteristics of congenital cataract: A systematic review and meta-analysis. Sci. Rep. 2016, 6, 28564. [Google Scholar] [CrossRef]
- Haargaard, B.; Wohlfahrt, J.; Fledelius, H.C.; Rosenberg, T.; Melbye, M. A nationwide Danish study of 1027 cases of congenital/infantile cataracts. Ophthalmology 2004, 111, 2292–2298. [Google Scholar] [CrossRef] [PubMed]
- Kakrana, A.; Yang, A.; Anand, D.; Djordjevic, D.; Ramachandruni, D.; Singh, A.; Huang, H.; Ho, J.; Lachke, S.A. iSyTE 2.0: A database for expression-based gene discovery in the eye. Nucleic Acids Res. 2018, 46, D875–D885. [Google Scholar] [CrossRef]
- Anand, D.; Kakrana, A.; Siddam, A.D.; Huang, H.; Saadi, I.; Lachke, S.A. RNA sequencing-based transcriptomic profiles of embryonic lens development for cataract gene discovery. Hum. Genet. 2018, 137, 941–954. [Google Scholar] [CrossRef]
- Lachke, S.A.; Ho, J.; Kryukov, G.V.; O’Connell, D.J.; Aboukhalil, A.; Bulyk, M.L.; Park, P.J.; Maas, R.L. iSyTE: Integrated Systems Tool for Eye Gene Discovery. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1617–1627. [Google Scholar] [CrossRef] [Green Version]
- You, C.; Wu, X.; Zhang, Y.; Dai, Y.; Huang, Y.; Xie, L. Visual Impairment and Delay in Presentation for Surgery in Chinese Pediatric Patients with Cataract. Ophthalmology 2011, 118, 17–23. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Barnes, J.; Pedrosa, E.; Herman, N.S.; Salas, F.; Wang, P.; Zheng, D.; Lachman, H.M. Transcriptome analysis of neural progenitor cells derived from Lowe syndrome induced pluripotent stem cells: Identification of candidate genes for the neurodevelopmental and eye manifestations. J. Neurodev. Disord. 2020, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Shiels, A.; Bennett, T.M.; Hejtmancik, J.F. Cat-Map: Putting cataract on the map. Mol. Vis. 2010, 16, 2007–2015. [Google Scholar] [PubMed]
- Anand, D.; Lachke, S.A. Systems biology of lens development: A paradigm for disease gene discovery in the eye. Exp. Eye Res. 2017, 156, 22–33. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, S.A.; Anand, D.; Siddam, A.D.; Kakrana, A.; Dash, S.; Scheiblin, D.A.; Dang, C.A.; Terrell, A.M.; Waters, S.M.; Singh, A.; et al. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum. Genet. 2015, 134, 717–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, D.; Agrawal, S.A.; Siddam, A.D.; Motohashi, H.; Yamamoto, M.; Lachke, S.A. An integrative approach to analyze microarray datasets for prioritization of genes relevant to lens biology and disease. Genom. Data 2015, 5, 223–227. [Google Scholar] [CrossRef]
- Choudhary, R.; Bodakhe, S.H. Olmesartan, an angiotensin II receptor blocker inhibits the progression of cataract formation in cadmium chloride induced hypertensive albino rats. Life Sci. 2016, 167, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Calva, J.A.; Pérez-Vázquez, M.L.; Serebryany, E.; King, J.A.; Quintanar, L. Mercury-induced aggregation of human lens γ-crystallins reveals a potential role in cataract disease. J. Biol. Inorg. Chem. 2018, 23, 1105–1118. [Google Scholar] [CrossRef]
- Langford-Smith, A.; Tilakaratna, V.; Lythgoe, P.R.; Clark, S.J.; Bishop, P.N.; Day, A.J. Age and Smoking Related Changes in Metal Ion Levels in Human Lens: Implications for Cataract Formation. PLoS ONE 2016, 11, e147576. [Google Scholar] [CrossRef] [Green Version]
- Shiels, A.; Hejtmancik, J.F. Mutations and mechanisms in congenital and age-related cataracts. Exp. Eye Res. 2017, 156, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Hawse, J.R.; Hejtmancik, J.F.; Horwitz, J.; Kantorow, M. Identification and functional clustering of global gene expression differences between age-related cataract and clear human lenses and aged human lenses. Exp. Eye Res. 2004, 79, 935–940. [Google Scholar] [CrossRef] [Green Version]
- Bell, S.J.; Oluonye, N.; Harding, P.; Moosajee, M. Congenital cataract: A guide to genetic and clinical management. Ther. Adv. Rare Dis. 2020, 1, 1726979822. [Google Scholar] [CrossRef]
- Shiels, A.; Hejtmancik, J.F. Molecular Genetics of Cataract. Prog. Mol. Biol. Transl. Sci. 2015, 134, 203–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaltner, H.; García, C.G.; Ludwig, A.K.; Manning, J.C.; Gabius, H.J. From glycophenotyping by (plant) lectin histochemistry to defining functionality of glycans by pairing with endogenous lectins. Histochem. Cell Biol. 2018, 149, 547–568. [Google Scholar] [CrossRef]
- Kaltner, H.; Toegel, S.; Caballero, G.G.; Manning, J.C.; Ledeen, R.W.; Gabius, H.J. Galectins: Their network and roles in immunity/tumor growth control. Histochem. Cell Biol. 2017, 147, 239–256. [Google Scholar] [CrossRef]
- García Caballero, G.; Schmidt, S.; Schnölzer, M.; Schlötzer-Schrehardt, U.; Knospe, C.; Ludwig, A.; Manning, J.C.; Muschler, P.; Kaltner, H.; Kopitz, J.; et al. Chicken GRIFIN: Binding partners, developmental course of localization and activation of its lens-specific gene expression by L-Maf/Pax6. Cell Tissue Res. 2019, 375, 665–683. [Google Scholar] [CrossRef] [PubMed]
- García, C.G.; Kaltner, H.; Michalak, M.; Shilova, N.; Yegres, M.; André, S.; Ludwig, A.K.; Manning, J.C.; Schmidt, S.; Schnölzer, M.; et al. Chicken GRIFIN: A homodimeric member of the galectin network with canonical properties and a unique expression profile. Biochimie 2016, 128–129, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Barton, K.A.; Hsu, C.; Petrash, J.M. Interactions between Small Heat Shock Protein α-Crystallin and Galectin-Related Interfiber Protein (GRIFIN) in the Ocular Lens. Biochemistry 2009, 48, 3956–3966. [Google Scholar] [CrossRef] [Green Version]
- Taiyab, A.; Holms, J.; West-Mays, J.A. β-Catenin/Smad3 Interaction Regulates Transforming Growth Factor-β-Induced Epithelial to Mesenchymal Transition in the Lens. Int. J. Mol. Sci. 2019, 20, 2078. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Guo, D.; Wu, Q.; Liu, D.; Bi, H. Zinc Chloride Inhibits Human Lens Epithelial Cell Migration and Proliferation Involved in TGF-β1 and TNF-α Signaling Pathways in HLE B-3 Cells. Biol. Trace Elem. Res. 2014, 159, 425–433. [Google Scholar] [CrossRef]
- Kubo, E.; Shibata, T.; Singh, D.; Sasaki, H. Roles of TGF β and FGF Signals in the Lens: Tropomyosin Regulation for Posterior Capsule Opacity. Int. J. Mol. Sci. 2018, 19, 3093. [Google Scholar] [CrossRef] [Green Version]
- de Iongh, R.U.; Lovicu, F.J.; Overbeek, P.A.; Schneider, M.D.; Joya, J.; Hardeman, E.D.; Mcavoy, J.W. Requirement for TGFbeta receptor signaling during terminal lens fiber differentiation. Development 2001, 128, 3995–4010. [Google Scholar] [CrossRef] [PubMed]
- Launay, S.; Maubert, E.; Lebeurrier, N.; Tennstaedt, A.; Campioni, M.; Docagne, F.; Gabriel, C.; Dauphinot, L.; Potier, M.C.; Ehrmann, M.; et al. HtrA1-dependent proteolysis of TGF-beta controls both neuronal maturation and developmental survival. Cell Death Differ. 2008, 15, 1408–1416. [Google Scholar] [CrossRef] [PubMed]
- Canfield, A.E.; Hadfield, K.D.; Rock, C.F.; Wylie, E.C.; Wilkinson, F.L. HtrA1: A novel regulator of physiological and pathological matrix mineralization? Biochem. Soc. Trans. 2007, 35, 669–671. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, M.J.; Marsh, J.A.; Morrison, M.; Kazlauskas, A.; Khadka, A.; Rosenkranz, S.; Deangelis, M.M.; Saint-Geniez, M.; Jacobo, S.M.P. ER stress-induced aggresome trafficking of HtrA1 protects against proteotoxicity. J. Mol. Cell Biol. 2017, 9, 516–532. [Google Scholar] [CrossRef] [PubMed]
- May, A.; Su, F.; Dinh, B.; Ehlen, R.; Tran, C.; Adivikolanu, H.; Shaw, P.X. Ongoing controversies and recent insights of the ARMS2-HTRA1 locus in age-related macular degeneration. Exp. Eye Res. 2021, 210, 108605. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, J.; Rothzerg, E.; Wu, X.; Xu, H.; Zhu, S.; Xu, J. Molecular structure and the role of high-temperature requirement protein 1 in skeletal disorders and cancers. Cell Proliferat. 2020, 53, e12746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Ban, G.; Al-Shameri, B.; He, X.; Liang, D.; Chen, W. High-temperature Requirement Protein A1 Regulates Odontoblastic Differentiation of Dental Pulp Cells via the Transforming Growth Factor Beta 1/Smad Signaling Pathway. J. Endodont. 2018, 44, 765–772. [Google Scholar] [CrossRef]

| Target | F/R | Primer Sequence (5′→3′) | Amplicon Size (BP) |
|---|---|---|---|
| CRYGC | Forward | TCCCAACTACCAAGGTCAACA | 232 |
| Reverse | GTGGAGGGAACGGATCTCG | ||
| CRYGB | Forward | CACCTCACTGAAATTCACTCCC | 125 |
| Reverse | CCCCAATCAAGAAACCTCCTGT | ||
| CRYGA | Forward | GGTCCAATCCTGCCGTATAATTC | 109 |
| Reverse | ACAGGCGCAGTCATCAGTG | ||
| CRYBB1 | Forward | GTGCTCAAATCTGGCAGACC | 92 |
| Reverse | GGAAGTTGGACTGCTCAAAGG | ||
| CRYBA1 | Forward | GGGCAAGAGGATGGAGTTCAC | 122 |
| Reverse | CACAGAAGCTGGTATGCTCATAA | ||
| HBA2 | Forward | CAAGACCTACTTCCCGCACTTCG | 341 |
| Reverse | GGGCAGGAGGAACGGCTACC | ||
| HBA1 | Forward | TCAACTTCAAGCTCCTAAGCCACTG | 110 |
| Reverse | CACAGAAGCCAGGAACTTGTCCAG | ||
| HBB | Forward | AGGAGAAGTCTGCCGTTACTG | 190 |
| Reverse | CCGAGCACTTTCTTGCCATGA | ||
| S100A9 | Forward | CTGTGTGGCTCCTCGGCTTTG | 98 |
| Reverse | TGGTGGAAGGTGTTGATGATGGTC | ||
| HBG2 | Forward | GTGGAAGATGCTGGAGGAGAAACC | 106 |
| Reverse | TGATGGCAGAGGCAGAGGACAG | ||
| APOE | Forward | GTTGCTGGTCACATTCCTGG | 146 |
| Reverse | GCAGGTAATCCCAAAAGCGAC | ||
| TSPAN12 | Forward | CCAGAGAAGATTCCGTGAAGTG | 107 |
| Reverse | GTCCCTCATCCAAGCAGAAAC | ||
| PRSS35 | Forward | CATCGAATGCCAGAAAGAACTCC | 135 |
| Reverse | GTCGGCTCAAGAACCAAATCT | ||
| ASS1 | Forward | TCCGTGGTTCTGGCCTACA | 126 |
| Reverse | GGCTTCCTCGAAGTCTTCCTT | ||
| SULF1 | Forward | GATCCCCGAGGTTCAGAGGA | 178 |
| Reverse | GGTGTAGTCACAAAGGCATTGA | ||
| METRN | Forward | CACAGACACCGCCAGGCAAG | 103 |
| Reverse | CCACTCCTCTCCCGTCCACAC | ||
| PRAP1 | Forward | ACAGCCTGTACCACCCTCC | 82 |
| Reverse | AGCACCTGGTGATTTGGCATC | ||
| RAMP1 | Forward | CCAGGAGGCTAACTACGGTG | 297 |
| Reverse | GGGACCACGATGAAGGGGTA | ||
| TMEM54 | Forward | TGGGCCATGTGAGCTTCATC | 176 |
| Reverse | GAGGTAGCGTGACAACACGAT | ||
| GAPDH | Forward | GGAGCGAGATCCCTCCAAAAT | 197 |
| Reverse | GGCTGTTGTCATACTTCTCATGG |
| Sample | Tissue | Raw Reads | Clean Reads | Clean Read-Pairs | Average Length | Mapped Ratio (%) |
|---|---|---|---|---|---|---|
| PSC patient 1 | Capsules | 79,150,688 | 23,475,256 | 11,737,628 | 138.3 | 88.80% |
| PSC patient 2 | Capsules | 92,790,046 | 36,750,674 | 18,375,337 | 141.9 | 93.40% |
| PSC patient 3 | Capsules | 102,532,016 | 47,760,460 | 23,880,230 | 142.0 | 91.70% |
| PSC patient 4 | Capsules | 100,012,388 | 32,018,416 | 16,009,208 | 142.4 | 92.60% |
| PSC patient 5 | Capsules | 98,193,322 | 36,350,310 | 18,175,155 | 134.7 | 79.10% |
| PSC patient 6 | Capsules | 100,226,088 | 40,541,834 | 20,270,917 | 134.1 | 79.90% |
| NC 1 | Capsules | 127,355,192 | 60,141,116 | 30,070,558 | 144.6 | 95.60% |
| NC 2 | Capsules | 100,869,818 | 37,378,548 | 18,689,274 | 138.0 | 86.40% |
| NC 3 | Capsules | 111,902,712 | 47,398,990 | 23,699,495 | 141.5 | 92.20% |
| NC 4 | Capsules | 127,358,610 | 48,182,942 | 24,091,471 | 144.2 | 94% |
| NC 5 | Capsules | 120,259,640 | 52,969,554 | 26,484,777 | 134.6 | 90.60% |
| NC 6 | Capsules | 91,405,500 | 31,710,346 | 15,855,173 | 133.6 | 84.20% |
| NC 7 | Capsules | 125,882,600 | 56,962,562 | 28,481,281 | 136.5 | 93% |
| NC 8 | Capsules | 131,778,762 | 54,214,656 | 27,107,328 | 135.5 | 91.70% |
| PSC patient 1 | Cortex | 123,977,234 | 45,747,380 | 22,873,690 | 141.6 | 93.40% |
| PSC patient 3 | Cortex | 91,869,478 | 25,684,820 | 12,842,410 | 137.9 | 85.90% |
| PSC patient 4 | Cortex | 110,104,320 | 109,339,070 | 54,669,535 | 150.0 | 97.60% |
| PSC patient 5 | Cortex | 128,652,528 | 48,532,586 | 24,266,293 | 138.7 | 89.40% |
| PSC patient 6 | Cortex | 116,294,432 | 47,393,278 | 23,696,639 | 136.5 | 84.80% |
| NC 1 | Cortex | 98,977,182 | 32,642,774 | 16,321,387 | 141.8 | 92.40% |
| NC 4 | Cortex | 167,149,114 | 67,734,538 | 33,867,269 | 142.4 | 95.10% |
| NC 5 | Cortex | 94,542,668 | 93,252,442 | 46,626,221 | 148.9 | 96.90% |
| NC 6 | Cortex | 114,900,736 | 113,734,102 | 56,867,051 | 148.8 | 97.90% |
| NC 7 | Cortex | 92,822,214 | 92,085,706 | 46,042,853 | 149.5 | 97.90% |
| DEG | Symbol | Description | Log2 Fold Change | p-Value | False Discovery Rate (FDR) Value |
|---|---|---|---|---|---|
| Downregulated | CRYGC | Crystallin gamma C | −8.117 | 3.04 × 10−15 | 6.00 × 10−12 |
| CRYGB | Crystallin gamma B | −7.952 | 4.24 × 10−14 | 6.30 × 10−11 | |
| CRYGA | Crystallin gamma A | −7.818 | 4.67 × 10−18 | 1.61 × 10−14 | |
| CRYBB1 | Crystallin beta B1 | −7.695 | 3.07 × 10−22 | 2.11 × 10−18 | |
| CRYBA1 | Crystallin beta A1 | −7.667 | 7.63 × 10−24 | 7.00 × 10−20 | |
| Upregulated | HBA2 | Hemoglobin subunit alpha 2 | 10.182 | 1.20 × 10−41 | 2.21 × 10−37 |
| HBA1 | Hemoglobin subunit alpha 1 | 9.838 | 1.27 × 10−44 | 3.48 × 10−40 | |
| HBB | Hemoglobin subunit beta | 9.414 | 4.16 × 10−49 | 2.29 × 10−44 | |
| S100A9 | S100 calcium binding protein A9 | 5.087 | 3.54 × 10−15 | 6.71 × 10−12 | |
| HBG2 | Hemoglobin subunit gamma 2 | 4.863 | 2.18 × 10−6 | 4.59 × 10−4 |
| DEGs | Symbol | Description | Log2 Fold Change | p-Value | FDR Value |
|---|---|---|---|---|---|
| Downregulated | APOE | Apolipoprotein E | −5.889 | 1.47 × 10−11 | 9.71 × 10−8 |
| TSPAN12 | Tetraspanin 12 | −4.435 | 3.64 × 10−7 | 0.001 | |
| PRSS35 | Protease, serine 35 | −4.249 | 2.21 × 10−6 | 0.004 | |
| ASS1 | Argininosuccinate Synthase 1 | −3.786 | 6.89 × 10−7 | 0.002 | |
| SULF1 | Sulfatase 1 | −3.338 | 3.63 × 10−5 | 0.028 | |
| Upregulated | METRN | Meteorin, glial cell differentiation regulator | 2.872 | 5.07 × 10−6 | 0.007 |
| PRAP1 | Proline rich acidic Protein 1 | 2.852 | 2.85 × 10−5 | 0.024 | |
| RAMP1 | Receptor activity-modifying protein 1 | 2.311 | 1.56 × 10−5 | 0.016 | |
| TMEM54 | Transmembrane protein 54 | 1.921 | 1.37 × 10−6 | 0.003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Li, H.; Yang, T.; Liu, X.; Fan, F.; Zhou, X.; Luo, Y. Transcriptomics Analysis of Lens from Patients with Posterior Subcapsular Congenital Cataract. Genes 2021, 12, 1904. https://doi.org/10.3390/genes12121904
Lin X, Li H, Yang T, Liu X, Fan F, Zhou X, Luo Y. Transcriptomics Analysis of Lens from Patients with Posterior Subcapsular Congenital Cataract. Genes. 2021; 12(12):1904. https://doi.org/10.3390/genes12121904
Chicago/Turabian StyleLin, Xiaolei, Hongzhe Li, Tianke Yang, Xin Liu, Fan Fan, Xiyue Zhou, and Yi Luo. 2021. "Transcriptomics Analysis of Lens from Patients with Posterior Subcapsular Congenital Cataract" Genes 12, no. 12: 1904. https://doi.org/10.3390/genes12121904
APA StyleLin, X., Li, H., Yang, T., Liu, X., Fan, F., Zhou, X., & Luo, Y. (2021). Transcriptomics Analysis of Lens from Patients with Posterior Subcapsular Congenital Cataract. Genes, 12(12), 1904. https://doi.org/10.3390/genes12121904






