Nucleic Acid-Sensing and Interferon-Inducible Pathways Show Differential Methylation in MZ Twins Discordant for Lupus and Overexpression in Independent Lupus Samples: Implications for Pathogenic Mechanism and Drug Targeting
Abstract
:1. Introduction
2. Materials and Methods
2.1. Discovery Cohort
2.2. Replication Cohort
2.3. Genome-Wide DNA Methylation Assay and Array Validation in LFRR Twins
2.4. Collection of Gene Expression Experiments from SLE Patient Datasets
2.5. Data Analysis
3. Results
3.1. Characteristics of the MZ Twins
3.2. Identification of Differentially Methylated Regions in Twins Discordant for SLE
| CpG * | Chr | Pos (bp) † | Gene | Δβ | p-Value | Interferon-Regulated ‡ | Relation to CpG †† | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Pair1 | Pair2 | Pair3 | Mean | |||||||
| cg13304609 | 1 | 79085162 | IFI44L | −0.24 | −0.27 | −0.37 | −0.29 | 1.58 × 10−14 | IRG | |
| cg06872964 | 1 | 79085250 | IFI44L | −0.26 | −0.21 | −0.24 | 1.05 × 10−71 | IRG | ||
| cg03607951 | 1 | 79085586 | IFI44L | −0.27 | −0.3 | −0.21 | −0.26 | 7.23 × 10−22 | IRG | |
| cg17515347 | 1 | 159047163 | AIM2 | −0.09 | −0.11 | −0.07 | −0.09 | 3.01 × 10−12 | IRG | |
| cg08272268 | 1 | 200380059 | ZNF281 | −0.08 | −0.07 | −0.11 | −0.09 | 4.33 × 10−15 | S_Shore | |
| cg01028142 | 2 | 7004578 | CMPK2 | −0.22 | −0.36 | −0.43 | −0.33 | 7.98 × 10−8 | IRG | N_Shore |
| cg10959651 | 2 | 7018020 | RSAD2 | −0.13 | −0.1 | −0.16 | −0.13 | 3.14 × 10−14 | IRG | |
| cg10549986 | 2 | 7018153 | RSAD2 | −0.08 | −0.09 | −0.1 | −0.09 | 1.95 × 10−91 | IRG | |
| cg14126601 | 2 | 37384708 | EIF2AK2 | −0.08 | −0.1 | −0.12 | −0.1 | 5.55 × 10−16 | IRG | S_Shore |
| cg26337070 | 2 | 85999873 | ATOH8 | −0.06 | −0.12 | −0.11 | −0.1 | 7.55 × 10−9 | ||
| cg04781494 | 2 | 202047246 | CASP10 | −0.07 | −0.13 | −0.08 | −0.09 | 8.39 × 10−8 | IRG | |
| cg15768138 | 2 | 219030752 | CXCR1 | −0.09 | −0.12 | −0.11 | −0.11 | 7.38 × 10−27 | ||
| cg13411554 | 3 | 53700276 | CACNA1D | −0.06 | −0.12 | −0.09 | −0.09 | 8.66 × 10−8 | ||
| cg22930808 | 3 | 122281881 | PARP9-DTX3L | −0.36 | −0.34 | −0.4 | −0.37 | 6.74 × 10−126 | IRG | N_Shore |
| cg08122652 | 3 | 122281939 | PARP9-DTX3L | −0.34 | −0.31 | −0.51 | −0.38 | 1.11 × 10−9 | IRG | N_Shore |
| cg00959259 | 3 | 122281975 | PARP9-DTX3L | −0.37 | −0.3 | −0.34 | −0.34 | 1.32 × 10−56 | IRG | N_Shore |
| cg06981309 | 3 | 146260954 | PLSCR1 | −0.24 | −0.28 | −0.21 | −0.24 | 6.41 × 10−31 | IRG | N_Shore |
| cg02556393 | 3 | 168866705 | MECOM | −0.08 | −0.09 | −0.1 | −0.09 | 3.14 × 10−95 | N_Shore | |
| cg07809027 | 4 | 15007205 | CPEB2 | −0.07 | −0.1 | −0.12 | −0.1 | 2.08 × 10−14 | S_Shore | |
| cg02215171 | 4 | 89379156 | HERC5 | −0.08 | −0.09 | −0.11 | −0.09 | 4.48 × 10−18 | IRG | S_Shore |
| cg17786255 | 4 | 108814389 | SGMS2 | −0.07 | −0.09 | −0.11 | −0.09 | 2.01 × 10−16 | IRG | |
| cg21873524 | 4 | 190942744 | −0.1 | −0.1 | −0.12 | −0.11 | 1.03 × 10−55 | Island | ||
| cg24740632 | 5 | 134486678 | −0.11 | −0.12 | −0.14 | −0.12 | 2.26 × 10−60 | |||
| cg06012695 | 6 | 28770593 | −0.1 | −0.13 | −0.11 | 3.59 × 10−16 | ||||
| cg25138053 | 6 | 31368016 | −0.11 | −0.09 | −0.07 | −0.09 | 3.67 × 10−15 | S_Shore | ||
| cg22708150 | 6 | 31649619 | LY6G5C | −0.12 | −0.14 | −0.17 | −0.14 | 1.05 × 10−19 | N_Shore | |
| cg07292773 | 6 | 156718177 | 0.07 | 0.1 | 0.11 | 0.1 | 2.22 × 10−17 | Island | ||
| cg12013713 | 7 | 139760671 | PARP12 | −0.12 | −0.14 | −0.09 | −0.12 | 1.44 × 10−16 | IRG | N_Shore |
| cg20190772 | 8 | 48572496 | KIAA0146 | −0.08 | −0.07 | −0.13 | −0.09 | 1.40 × 10−8 | ||
| cg14864167 | 8 | 66751182 | PDE7A | −0.25 | −0.35 | −0.45 | −0.35 | 1.21 × 10−9 | N_Shelf | |
| cg06102678 | 8 | 81491328 | −0.08 | −0.12 | −0.07 | −0.09 | 1.00 × 10−8 | Island | ||
| cg12110437 | 8 | 144098888 | LY6E | −0.16 | −0.17 | −0.27 | −0.2 | 3.14 × 10−9 | IRG | N_Shore |
| cg17555806 | 10 | 74448117 | −0.08 | −0.12 | −0.07 | −0.09 | 1.51 × 10−8 | N_Shelf | ||
| cg02314339 | 10 | 91020653 | −0.08 | −0.14 | −0.11 | −0.11 | 1.72 × 10−8 | |||
| cg06188083 | 10 | 91093005 | IFIT3 | −0.29 | −0.16 | −0.31 | −0.25 | 6.18 × 10−8 | IRG | |
| cg05552874 | 10 | 91153143 | IFIT1 | −0.2 | −0.28 | −0.3 | −0.26 | 6.01 × 10−16 | IRG | |
| cg14910175 | 10 | 131840954 | −0.07 | −0.11 | −0.08 | −0.09 | 1.56 × 10−11 | N_Shelf | ||
| cg10552523 | 11 | 313478 | IFITM1 | −0.14 | −0.12 | −0.14 | −0.13 | 5.90 × 10−115 | IRG | N_Shelf |
| cg20566897 | 11 | 313527 | IFITM1 | −0.11 | −0.11 | −0.09 | −0.1 | 7.00 × 10−62 | IRG | N_Shelf |
| cg23570810 | 11 | 315102 | IFITM1 | −0.24 | −0.25 | −0.34 | −0.27 | 1.43 × 10−18 | IRG | N_Shore |
| cg03038262 | 11 | 315262 | IFITM1 | −0.24 | −0.22 | −0.29 | −0.25 | 4.41 × 10−40 | IRG | N_Shore |
| cg20045320 | 11 | 319555 | −0.19 | −0.13 | −0.2 | −0.18 | 4.85 × 10−17 | S_Shore | ||
| cg17990365 | 11 | 319718 | IFITM3 | −0.16 | −0.15 | −0.15 | −0.16 | 8.78 × 10−295 | IRG | S_Shore |
| cg08926253 | 11 | 614761 | IRF7 | −0.15 | −0.14 | −0.23 | −0.17 | 2.01 × 10−9 | IRG | Island |
| cg12461141 | 11 | 5710654 | TRIM22 | −0.1 | −0.08 | −0.12 | −0.1 | 6.35 × 10−25 | IRG | |
| cg23571857 | 17 | 6658898 | XAF1 | −0.07 | −0.13 | −0.11 | −0.1 | 1.46 × 10−8 | IRG | |
| cg04927537 | 17 | 76976091 | LGALS3BP | −0.14 | −0.11 | −0.2 | −0.15 | 2.77 × 10−10 | IRG | |
| cg25178683 | 17 | 76976267 | LGALS3BP | −0.15 | −0.11 | −0.21 | −0.16 | 2.01 × 10−8 | IRG | |
| cg16503797 | 18 | 19476805 | −0.08 | −0.12 | −0.08 | −0.09 | 5.39 × 10−12 | N_Shore | ||
| cg15871086 | 18 | 56526595 | −0.07 | −0.11 | −0.08 | −0.09 | 2.08 × 10−11 | N_Shelf | ||
| cg23352030 | 20 | 62198469 | PRIC285 | 0.13 | 0.19 | 0.11 | 0.14 | 2.36 × 10−11 | Island | |
| cg16785077 | 21 | 42791867 | MX1 | −0.11 | −0.09 | −0.12 | −0.11 | 8.45 × 10−27 | IRG | N_Shore |
| cg22862003 | 21 | 42797588 | MX1 | −0.31 | −0.25 | −0.35 | −0.31 | 1.62 × 10−25 | IRG | N_Shore |
| cg26312951 | 21 | 42797847 | MX1 | −0.26 | −0.17 | −0.2 | −0.21 | 6.28 × 10−15 | IRG | N_Shore |
| cg21549285 | 21 | 42799141 | MX1 | −0.5 | −0.35 | −0.57 | −0.47 | 6.59 × 10−13 | IRG | S_Shore |
| cg05543864 | 22 | 24979755 | GGT1 | −0.08 | −0.08 | −0.1 | −0.09 | 1.44 × 10−45 | ||
| cg20098015 | 22 | 50971140 | ODF3B | −0.19 | −0.22 | −0.21 | −0.21 | 9.88 × 10−83 | IRG | S_Shore |
| cg05523603 | 22 | 50973101 | −0.17 | −0.23 | −0.27 | −0.22 | 5.51 × 10−14 | S_Shelf | ||
| cg02247863 | 22 | 50983415 | −0.07 | −0.1 | −0.11 | −0.09 | 2.51 × 10−13 | N_Shore | ||
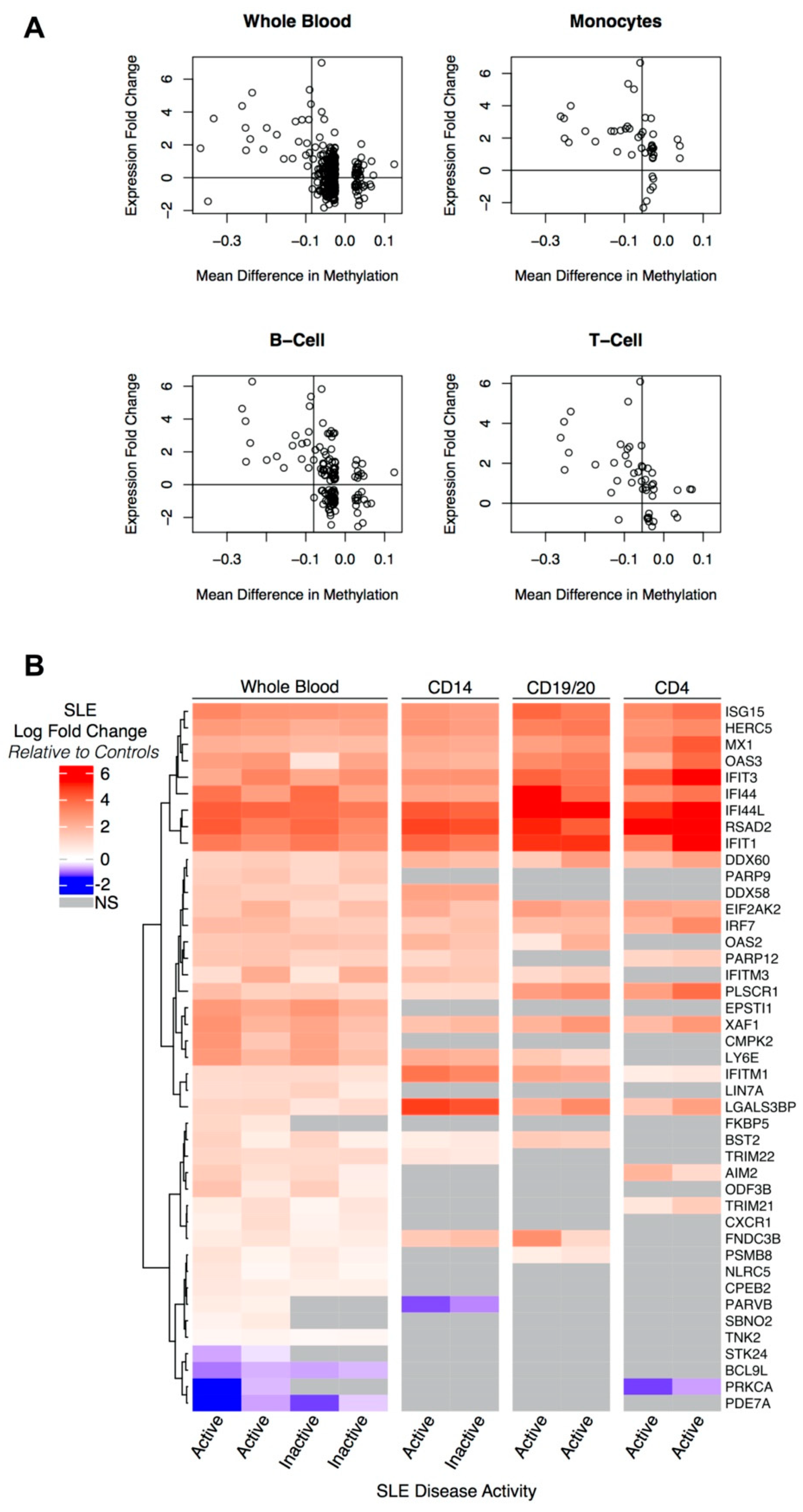
3.3. Hypomethylated Genes Are Overexpressed in Independent Cohorts
| Active SLE § | Inactive SLE § | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CpG * | Chr | Pos (bp) † | Gene | Mean Δβ | Methylation p-Value | Interferon-Regulated ‡ | Log FC Expt 1 | Log FC Expt 2 | Log FC Expt 1 | Log FC Expt 2 |
| cg16526047 | 1 | 949893 | ISG15 | −0.11 | 1.28 × 10−4 | IRG | 3.1 | 2.77 | 2.74 | 2.59 |
| cg05696877 | 1 | 79088769 | IFI44L | −0.3 | 6.60 × 10−6 | IRG | 3.98 | 3.8 | 3.64 | 3.4 |
| cg01079652 | 1 | 79118191 | IFI44 | −0.34 | 5.34 × 10−4 | IRG | 3.54 | 2.53 | 3.7 | 2.33 |
| cg17515347 | 1 | 159047163 | AIM2 | −0.09 | 3.01 × 10−12 | IRG | 1.39 | 0.86 | 1.08 | 0.49 |
| cg01028142 | 2 | 7004578 | CMPK2 | −0.33 | 7.98 × 10−8 | IRG | 2.76 | 1.5 | 2.43 | 1.51 |
| cg10959651 | 2 | 7018020 | RSAD2 | −0.13 | 3.14 × 10−14 | IRG | 4.04 | 3.32 | 3.76 | 3.04 |
| cg14126601 | 2 | 37384708 | EIF2AK2 | −0.1 | 5.55 × 10−16 | IRG | 1.47 | 2.02 | 1.08 | 1.68 |
| cg15768138 | 2 | 219030752 | CXCR1 | −0.11 | 7.38 × 10−27 | 0.43 | 0.96 | 0.38 | 0.66 | |
| cg08122652 | 3 | 122281939 | PARP9-DTX3L | −0.38 | 1.11 × 10−9 | IRG | 1.36 | 1.56 | 1.07 | 1.55 |
| cg06981309 | 3 | 146260954 | PLSCR1 | −0.24 | 6.41 × 10−31 | IRG | 1.77 | 1.25 | 1.38 | 1.07 |
| cg02694620 | 3 | 172109284 | FNDC3B | −0.11 | 3.80 × 10−3 | 0.57 | 0.82 | 0.41 | 0.52 | |
| cg15065340 | 3 | 195632915 | TNK2 | −0.16 | 4.04 × 10−3 | 0.22 | 0.31 | 0.2 | 0.25 | |
| cg07809027 | 4 | 15007205 | CPEB2 | −0.1 | 2.08 × 10−14 | 0.66 | 0.52 | 0.42 | 0.45 | |
| cg02215171 | 4 | 89379156 | HERC5 | −0.09 | 4.48 × 10−18 | IRG | 2.62 | 2.48 | 2.14 | 2.36 |
| cg05883128 | 4 | 169239131 | DDX60 | −0.25 | 2.13 × 10−5 | IRG | 1.24 | 1.38 | 1.06 | 1.46 |
| cg08099136 | 6 | 32811251 | PSMB8 | −0.11 | 1.43 × 10−4 | IRG | −0.39 | −0.13 | NS | NS |
| cg00052684 | 6 | 35694245 | FKBP5 | −0.16 | 1.65 × 10−3 | 1.11 | 0.71 | NS | NS | |
| cg05994974 | 7 | 139761087 | PARP12 | −0.15 | 6.89 × 10−5 | IRG | 1.52 | 1.57 | 1.14 | 1.25 |
| cg14864167 | 8 | 66751182 | PDE7A | −0.35 | 1.21 × 10−9 | −1.24 | −0.41 | −0.82 | −0.23 | |
| cg12110437 | 8 | 144098888 | LY6E | −0.2 | 3.14 × 10−9 | IRG | 2.66 | 1.92 | 2.43 | 1.7 |
| cg03848588 | 9 | 32525008 | DDX58 | −0.1 | 4.34 × 10−4 | IRG | 1.48 | 1.3 | 1.32 | 1.07 |
| cg06188083 | 10 | 91093005 | IFIT3 | −0.25 | 6.18 × 10−8 | IRG | 2.25 | 3.15 | 2.3 | 2.87 |
| cg05552874 | 10 | 91153143 | IFIT1 | −0.26 | 6.01 × 10−16 | IRG | 3.39 | 2.94 | 3.42 | 2.81 |
| cg23570810 | 11 | 315102 | IFITM1 | −0.27 | 1.43 × 10−18 | IRG | 1 | 1.03 | 1.03 | 0.81 |
| cg17990365 | 11 | 319718 | IFITM3 | −0.16 | 8.78 × 10−295 | IRG | 0.92 | 2.23 | 0.71 | 2.13 |
| cg08926253 | 11 | 614761 | IRF7 | −0.17 | 2.01 × 10−9 | IRG | 1.84 | 1.79 | 1.4 | 1.37 |
| cg08577913 | 11 | 4415193 | TRIM21 | −0.1 | 1.74 × 10−3 | IRG | 0.56 | 0.93 | 0.28 | 0.75 |
| cg12461141 | 11 | 5710654 | TRIM22 | −0.1 | 6.35 × 10−25 | IRG | 1.14 | 1 | 0.99 | 1.05 |
| cg26811705 | 11 | 118781408 | BCL9L | −0.09 | 1.64 × 10−3 | −0.6 | −0.35 | −0.41 | −0.32 | |
| cg19347790 | 12 | 81332050 | LIN7A | −0.09 | 1.87 × 10−4 | 0.93 | 0.99 | 1.24 | 0.61 | |
| cg25800166 | 12 | 113375896 | OAS3 | −0.13 | 5.36 × 10−5 | IRG | 2.52 | 2.69 | 0.73 | 2.35 |
| cg19371652 | 12 | 113415883 | OAS2 | −0.11 | 2.24 × 10−5 | IRG | 1.48 | 1.56 | 1.64 | 1.53 |
| cg03753191 | 13 | 43566902 | EPSTI1 | −0.1 | 9.23 × 10−5 | IRG | 2.65 | 2.26 | 2.71 | 2.02 |
| cg00246969 | 13 | 99159656 | STK24 | −0.11 | 6.26 × 10−6 | 0.81 | 0.32 | 0.66 | 0.36 | |
| cg07839457 | 16 | 57023022 | NLRC5 | −0.23 | 6.10 × 10−6 | IRG | 0.7 | 0.23 | 0.53 | 0.27 |
| cg23571857 | 17 | 6658898 | XAF1 | −0.1 | 1.46 × 10−8 | IRG | 2.85 | 1.96 | 2.35 | 1.68 |
| cg23378941 | 17 | 64361956 | PRKCA | −0.11 | 6.89 × 10−5 | IRG | −1.11 | −0.3 | NS | NS |
| cg25178683 | 17 | 76976267 | LGALS3BP | −0.16 | 2.0 × 10−8 | IRG | 1.16 | 1.21 | 0.72 | 1.05 |
| cg07573872 | 19 | 1126342 | SBNO2 | −0.15 | 2.77 × 10−3 | IRG | 0.38 | 0.58 | NS | NS |
| cg07839313 | 19 | 17514600 | BST2 | −0.12 | 3.48 × 10−3 | IRG | 1.24 | 0.49 | 1.17 | 0.41 |
| cg21549285 | 21 | 42799141 | MX1 | −0.47 | 6.59 × 10−13 | IRG | 2.12 | 2 | 1.86 | 1.79 |
| cg19460508 | 22 | 44422195 | PARVB | −0.1 | 1.64 × 10−3 | 0.54 | 0.39 | NS | NS | |
| cg20098015 | 22 | 50971140 | ODF3B | −0.21 | 9.88 × 10−83 | IRG | 1.61 | 0.61 | 1.36 | 0.47 |
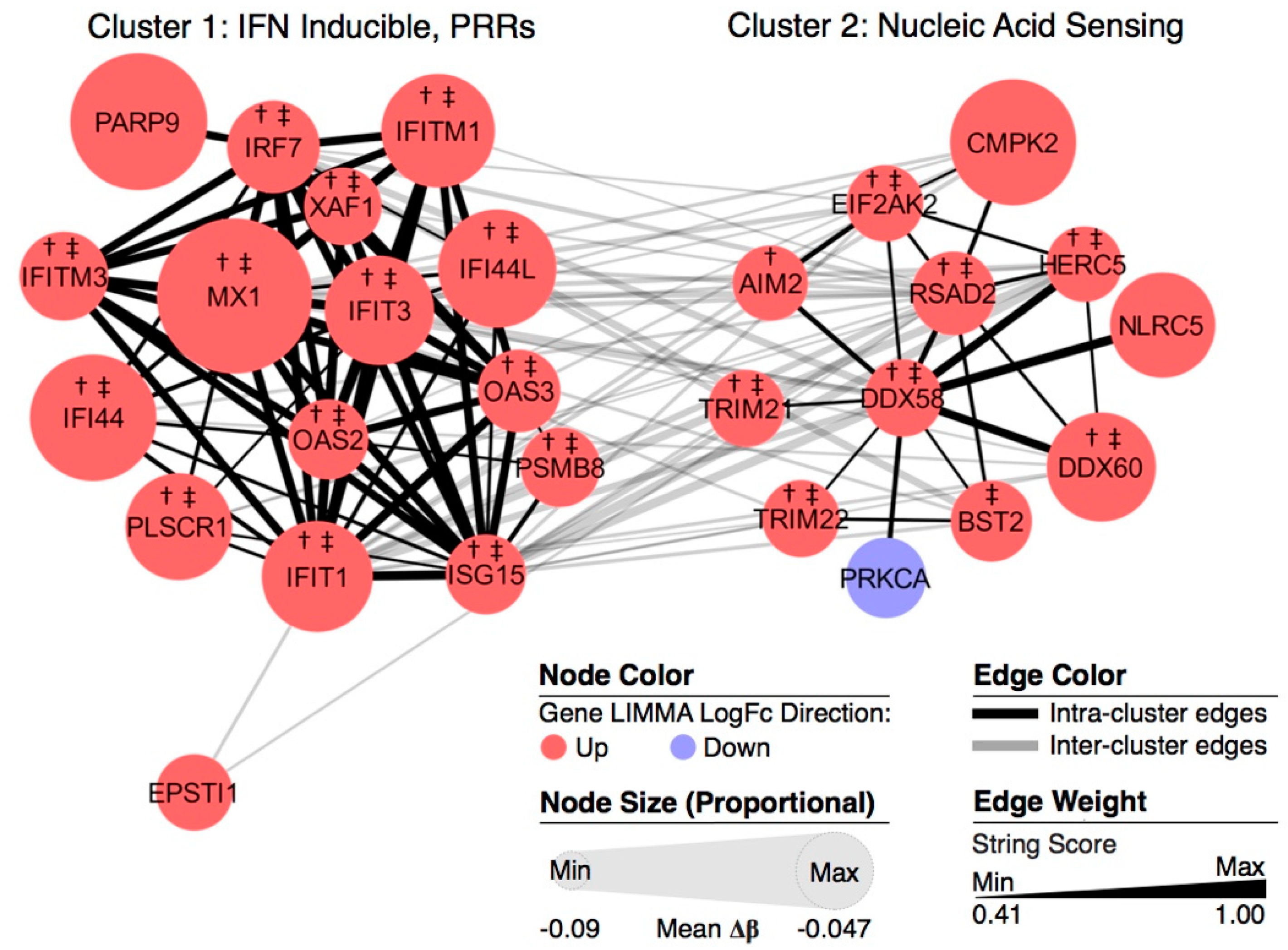
3.4. Pathway Analysis of DM-DE Genes
| CpG * | Chr | Pos (bp) † | Gene | Mean Δβ | Methylation p-Value | Interferon-Regulated ‡ | Log FC Glomerulus | Log FC Tubulointerstitium |
|---|---|---|---|---|---|---|---|---|
| cg16526047 | 1 | 949893 | ISG15 ‖ | −0.11 | 1.28 × 10−4 | IRG | 3.32 | 4.7 |
| cg05696877 | 1 | 79088769 | IFI44L Δ | −0.3 | 6.60 × 10−6 | IRG | 5.14 | 5.94 |
| cg01079652 | 1 | 79118191 | IFI44 ‖ | −0.34 | 5.34 × 10−4 | IRG | 3.94 | 4.76 |
| cg17515347 | 1 | 159047163 | AIM2 | −0.09 | 3.01 × 10−12 | IRG | 0.58 | NS |
| cg01028142 | 2 | 7004578 | CMPK2 Δ | −0.33 | 7.98 × 10−8 | IRG | NS | NS |
| cg10959651 | 2 | 7018020 | RSAD2 | −0.13 | 3.14 × 10−14 | IRG | 4.31 | 3.36 |
| cg14126601 | 2 | 37384708 | EIF2AK2 Δ | −0.1 | 5.55 × 10−16 | IRG | 1.54 | 1.72 |
| cg15768138 | 2 | 219030752 | CXCR1 | −0.11 | 7.38 × 10−27 | 0.68 | −0.17 | |
| cg08122652 | 3 | 122281939 | PARP9-DTX3L Δ | −0.38 | 1.11 × 10−9 | IRG | NS | NS |
| cg06981309 | 3 | 146260954 | PLSCR1 Δ | −0.24 | 6.41 × 10−31 | IRG | 1.92 | 2.07 |
| cg02694620 | 3 | 172109284 | FNDC3B | −0.11 | 3.80 × 10−3 | NS | 0.47 | |
| cg15065340 | 3 | 195632915 | TNK2 ‖ | −0.16 | 4.04 × 10−3 | 0.38 | −0.4 | |
| cg07809027 | 4 | 15007205 | CPEB2 | −0.1 | 2.08 × 10−14 | NS | NS | |
| cg02215171 | 4 | 89379156 | HERC5 ¶ | −0.09 | 4.48 × 10−18 | IRG | 3.16 | 1.96 |
| cg05883128 | 4 | 169239131 | DDX60 | −0.25 | 2.13 × 10−5 | IRG | 1.11 | 2.31 |
| cg08099136 | 6 | 32811251 | PSMB8 | −0.11 | 1.43 × 10−4 | IRG | 0.76 | 2.51 |
| cg00052684 | 6 | 35694245 | FKBP5 § | −0.16 | 1.65 × 10−3 | −1.27 | −2.77 | |
| cg05994974 | 7 | 139761087 | PARP12 ‖ | −0.15 | 6.89 × 10−5 | IRG | 2.26 | 1.86 |
| cg14864167 | 8 | 66751182 | PDE7A | −0.35 | 1.21 × 10−9 | NS | NS | |
| cg12110437 | 8 | 144098888 | LY6E ◊ | −0.2 | 3.14 × 10−9 | IRG | 1.28 | 1.23 |
| cg03848588 | 9 | 32525008 | DDX58 ◊ | −0.1 | 4.34 × 10−4 | IRG | 2.89 | 2.59 |
| cg06188083 | 10 | 91093005 | IFIT3 ◊ | −0.25 | 6.18 × 10−8 | IRG | 2.59 | 3.14 |
| cg05552874 | 10 | 91153143 | IFIT1 ◊ | −0.26 | 6.01 × 10−16 | IRG | 2.24 | 2.77 |
| cg23570810 | 11 | 315102 | IFITM1 | −0.27 | 1.43 × 10−18 | IRG | 2.24 | 3.29 |
| cg17990365 | 11 | 319718 | IFITM3 | −0.16 | 8.78 × 10−295 | IRG | 2.24 | 2 |
| cg08926253 | 11 | 614761 | IRF7 ‖ | −0.17 | 2.01 × 10−9 | IRG | 2.8 | 1 |
| cg08577913 | 11 | 4415193 | TRIM21 | −0.1 | 1.74 × 10−3 | IRG | 1.35 | 0.77 |
| cg12461141 | 11 | 5710654 | TRIM22 | −0.1 | 6.35 × 10−25 | IRG | 1.73 | 2.86 |
| cg26811705 | 11 | 118781408 | BCL9L | −0.09 | 1.64 × 10−3 | NS | NS | |
| cg19347790 | 12 | 81332050 | LIN7A | −0.09 | 1.87 × 10−4 | NS | −0.57 | |
| cg25800166 | 12 | 113375896 | OAS3 | −0.13 | 5.36 × 10−5 | IRG | 3.77 | 1.1 |
| cg19371652 | 12 | 113415883 | OAS2 | −0.11 | 2.24 × 10−5 | IRG | 4.86 | 1.74 |
| cg03753191 | 13 | 43566902 | EPSTI1 ¶ | −0.1 | 9.23 × 10−5 | IRG | NS | NS |
| cg00246969 | 13 | 99159656 | STK24 | −0.11 | 6.26 × 10−6 | NS | 0.28 | |
| cg07839457 | 16 | 57023022 | NLRC5 | −0.23 | 6.10 × 10−6 | IRG | NS | NS |
| cg23571857 | 17 | 6658898 | XAF1 | −0.1 | 1.46 × 10−8 | IRG | 3.14 | 3.05 |
| cg23378941 | 17 | 64361956 | PRKCA | −0.11 | 6.89 × 10−5 | IRG | −0.48 | −0.08 |
| cg25178683 | 17 | 76976267 | LGALS3BP | −0.16 | 2.0 × 10−8 | IRG | 0.57 | 1.49 |
| cg07573872 | 19 | 1126342 | SBNO2 | −0.15 | 2.77 × 10−3 | IRG | NS | NS |
| cg07839313 | 19 | 17514600 | BST2 ‖ | −0.12 | 3.48 × 10−3 | IRG | NS | 2.91 |
| cg21549285 | 21 | 42799141 | MX1 Δ | −0.47 | 6.59 × 10−13 | IRG | 4.05 | 4.64 |
| cg19460508 | 22 | 44422195 | PARVB | −0.1 | 1.64 × 10−3 | 0.28 | NS | |
| cg20098015 | 22 | 50971140 | ODF3B | −0.21 | 9.88 × 10−83 | IRG | NS | NS |
3.5. Potential Drug Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN); Harley, J.B.; Alarcón-Riquelme, M.E.; Criswell, L.A.; Jacob, C.O.; Kimberly, R.P.; Moser, K.L.; Tsao, B.P.; Vyse, T.J.; Langefeld, C.D.; et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008, 40, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.L.; Sheng, Y.; Zhang, Y.; Wang, Y.-F.; Zhu, Z.; Tombleson, P.; Chen, L.; Cunninghame Graham, D.S.; Bentham, J.; Roberts, A.L.; et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 2016, 48, 940–946. [Google Scholar] [CrossRef]
- Lessard, C.J.; Sajuthi, S.; Zhao, J.; Kim, K.; Ice, J.A.; Li, H.; Ainsworth, H.; Rasmussen, A.; Kelly, J.A.; Marion, M.; et al. Identification of a Systemic Lupus Erythematosus Risk Locus Spanning ATG16L2, FCHSD2, and P2RY2 in Koreans. Arthritis Rheumatol. 2016, 68, 1197–1209. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Molineros, J.E.; Looger, L.L.; Zhou, X.-J.; Kim, K.; Okada, Y.; Ma, J.; Qi, Y.-Y.; Kim-Howard, X.; Motghare, P.; et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat. Genet. 2016, 48, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Alarcón-Riquelme, M.E.; Ziegler, J.T.; Molineros, J.; Howard, T.D.; Moreno-Estrada, A.; Sánchez-Rodríguez, E.; Ainsworth, H.C.; Ortiz-Tello, P.; Comeau, M.E.; Rasmussen, A.; et al. Genome-Wide Association Study in an Amerindian Ancestry Population Reveals Novel Systemic Lupus Erythematosus Risk Loci and the Role of European Admixture. Arthritis Rheumatol. 2016, 68, 932–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langefeld, C.D.; Ainsworth, H.C.; Cunninghame Graham, D.S.; Kelly, J.A.; Comeau, M.E.; Marion, M.C.; Howard, T.D.; Ramos, P.S.; Croker, J.A.; Morris, D.L.; et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat. Commun. 2017, 8, 16021. [Google Scholar] [CrossRef]
- Deng, Y.; Tsao, B.P. Updates in Lupus Genetics. Curr. Rheumatol. Rep. 2017, 19, 68. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Morris, D.L.; Vyse, T.J. Genetic advances in systemic lupus erythematosus: An update. Curr. Opin. Rheumatol. 2017, 29, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Deapen, D.; Escalante, A.; Weinrib, L.; Horwitz, D.; Bachman, B.; Roy-Burman, P.; Walker, A.; Mack, T.M. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992, 35, 311–318. [Google Scholar]
- Hughes, T.; Sawalha, A.H. The role of epigenetic variation in the pathogenesis of systemic lupus erythematosus. Arthritis Res. Ther. 2011, 13, 245. [Google Scholar] [CrossRef] [Green Version]
- Jeffries, M.A.; Dozmorov, M.; Tang, Y.; Merrill, J.T.; Wren, J.D.; Sawalha, A.H. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics 2011, 6, 593–601. [Google Scholar] [CrossRef] [Green Version]
- Coit, P.; Renauer, P.; Jeffries, M.A.; Merrill, J.T.; McCune, W.J.; Maksimowicz-McKinnon, K.; Sawalha, A.H. Renal involvement in lupus is characterized by unique DNA methylation changes in naïve CD4+ T cells. J. Autoimmun. 2015, 61, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Renauer, P.; Coit, P.; Jeffries, M.A.; Merrill, J.T.; McCune, W.J.; Maksimowicz-McKinnon, K.; Sawalha, A.H. DNA methylation patterns in naïve CD4+ T cells identify epigenetic susceptibility loci for malar rash and discoid rash in systemic lupus erythematosus. Lupus Sci. Med. 2015, 2, e000101. [Google Scholar] [CrossRef] [Green Version]
- Ulff-Møller, C.J.; Asmar, F.; Liu, Y.; Svendsen, A.J.; Busato, F.; Grønbaek, K.; Tost, J.; Jacobsen, S. Twin DNA Methylation Profiling Reveals Flare-Dependent Interferon Signature and B Cell Promoter Hypermethylation in Systemic Lupus Erythematosus. Arthritis Rheumatol. 2018, 70, 878–890. [Google Scholar] [CrossRef] [Green Version]
- Coit, P.; Jeffries, M.; Altorok, N.; Dozmorov, M.G.; Koelsch, K.A.; Wren, J.D.; Merrill, J.T.; McCune, W.J.; Sawalha, A.H. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naïve CD4+ T cells from lupus patients. J. Autoimmun. 2013, 43, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Coit, P.; Ognenovski, M.; Gensterblum, E.; Maksimowicz-McKinnon, K.; Wren, J.D.; Sawalha, A.H. Ethnicity-specific epigenetic variation in naïve CD4+ T cells and the susceptibility to autoimmunity. Epigenetics Chromatin 2015, 8, 49. [Google Scholar] [CrossRef] [Green Version]
- Absher, D.M.; Li, X.; Waite, L.L.; Gibson, A.; Roberts, K.; Edberg, J.; Chatham, W.W.; Kimberly, R.P. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013, 9, e1003678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coit, P.; Yalavarthi, S.; Ognenovski, M.; Zhao, W.; Hasni, S.; Wren, J.D.; Kaplan, M.J.; Sawalha, A.H. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J. Autoimmun. 2015, 58, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mok, A.; Solomon, O.; Nayak, R.R.; Coit, P.; Quach, H.L.; Nititham, J.; Sawalha, A.H.; Barcellos, L.F.; Criswell, L.A.; Chung, S.A. Genome-wide profiling identifies associations between lupus nephritis and differential methylation of genes regulating tissue hypoxia and type 1 interferon responses. Lupus Sci. Med. 2016, 3, e000183. [Google Scholar] [CrossRef] [Green Version]
- Chung, S.A.; Nititham, J.; Elboudwarej, E.; Quach, H.L.; Taylor, K.E.; Barcellos, L.F.; Criswell, L.A. Genome-Wide Assessment of Differential DNA Methylation Associated with Autoantibody Production in Systemic Lupus Erythematosus. PLoS ONE 2015, 10, e0129813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeung, K.S.; Chung, B.H.-Y.; Choufani, S.; Mok, M.Y.; Wong, W.L.; Mak, C.C.Y.; Yang, W.; Lee, P.P.W.; Wong, W.H.S.; Chen, Y.-A.; et al. Genome-Wide DNA Methylation Analysis of Chinese Patients with Systemic Lupus Erythematosus Identified Hypomethylation in Genes Related to the Type I Interferon Pathway. PLoS ONE 2017, 12, e0169553. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Mi, W.; Luo, H.; Chen, T.; Liu, S.; Raman, I.; Zuo, X.; Li, Q.-Z. Whole-genome transcription and DNA methylation analysis of peripheral blood mononuclear cells identified aberrant gene regulation pathways in systemic lupus erythematosus. Arthritis Res. Ther. 2016, 18, 162. [Google Scholar] [CrossRef] [Green Version]
- Imgenberg-Kreuz, J.; Carlsson Almlöf, J.; Leonard, D.; Alexsson, A.; Nordmark, G.; Eloranta, M.-L.; Rantapää-Dahlqvist, S.; Bengtsson, A.A.; Jönsen, A.; Padyukov, L.; et al. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2018, 77, 736–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, S.; George, N.I.; Green-Knox, B.; Treadwell, E.L.; Word, B.; Yim, S.; Lyn-Cook, B. Epigenome-wide association study of peripheral blood mononuclear cells in systemic lupus erythematosus: Identifying DNA methylation signatures associated with interferon-related genes based on ethnicity and SLEDAI. J. Autoimmun. 2019, 96, 147–157. [Google Scholar] [CrossRef]
- Breitbach, M.E.; Ramaker, R.C.; Roberts, K.; Kimberly, R.P.; Absher, D. Population-Specific Patterns of Epigenetic Defects in the B Cell Lineage in Patients with Systemic Lupus Erythematosus. Arthritis Rheumatol. 2020, 72, 282–291. [Google Scholar] [CrossRef]
- Yeung, K.S.; Lee, T.L.; Mok, M.Y.; Mak, C.C.Y.; Yang, W.; Chong, P.C.Y.; Lee, P.P.W.; Ho, M.H.K.; Choufani, S.; Lau, C.S.; et al. Cell Lineage-specific Genome-wide DNA Methylation Analysis of Patients with Paediatric-onset Systemic Lupus Erythematosus. Epigenetics 2019, 14, 314–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weeding, E.; Sawalha, A.H. Deoxyribonucleic Acid Methylation in Systemic Lupus Erythematosus: Implications for Future Clinical Practice. Front. Immunol. 2018, 9, 875. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Fernandez, J.E.; Spector, T.D.; Bell, J.T. Epigenetics of discordant monozygotic twins: Implications for disease. Genome Med. 2014, 6, 60. [Google Scholar] [CrossRef]
- Javierre, B.M.; Fernandez, A.F.; Richter, J.; Al-Shahrour, F.; Martin-Subero, J.I.; Rodriguez-Ubreva, J.; Berdasco, M.; Fraga, M.F.; O’Hanlon, T.P.; Rider, L.G.; et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010, 20, 170–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakyan, V.K.; Beyan, H.; Down, T.A.; Hawa, M.I.; Maslau, S.; Aden, D.; Daunay, A.; Busato, F.; Mein, C.A.; Manfras, B.; et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011, 7, e1002300. [Google Scholar] [CrossRef]
- Gervin, K.; Vigeland, M.D.; Mattingsdal, M.; Hammerø, M.; Nygård, H.; Olsen, A.O.; Brandt, I.; Harris, J.R.; Undlien, D.E.; Lyle, R. DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: Identification of epigenetically dysregulated genes. PLoS Genet. 2012, 8, e1002454. [Google Scholar] [CrossRef]
- Häsler, R.; Feng, Z.; Bäckdahl, L.; Spehlmann, M.E.; Franke, A.; Teschendorff, A.; Rakyan, V.K.; Down, T.A.; Wilson, G.A.; Feber, A.; et al. A functional methylome map of ulcerative colitis. Genome Res. 2012, 22, 2130–2137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, A.; Sevier, S.; Kelly, J.A.; Glenn, S.B.; Aberle, T.; Cooney, C.M.; Grether, A.; James, E.; Ning, J.; Tesiram, J.; et al. The lupus family registry and repository. Rheumatology 2011, 50, 47–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochberg, M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997, 40, 1725. [Google Scholar] [CrossRef] [PubMed]
- Bombardier, C.; Gladman, D.D.; Urowitz, M.B.; Caron, D.; Chang, C.H. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992, 35, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika 1988, 75, 800–802. [Google Scholar] [CrossRef]
- Rusinova, I.; Forster, S.; Yu, S.; Kannan, A.; Masse, M.; Cumming, H.; Chapman, R.; Hertzog, P.J. Interferome v2.0: An updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013, 41, D1040–D1046. [Google Scholar] [CrossRef] [PubMed]
- Hachiya, T.; Furukawa, R.; Shiwa, Y.; Ohmomo, H.; Ono, K.; Katsuoka, F.; Nagasaki, M.; Yasuda, J.; Fuse, N.; Kinoshita, K.; et al. Genome-wide identification of inter-individually variable DNA methylation sites improves the efficacy of epigenetic association studies. NPJ Genom Med. 2017, 2, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Irizarry, R.; MacDonald, J.; Gentry, J. Gcrma: Background adjustment using sequence information. R Package Version 2012, 2200, 3–10. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Griffith, O.L.; Coffman, A.C.; Weible, J.V.; McMichael, J.F.; Spies, N.C.; Koval, J.; Das, I.; Callaway, M.B.; Eldred, J.M.; et al. DGIdb: Mining the druggable genome. Nat. Methods 2013, 10, 1209–1210. [Google Scholar] [CrossRef] [PubMed]
- Grammer, A.C.; Ryals, M.M.; Heuer, S.E.; Robl, R.D.; Madamanchi, S.; Davis, L.S.; Lauwerys, B.; Catalina, M.D.; Lipsky, P.E. Drug repositioning in SLE: Crowd-sourcing, literature-mining and Big Data analysis. Lupus 2016, 25, 1150–1170. [Google Scholar] [CrossRef]
- Mallya, M.; Campbell, R.D.; Aguado, B. Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein Sci. 2006, 15, 2244–2256. [Google Scholar] [CrossRef] [Green Version]
- Alaridah, N.; Winqvist, N.; Håkansson, G.; Tenland, E.; Rönnholm, A.; Sturegård, E.; Björkman, P.; Godaly, G. Impaired CXCR1-dependent oxidative defence in active tuberculosis patients. Tuberculosis 2015, 95, 744–750. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Bao, C.; Huang, H.; Lin, F.; Yuan, Y.; Wang, S.; Jin, L.; Yang, T.; Shi, M.; Zhang, Z.; et al. Low expression of CXCR1/2 on neutrophils predicts poor survival in patients with hepatitis B virus-related acute-on-chronic liver failure. Sci. Rep. 2016, 6, 38714. [Google Scholar] [CrossRef] [Green Version]
- Almajhdi, F.N.; Al-Ahdal, M.; Abdo, A.A.; Sanai, F.M.; Al-Anazi, M.; Khalaf, N.; Viswan, N.A.; Al-Ashgar, H.; Al-Kahtani, K.; Al-Humaidan, H.; et al. Single nucleotide polymorphisms in CXCR1 gene and its association with hepatitis B infected patients in Saudi Arabia. Ann. Hepatol. 2013, 12, 220–227. [Google Scholar] [CrossRef]
- Swamydas, M.; Gao, J.-L.; Break, T.J.; Johnson, M.D.; Jaeger, M.; Rodriguez, C.A.; Lim, J.K.; Green, N.M.; Collar, A.L.; Fischer, B.G.; et al. CXCR1-mediated neutrophil degranulation and fungal killing promote Candida clearance and host survival. Sci. Transl. Med. 2016, 8, 322ra10. [Google Scholar] [CrossRef] [Green Version]
- Gardiner-Garden, M.; Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987, 196, 261–282. [Google Scholar] [CrossRef]
- Zhao, M.; Zhou, Y.; Zhu, B.; Wan, M.; Jiang, T.; Tan, Q.; Liu, Y.; Jiang, J.; Luo, S.; Tan, Y.; et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 2016, 75, 1998–2006. [Google Scholar] [CrossRef] [Green Version]
- Kiso, K.; Yoshifuji, H.; Oku, T.; Hikida, M.; Kitagori, K.; Hirayama, Y.; Nakajima, T.; Haga, H.; Tsuruyama, T.; Miyagawa-Hayashino, A. Transgelin-2 is upregulated on activated B-cells and expressed in hyperplastic follicles in lupus erythematosus patients. PLoS ONE 2017, 12, e0184738. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-R.; Lee, H.-S.; Lee, K.-S.; Jung, I.D.; Kwon, M.-S.; Kim, C.-H.; Kim, S.-M.; Yoon, M.-H.; Park, Y.-M.; Lee, S.-M.; et al. An Essential Role for TAGLN2 in Phagocytosis of Lipopolysaccharide-activated Macrophages. Sci. Rep. 2017, 7, 8731. [Google Scholar] [CrossRef] [Green Version]
- Dragovich, M.A.; Mor, A. The SLAM family receptors: Potential therapeutic targets for inflammatory and autoimmune diseases. Autoimmun. Rev. 2018, 17, 674–682. [Google Scholar] [CrossRef]
- Li, F.J.; Won, W.J.; Becker, E.J.; Easlick, J.L.; Tabengwa, E.M.; Li, R.; Shakhmatov, M.; Honjo, K.; Burrows, P.D.; Davis, R.S. Emerging roles for the FCRL family members in lymphocyte biology and disease. Curr. Top. Microbiol. Immunol. 2014, 382, 29–50. [Google Scholar] [CrossRef] [Green Version]
- Schreeder, D.M.; Cannon, J.P.; Wu, J.; Li, R.; Shakhmatov, M.A.; Davis, R.S. Cutting edge: FcR-like 6 is an MHC class II receptor. J. Immunol. 2010, 185, 23–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balada, E.; Felip, L.; Ordi-Ros, J.; Vilardell-Tarrés, M. DUSP23 is over-expressed and linked to the expression of DNMTs in CD4+ T cells from systemic lupus erythematosus patients. Clin. Exp. Immunol. 2017, 187, 242–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavlar, T.; Ablasser, A.; Hornung, V. Induction of type I IFNs by intracellular DNA-sensing pathways. Immunol. Cell Biol. 2012, 90, 474–482. [Google Scholar] [CrossRef]
- Suárez-Calvet, X.; Gallardo, E.; Nogales-Gadea, G.; Querol, L.; Navas, M.; Díaz-Manera, J.; Rojas-Garcia, R.; Illa, I. Altered RIG-I/DDX58-mediated innate immunity in dermatomyositis. J. Pathol. 2014, 233, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Crow, M.K. Collaboration, genetic associations, and lupus erythematosus. N. Engl. J. Med. 2008, 358, 956–961. [Google Scholar] [CrossRef]
- Ambrosi, C.; Manzo, M.; Baubec, T. Dynamics and Context-Dependent Roles of DNA Methylation. J. Mol. Biol. 2017, 429, 1459–1475. [Google Scholar] [CrossRef]
- Crowl, J.T.; Gray, E.E.; Pestal, K.; Volkman, H.E.; Stetson, D.B. Intracellular Nucleic Acid Detection in Autoimmunity. Annu. Rev. Immunol. 2017, 35, 313–336. [Google Scholar] [CrossRef]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, M.; Niewold, T.B. Nucleic Acid sensors and type I interferon production in systemic lupus erythematosus. Front. Immunol. 2013, 4, 319. [Google Scholar] [CrossRef] [Green Version]
- Rathinam, V.A.K.; Jiang, Z.; Waggoner, S.N.; Sharma, S.; Cole, L.E.; Waggoner, L.; Vanaja, S.K.; Monks, B.G.; Ganesan, S.; Latz, E.; et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 2010, 11, 395–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Man, S.M.; Karki, R.; Kanneganti, T.-D. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 2016, 46, 269–280. [Google Scholar] [CrossRef] [Green Version]
- Mustelin, T.; Lood, C.; Giltiay, N.V. Sources of Pathogenic Nucleic Acids in Systemic Lupus Erythematosus. Front. Immunol. 2019, 10, 1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, K.; Kristiansen, M.; Hagen-Larsen, H.; Skytthe, A.; Bathum, L.; Jeune, B.; Andersen-Ranberg, K.; Vaupel, J.W.; Orstavik, K.H. X-linked genetic factors regulate hematopoietic stem-cell kinetics in females. Blood 2000, 95, 2449–2451. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Parfitt, A.; Grennan, D.M.; Manolios, N. X-chromosome inactivation in monozygotic twins with systemic lupus erythematosus. Autoimmunity 1997, 26, 85–93. [Google Scholar] [CrossRef] [PubMed]

| CpG * | Chr | Pos(bp) † | Gene | Mean ∆β | p-Value | STITCH [43] | IPA ‡ | DGIdb [44] |
|---|---|---|---|---|---|---|---|---|
| cg16526047 | 1 | 949893 | ISG15 | −0.11 | 1.28 × 10−4 | Irinotecan F | ||
| cg10959651 | 2 | 7018020 | RSAD2 | −0.13 | 3.14 × 10−14 | Fludarabine F | ||
| cg14126601 | 2 | 37384708 | EIF2AK2 | −0.1 | 5.55 × 10−16 | Indirubin derivative E804 | ||
| cg15768138 | 2 | 219030752 | CXCR1 | −0.11 | 7.38 × 10−27 | Reparixin D | Reparixin D | SCH-527123, Ketoprofen F |
| cg06981309 | 3 | 146260954 | PLSCR1 | −0.24 | 6.41 × 10−31 | Wogonin G | ||
| cg15065340 | 3 | 195632915 | TNK2 | −0.16 | 4.04 × 10−3 | Dasatinib−1 F | Osimertinib F, VemurafenibF | Debromohymenialdisine |
| cg08099136 | 6 | 32811251 | PSMB8 | −0.11 | 1.43 × 10−4 | Carfilzomib4 F,Oprozomib D, Bortezomib6 F | Carfilzomib4 F | Carfilzomib4 F, |
| cg00052684 | 6 | 35694245 | FKBP5 | −0.16 | 1.65 × 10−3 | Rapamycin/Sirolimus2 F, Tacrolimus5 F | Venlafaxine F, Clomipramine F | |
| cg14864167 | 8 | 66751182 | PDE7A | −0.35 | 1.21 × 10−9 | Ketotifen F, Dyphylline F | ||
| cg12110437 | 8 | 144098888 | LY6E | −0.2 | 3.14 × 10−9 | DLYE5953AD | ||
| cg06188083 | 10 | 91093005 | IFIT3 | −0.25 | 6.18 × 10−8 | Imidazoles D | ||
| cg08926253 | 11 | 614761 | IRF7 | −0.17 | 2.01 × 10−9 | Hesperidin D | ||
| cg03753191 | 13 | 43566902 | EPSTI1 | −0.1 | 9.23 × 10−5 | Methotrexate F T, Vinblastine F, Doxorubicin F, Cisplatin F | ||
| cg00246969 | 13 | 99159656 | STK24 | −0.11 | 6.26 × 10−6 | Staurosporine D | ||
| cg23378941 | 17 | 64361956 | PRKCA | −0.11 | 6.89 × 10−5 | Staurosporine D | Aprinocarsen | Midostaurin F, Enzastaurin D, Quercetin D G, Aprinocarsen, Ruboxistaurin D, Ingenol Mebutate FW, Bryostatin D, Sotrastaurin Acetate D, Tamoxifen2 F |
| cg07839313 | 19 | 17514600 | BST2 | −0.12 | 3.48 × 10−3 | Resveratrol6 D G | ||
| cg21549285 | 21 | 42799141 | MX1 | −0.47 | 6.59 × 10−13 | Mitomycin C F, Colchicine F | ||
| cg19460508 | 22 | 44422195 | PARVB | −0.1 | 1.64 × 10−3 | Lovastatin3 F | Bortezomib6 F |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marion, M.C.; Ramos, P.S.; Bachali, P.; Labonte, A.C.; Zimmerman, K.D.; Ainsworth, H.C.; Heuer, S.E.; Robl, R.D.; Catalina, M.D.; Kelly, J.A.; et al. Nucleic Acid-Sensing and Interferon-Inducible Pathways Show Differential Methylation in MZ Twins Discordant for Lupus and Overexpression in Independent Lupus Samples: Implications for Pathogenic Mechanism and Drug Targeting. Genes 2021, 12, 1898. https://doi.org/10.3390/genes12121898
Marion MC, Ramos PS, Bachali P, Labonte AC, Zimmerman KD, Ainsworth HC, Heuer SE, Robl RD, Catalina MD, Kelly JA, et al. Nucleic Acid-Sensing and Interferon-Inducible Pathways Show Differential Methylation in MZ Twins Discordant for Lupus and Overexpression in Independent Lupus Samples: Implications for Pathogenic Mechanism and Drug Targeting. Genes. 2021; 12(12):1898. https://doi.org/10.3390/genes12121898
Chicago/Turabian StyleMarion, Miranda C., Paula S. Ramos, Prathyusha Bachali, Adam C. Labonte, Kip D. Zimmerman, Hannah C. Ainsworth, Sarah E. Heuer, Robert D. Robl, Michelle D. Catalina, Jennifer A. Kelly, and et al. 2021. "Nucleic Acid-Sensing and Interferon-Inducible Pathways Show Differential Methylation in MZ Twins Discordant for Lupus and Overexpression in Independent Lupus Samples: Implications for Pathogenic Mechanism and Drug Targeting" Genes 12, no. 12: 1898. https://doi.org/10.3390/genes12121898
APA StyleMarion, M. C., Ramos, P. S., Bachali, P., Labonte, A. C., Zimmerman, K. D., Ainsworth, H. C., Heuer, S. E., Robl, R. D., Catalina, M. D., Kelly, J. A., Howard, T. D., Lipsky, P. E., Grammer, A. C., & Langefeld, C. D. (2021). Nucleic Acid-Sensing and Interferon-Inducible Pathways Show Differential Methylation in MZ Twins Discordant for Lupus and Overexpression in Independent Lupus Samples: Implications for Pathogenic Mechanism and Drug Targeting. Genes, 12(12), 1898. https://doi.org/10.3390/genes12121898






