Supranutritional Maternal Organic Selenium Supplementation during Different Trimesters of Pregnancy Affects the Muscle Gene Transcriptome of Newborn Beef Calves in a Time-Dependent Manner
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Tissue Collection
2.3. Whole-Blood and Muscle Collection and Selenium Analysis
2.4. Total RNA Isolation, Library Preparation, Sequencing, and Data Processing
2.5. Transcriptome Analysis
2.6. Analysis of Genes Encoding Transcription Factors with Regulatory Potential
3. Results
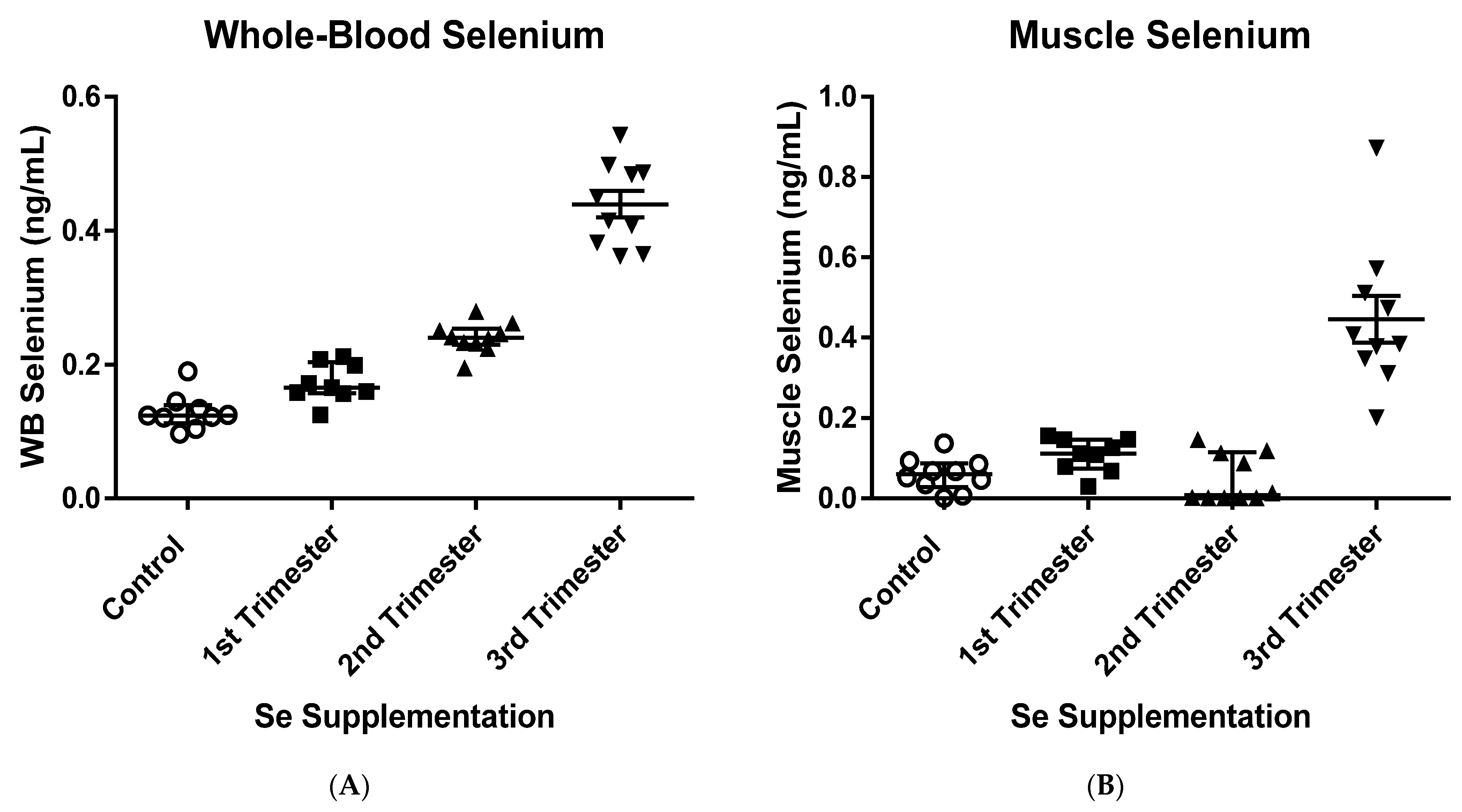
3.1. Muscle and Whole-Blood Selenium Concentrations and Body Weights of Newborn Calves
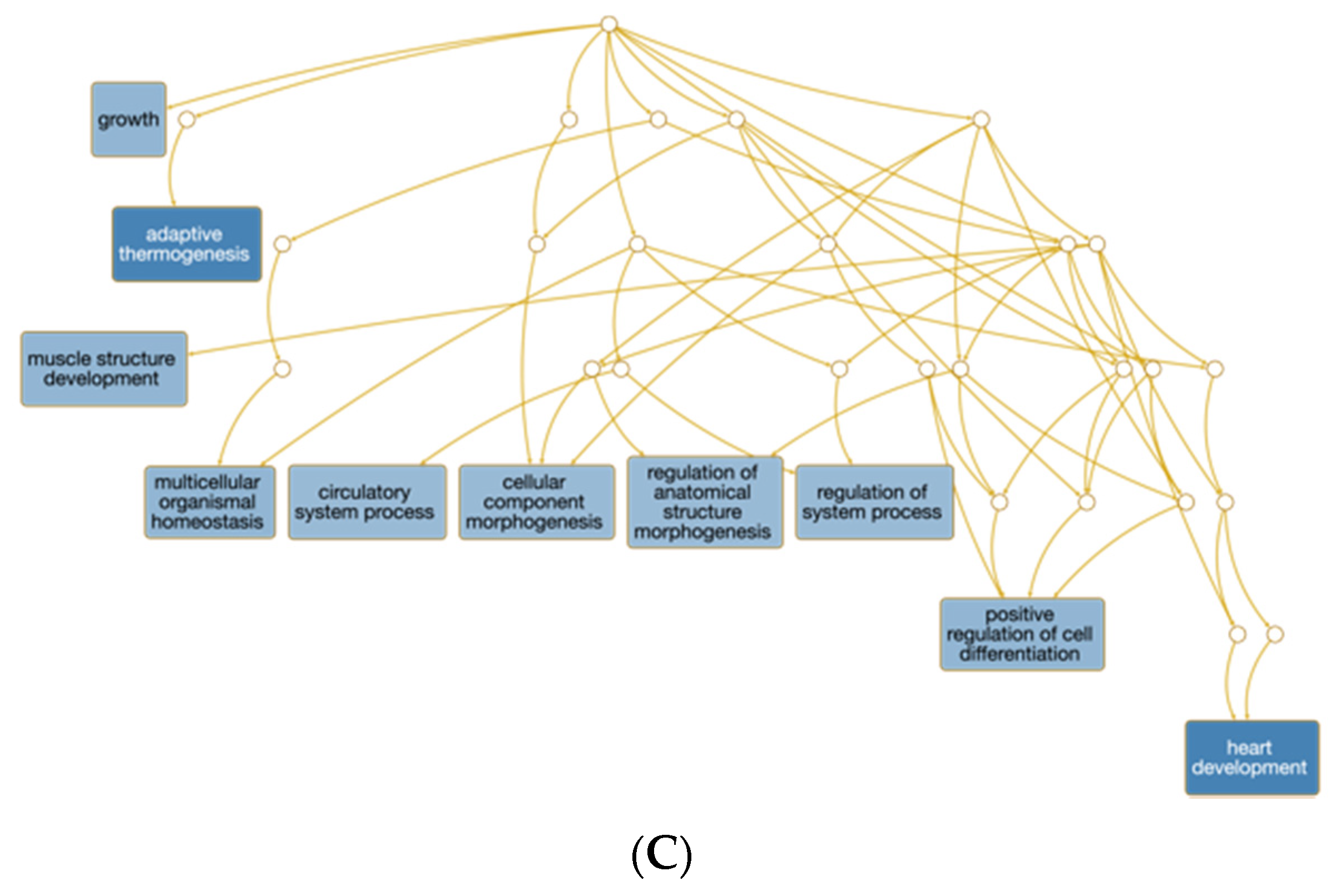
3.2. RNA-Seq Analysis of Muscle Biopsies of Newborn Calves
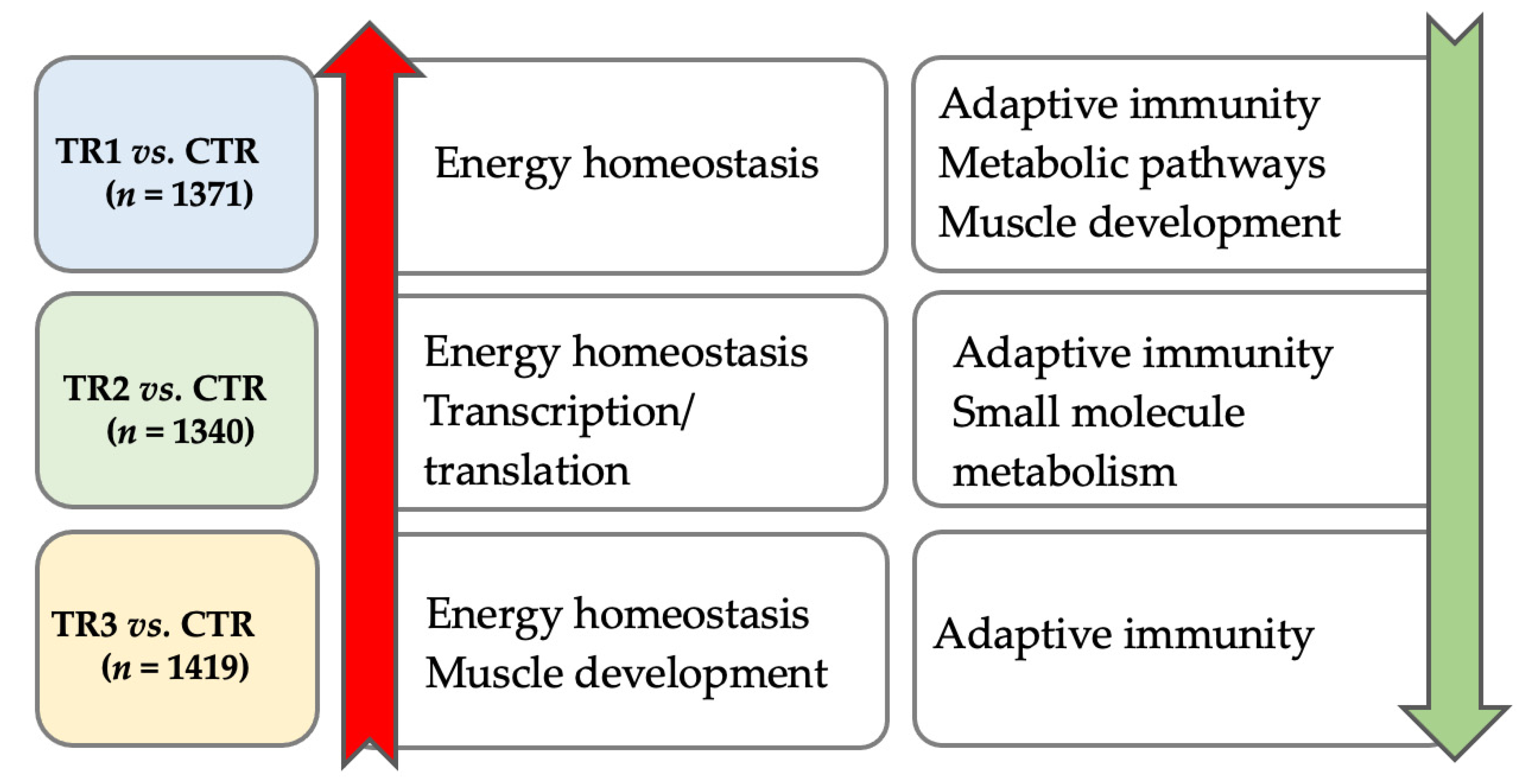
3.2.1. Comparison of the Three Se-Supplemented Groups versus Control
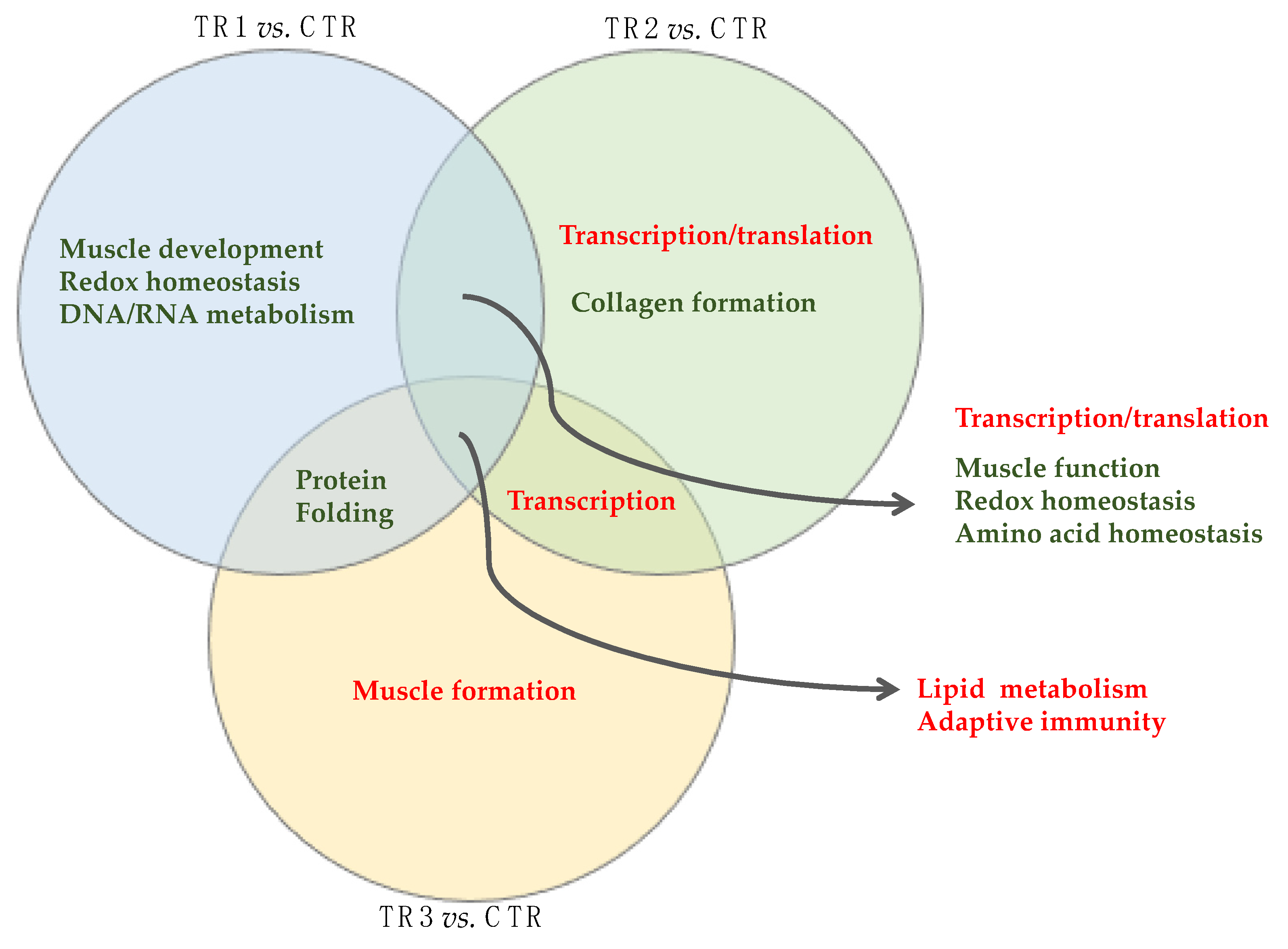
3.2.2. Unique and Shared Changes between the Se-Supplementation Groups versus Control
3.3. Transcription Factors Putatively Regulating the Differentially Expressed Genes
4. Discussion
4.1. Supranutritional Maternal Se-Yeast Supplementation Effects on the Longissimus Dorsi Muscle Transcriptome That Are Dependent on the Trimester of Supplementation
4.2. Supranutritional Maternal Se-Yeast Supplementation Effects on the Longissimus Dorsi Muscle Transcriptome That Are Independent of the Trimester of Supplementation
4.3. Effects of Supranutritional Maternal Se-Yeast Supplementation on Selenoproteins Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, J.; Nie, W.; Wang, F.; Guo, Y. Maternal selenium supplementation enhanced skeletal muscle development through increasing protein synthesis and SelW mRNA levels of their offspring. Biol. Trace Elem. Res. 2018, 186, 238–248. [Google Scholar] [CrossRef]
- Pappas, A.C.; Zoidis, E.; Chadio, S.E. Maternal selenium and developmental programming. Antioxidants 2019, 8, 145. [Google Scholar] [CrossRef] [Green Version]
- Lammi, M.J.; Qu, C. Selenium-Related transcriptional regulation of gene expression. Int. J. Mol. Sci. 2018, 19, 2665. [Google Scholar] [CrossRef] [Green Version]
- Wischhusen, P.; Saito, T.; Heraud, C.; Kaushik, S.J.; Fauconneau, B.; Prabhu, P.A.J.; Fontagné-Dicharry, S.; Skjærven, K.H. Parental selenium nutrition affects the one-carbon metabolism and the hepatic dna methylation pattern of rainbow trout (Oncorhynchus mykiss) in the progeny. Life 2020, 10, 121. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.A.; Bobe, G.; Vorachek, W.R.; Estill, C.T.; Mosher, W.D.; Pirelli, G.J.; Gamroth, M. Effect of supranutritional maternal and colostral selenium supplementation on passive absorption of immunoglobulin G in selenium-replete dairy calves. J. Dairy Sci. 2014, 97, 4379–4391. [Google Scholar] [CrossRef]
- Rederstorff, M.; Krol, A.; Lescure, A. Understanding the importance of selenium and selenoproteins in muscle function. Cell. Mol. Life Sci. 2006, 63, 52–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, A.; Juniper, D.T.; Sanil, M.; Morgan, L.; Clark, L.; Sies, H.; Rayman, M.P.; Steinbrenner, H. Supranutritional selenium induces alterations in molecular targets related to energy metabolism in skeletal muscle and visceral adipose tissue of pigs. J. Inorg. Biochem. 2012, 114, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, M.L.; Nogales, F.; Membrilla, A.; Carreras, O. Maternal selenium status is profoundly involved in metabolic fetal programming by modulating insulin resistance, oxidative balance and energy homeostasis. Eur. J. Nutr. 2019, 58, 3171–3181. [Google Scholar] [CrossRef]
- Caton, J.S.; Crouse, M.S.; McLean, K.J.; Dahlen, C.R.; Ward, A.K.; Cushman, R.A.; Grazul-Bilska, A.T.; Neville, B.W.; Borowicz, P.P.; Reynolds, L.P. Maternal periconceptual nutrition, early pregnancy, and developmental outcomes in beef cattle. J. Anim. Sci. 2020, 98, skaa358. [Google Scholar] [CrossRef]
- Thayer, Z.M.; Rutherford, J.; Kuzawa, C.W. Maternal nutritional buffering model: An evolutionary framework for pregnancy nutritional intervention. Evol. Med. Public Heal. 2020, 2020, 14–27. [Google Scholar] [CrossRef] [Green Version]
- Godfrey, K.M.; Barker, D.J.P. Fetal nutrition and adult disease. Am. J. Clin. Nutr. 2000, 71, 1344–1352. [Google Scholar] [CrossRef]
- Lekatz, L.A.; Caton, J.S.; Taylor, J.B.; Reynolds, L.P.; Redmer, D.A.; Vonnahme, K.A. Maternal selenium supplementation and timing of nutrient restriction in pregnant sheep: Effects on maternal endocrine status and placental characteristics. J. Anim. Sci. 2010, 88, 955–971. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.H.; Clish, C.B.; Gladyshev, V.N. Selenium deficiency is associated with pro-longevity mechanisms. Cell Rep. 2019, 27, 2785–2797.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, L.P.; Ward, A.K.; Caton, J.S. Epigenetics and developmental programming in ruminants: Long-Term impacts on growth and development. In Biology of Domestic Animals; Scanes, C.G., Hill, R.A., Eds.; CRC Press: Boca Raton, FL, USA, 2017; pp. 85–120. ISBN 9781315152080. [Google Scholar]
- Caton, J.S.; Crouse, M.S.; Reynolds, L.P.; Neville, T.L.; Dahlen, C.R.; Ward, A.K.; Swanson, K.C. Maternal nutrition and programming of offspring energy requirements. Transl. Anim. Sci. 2019, 3, 976–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, M.; Tong, J.; Zhao, J.; Underwood, K.R.; Zhu, M.; Ford, S.P.; Nathanielsz, P.W. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 2009, 88, E51–E60. [Google Scholar] [CrossRef] [Green Version]
- Vonnahme, K.A.; Luther, J.S.; Reynolds, L.P.; Hammer, C.J.; Carlson, D.B.; Redmer, D.A.; Caton, J.S. Impacts of maternal selenium and nutritional level on growth, adiposity, and glucose tolerance in female offspring in sheep. Domest. Anim. Endocrinol. 2010, 39, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Wallace, L.G.; Bobe, G.; Vorachek, W.R.; Dolan, B.P.; Estill, C.T.; Pirelli, G.J.; Hall, J.A. Effects of feeding pregnant beef cows selenium-enriched alfalfa hay on selenium status and antibody titers in their newborn calves. J. Anim. Sci. 2017, 95, 2408–2420. [Google Scholar] [CrossRef]
- Apperson, K.D.; Vorachek, W.R.; Dolan, B.P.; Bobe, G.; Pirelli, G.J.; Hall, J.A. Effects of feeding pregnant beef cows selenium-enriched alfalfa hay on passive transfer of ovalbumin in their newborn calves. J. Trace Elem. Med. Biol. 2018, 50, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.A.; Bobe, G.; Vorachek, W.R.; Kasper, K.; Traber, M.G.; Mosher, W.D.; Pirelli, G.J.; Gamroth, M. Effect of supranutritional organic selenium supplementation on postpartum blood micronutrients, antioxidants, metabolites, and inflammation biomarkers in selenium-replete dairy cows. Biol. Trace Elem. Res. 2014, 161, 272–287. [Google Scholar] [CrossRef]
- Paradis, F.; Wood, K.M.; Swanson, K.C.; Miller, S.P.; McBride, B.W.; Fitzsimmons, C. Maternal nutrient restriction in mid-to-late gestation influences fetal mRNA expression in muscle tissues in beef cattle. BMC Genomics 2017, 18, 632. [Google Scholar] [CrossRef]
- Diniz, W.J.S.; Crouse, M.S.; Cushman, R.A.; McLean, K.J.; Caton, J.S.; Dahlen, C.R.; Reynolds, L.P.; Ward, A.K. Cerebrum, liver, and muscle regulatory networks uncover maternal nutrition effects in developmental programming of beef cattle during early pregnancy. Sci. Rep. 2021, 11, 2771. [Google Scholar] [CrossRef] [PubMed]
- Miguel-Pacheco, G.G.; Curtain, L.D.; Rutland, C.; Knott, L.; Norman, S.T.; Phillips, N.J.; Perry, V.E.A. Increased dietary protein in the second trimester of gestation increases live weight gain and carcass composition in weaner calves to 6 months of age. Animal 2016, 2016. 11, 991–999. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Bobe, G.; Filley, S.J.; Pirelli, G.J.; Bohle, M.G.; Davis, T.Z.; Bañuelos, G.L.; Hall, J.A. Effects of springtime sodium selenate foliar application and NPKS fertilization on selenium concentrations and selenium species in forages across Oregon. Anim. Feed Sci. Technol. 2021, 276, 114944. [Google Scholar] [CrossRef]
- Hall, J.A.; Harwell, A.M.; Van Saun, R.J.; Vorachek, W.R.; Stewart, W.C.; Galbraith, M.L.; Hooper, K.J.; Hunter, J.K.; Mosher, W.D.; Pirelli, G.J. Agronomic biofortification with selenium: Effects on whole blood selenium and humoral immunity in beef cattle. Anim. Feed Sci. Technol. 2011, 164, 184–190. [Google Scholar] [CrossRef]
- National Research Council. Mineral Tolerance of Animals: Second Revised Edition, 2005; The National Academies Press: Washington, DC, USA, 2005; ISBN 978-0-309-09654-6. [Google Scholar]
- Hall, J.A.; Bobe, G.; Hunter, J.K.; Vorachek, W.R.; Stewart, W.C.; Vanegas, J.A.; Estill, C.T.; Mosher, W.D.; Pirelli, G.J. Effect of feeding selenium-fertilized alfalfa hay on performance of weaned beef calves. PLoS ONE 2013, 8, e58188. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.; FASTQC. A Quality Control Tool for High throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 11 January 2021).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, giaa021. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R. A Language and Environment for Statistical Computing, 2018; R Core Team R: Vienna, Austria, 2018. [Google Scholar]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.; Di Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A.; Mundt, F. Factoextra: Extract and visualize the results of multivariate data analyses. R Package Version 2019, 1, 337–354. [Google Scholar]
- Reverter, A.; Ingham, A.; Lehnert, S.A.; Tan, S.H.; Wang, Y.; Ratnakumar, A.; Dalrymple, B.P. Simultaneous identification of differential gene expression and connectivity in inflammation, adipogenesis and cancer. Bioinformatics 2006, 22, 2396–2404. [Google Scholar] [CrossRef] [Green Version]
- de Biagi, C.A.O., Jr.; Nociti, R.P.; Funicheli, B.O.; de Cássia Ruy, P.; Ximenez, J.P.B.; Silva, W.A., Jr. CeTF: An R package to coexpression for transcription factors using regulatory impact factors (RIF) and partial correlation and information (PCIT) analysis. BioRxiv 2020. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [Green Version]
- Reverter, A.; Hudson, N.J.; Nagaraj, S.H.; Pérez-Enciso, M.; Dalrymple, B.P. Regulatory impact factors: Unraveling the transcriptional regulation of complex traits from expression data. Bioinformatics 2010, 26, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Miao, Y.-R.; Jia, L.-H.; Yu, Q.-Y.; Zhang, Q.; Guo, A.-Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019, 47, D33–D38. [Google Scholar] [CrossRef]
- Brummer, F.A.; Pirelli, G.J.; Hall, J.A. Selenium supplementation strategies for livestock in Oregon. Oregon State Univ. Ext. Serv. 2014, 1–9, EM 9094. [Google Scholar]
- Stewart, W.C.; Bobe, G.; Pirelli, G.J.; Mosher, W.D.; Hall, J.A. Organic and inorganic selenium: III. Ewe and progeny performance. J. Anim. Sci. 2012, 90, 4536–4543. [Google Scholar] [CrossRef]
- Long, N.M.; Vonnahme, K.A.; Hess, B.W.; Nathanielsz, P.W.; Ford, S.P. Effects of early gestational undernutrition on fetal growth, organ development, and placentomal composition in the bovine. J. Anim. Sci. 2009, 87, 1950–1959. [Google Scholar] [CrossRef] [Green Version]
- Valberg, S.J. Nutritional myopathies. Myopathies of ruminants and pigs. In The Merck Veterinary Manual; Aiello, S.E., Ed.; Merck & Co, Inc.: Kenilworth, NJ, USA, 2016; pp. 1172–1173. ISBN 978-0-911-91061-2. [Google Scholar]
- Diniz, W.J.S.; Banerjee, P.; Regitano, L.C.A. Cross talk between mineral metabolism and meat quality: A systems biology overview. Physiol. Genom. 2019, 51, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Dong, H.H. FoxO integration of insulin signaling with glucose and lipid metabolism. J. Endocrinol. 2017, 233, R67–R79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.J.; Kim, M.; Park, H.; Kim, H.S.; Jeon, M.J.; Oh, K.S.; Koh, E.H.; Won, J.C.; Kim, M.-S.; Oh, G.T.; et al. AMPK activation increases fatty acid oxidation in skeletal muscle by activating PPARα and PGC-1. Biochem. Biophys. Res. Commun. 2006, 340, 291–295. [Google Scholar] [CrossRef]
- Englund, P.; Lindroos, E.; Nennesmo, I.; Klareskog, L.; Lundberg, I.E. Skeletal muscle fibers express major histocompatibility complex class II antigens independently of inflammatory infiltrates in inflammatory myopathies. Am. J. Pathol. 2001, 159, 1263–1273. [Google Scholar] [CrossRef] [Green Version]
- Keller, C.W.; Fokken, C.; Turville, S.G.; Lünemann, A.; Schmidt, J.; Münz, C.; Lünemann, J.D. TNF-α induces macroautophagy and regulates MHC class II expression in human skeletal muscle cells. J. Biol. Chem. 2011, 286, 3970–3980. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.M.; Schild, C.O.; Cantón, G.J.; Riet-Correa, F.; Armendano, J.I.; Caffarena, R.D.; Brambilla, E.C.; García, J.A.; Morrell, E.L.; Poppenga, R.; et al. White muscle disease in three selenium deficient beef and dairy calves in Argentina and Uruguay. Ciência Rural. 2018, 48, 5. [Google Scholar] [CrossRef] [Green Version]
- Hugejiletu, H.; Bobe, G.; Vorachek, W.R.; Gorman, M.E.; Mosher, W.D.; Pirelli, G.J.; Hall, J.A. Selenium supplementation alters gene expression profiles associated with innate immunity in whole-blood neutrophils of sheep. Biol. Trace Elem. Res. 2013, 154, 28–44. [Google Scholar] [CrossRef]
- Hall, J.A.; Bobe, G.; Vorachek, W.R.; Hugejiletu; Gorman, M.E.; Mosher, W.D.; Pirelli, G.J. Effects of feeding selenium-enriched alfalfa hay on immunity and health of weaned beef calves. Biol. Trace Elem. Res. 2013, 156, 96–110. [Google Scholar] [CrossRef]
- Zhang, K.; Han, Y.; Zhao, Q.; Zhan, T.; Li, Y.; Sun, W.; Li, S.; Sun, D.; Si, X.; Yu, X.; et al. Targeted metabolomics analysis reveals that dietary supranutritional selenium regulates sugar and acylcarnitine metabolism homeostasis in pig liver. J. Nutr. 2020, 150, 704–711. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Agriculture. United States Department of Agriculture G-1 Certified Angus Beef: G-1 Specification 2020, 2; United States Department of Agriculture: Washington, DC, USA, 2020.
- Juszczuk-Kubiak, E.; Bujko, K.; Cymer, M.; Wicińska, K.; Gabryszuk, M.; Pierzchała, M. Effect of inorganic dietary selenium supplementation on selenoprotein and lipid metabolism gene expression patterns in liver and loin muscle of growing lambs. Biol. Trace Elem. Res. 2016, 172, 336–345. [Google Scholar] [CrossRef] [Green Version]
- Mehdi, Y.; Dufrasne, I. Selenium in Cattle: A Review. Molecules 2016, 21, 545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burk, R.F.; Hill, K.E. Selenoprotein P—Expression, functions, and roles in mammals. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 1441–1447. [Google Scholar] [CrossRef] [Green Version]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Roh, Y.J.; Han, S.J.; Park, I.; Lee, H.M.; Ok, Y.S.; Lee, B.C.; Lee, S.R. Role of selenoproteins in redox regulation of signaling and the antioxidant system: A review. Antioxidants 2020, 9, 383. [Google Scholar] [CrossRef]
- Talbert, L.E.; Julian, R.R. Methionine and selenomethionine as energy transfer acceptors for biomolecular structure elucidation in the gas phase. J. Am. Soc. Mass Spectrom. 2019, 30, 1601–1608. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diniz, W.J.S.; Bobe, G.; Klopfenstein, J.J.; Gultekin, Y.; Davis, T.Z.; Ward, A.K.; Hall, J.A. Supranutritional Maternal Organic Selenium Supplementation during Different Trimesters of Pregnancy Affects the Muscle Gene Transcriptome of Newborn Beef Calves in a Time-Dependent Manner. Genes 2021, 12, 1884. https://doi.org/10.3390/genes12121884
Diniz WJS, Bobe G, Klopfenstein JJ, Gultekin Y, Davis TZ, Ward AK, Hall JA. Supranutritional Maternal Organic Selenium Supplementation during Different Trimesters of Pregnancy Affects the Muscle Gene Transcriptome of Newborn Beef Calves in a Time-Dependent Manner. Genes. 2021; 12(12):1884. https://doi.org/10.3390/genes12121884
Chicago/Turabian StyleDiniz, Wellison J. S., Gerd Bobe, Joseph J. Klopfenstein, Yunus Gultekin, T. Zane Davis, Alison K. Ward, and Jean A. Hall. 2021. "Supranutritional Maternal Organic Selenium Supplementation during Different Trimesters of Pregnancy Affects the Muscle Gene Transcriptome of Newborn Beef Calves in a Time-Dependent Manner" Genes 12, no. 12: 1884. https://doi.org/10.3390/genes12121884
APA StyleDiniz, W. J. S., Bobe, G., Klopfenstein, J. J., Gultekin, Y., Davis, T. Z., Ward, A. K., & Hall, J. A. (2021). Supranutritional Maternal Organic Selenium Supplementation during Different Trimesters of Pregnancy Affects the Muscle Gene Transcriptome of Newborn Beef Calves in a Time-Dependent Manner. Genes, 12(12), 1884. https://doi.org/10.3390/genes12121884








