Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content
Abstract
1. Introduction
2. Fruit Development
3. Fruit Ripening
3.1. Fruit Softening
| Crop | Source Seed | Non Coding RNAs Identified | Targets Identified | Response To | Reference |
|---|---|---|---|---|---|
| Mango | EST database of mango | miRNAs—3 | 94 | Fruit development, ripening | [95] |
| RNA-seq database | miRNAs—104 lncRNAs—7610 | 2347 | Low temperature stress | [96] | |
| Genome assembly | tRNA—598 rRNA 45 snoRNA—47 snRNA—200 miRNA—235 | — | - | [97] | |
| EST database of mango | miRNAs—18 | 44 | Ripening | [98] | |
| Banana | Transcriptome | miRNAs—59 | 120 | Salt stress tolerance | [99] |
| Transcriptome | miRNAs—82 | 815 | Ripening | [100] | |
| Transcriptome | miRNAs—22 | 12 | Ripening | [82] | |
| Transcriptome | miRNAs—46 | 944 | Fruit softening and aroma biosynthesis | [84] | |
| Transcriptome | lncRNAs—12,462 | — | Low temperature stress | [101] | |
| Citrus | Transcriptome | miRNAs—101 | 28 | Alkaline stress | [71] |
| Papaya | Transcriptome data | miRNAs—213 | 1741 | Ripening | [102] |
| Guava | Guava genome, miRbase database | miRNAs—40 | 49 | Salinity stress | [103] |
| Dragon fruit | Transcriptome data | lncRNAs—11,650 | — | Betalain biosynthesis | [76] |
| Genome assembly | miRNAs—4989 tRNAs—4857 rRNAs—5909 snRNAs—3877 | — | Betalain biosynthesis | [74] |
3.2. Sugar Metabolism
3.3. Flavour
3.4. Pulp and Peel Colour
3.5. Fruit Quality
3.6. Bearing Characters
4. Shelf Life
5. Abiotic Stress
6. Biotic Stress
7. Genomic Assisted Breeding Strategies in Tropical Fruit Crops
7.1. Diversity Analysis
7.2. QTLS/Genes Related to Fruit Traits
7.3. Application of GWAS/GS in Marker Assisted Breeding
8. Genetic Engineering in Fruit Crops
8.1. Transgenics
8.2. Genome Editing/Gene Editing/CRISPR/Cas9
9. Databases
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. Major Tropical Fruits Market Review; FAO: Rome, Italy, 2020. [Google Scholar]
- OECD/FAO. OECD-FAO Agricultural Outlook 2020–2029; FAO: Rome, Italy; OECD Publishing: Paris, France, 2020; Volume 13, p. 27. [Google Scholar]
- Acham, I.O.; Ahemen, S.; Ukeyima, M.T.; Girgih, A.T. Tropical fruits: Bioactive properties and health promoting benefits in chronic disease prevention and management. Asian Food Sci. J. 2018, 3, 1–13. [Google Scholar] [CrossRef]
- Laldinchhana; Lalrengpuii, J.; Ray, S.; Pachuau, L. Indian tropical fruits and their bioactive compounds against human diseases. Plant-Derived Bioact. Chem. Mode Action 2020, 455–494. [Google Scholar] [CrossRef]
- Nath, V.; Kumar, G.; Pandey, S.D.; Pandey, S. Impact of climate change on tropical fruit production systems and its mitigation strategies. Clim. Chang. Agric. India Impact Adapt. 2019, 129–146. [Google Scholar] [CrossRef]
- Rajapaksha, L.; Gunathilake, D.C.; Pathirana, S.; Fernando, T. Reducing post-harvest losses in fruits and vegetables for ensuring food security—Case of Sri Lanka. MOJ Food Process. Technol. 2021, 9, 7–16. [Google Scholar] [CrossRef]
- Bantayehu, M.; Alemaye, M. Estimation of pre and postharvest losses of tropical fruits in ethiopia estimation of pre and postharvest losses of tropical fruits in Ethiopia 47. Int. J. Postharvest Technol. Innov. 2019, 6, 46–56. [Google Scholar] [CrossRef]
- Kuhn, D.N.; Bally, I.S.E.; Dillon, N.L.; Innes, D.; Groh, A.M.; Rahaman, J.; Ophir, R.; Cohen, Y.; Sherman, A. Genetic map of mango: A tool for mango breeding. Front. Plant Sci. 2017, 8, 577. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Hao, Y.; Fan, S.; Cai, J.; Chen, W.; Li, X.; Zhu, X. Metabolomic and transcriptomic profiling provide novel insights into fruit ripening and ripening disorder caused by 1-MCP treatments in papaya. Int. J. Mol. Sci. 2021, 22, 916. [Google Scholar] [CrossRef]
- Lobato-gómez, M.; Hewitt, S.; Capell, T.; Christou, P.; Dhingra, A.; Girón-calva, P.S. Transgenic and genome-edited fruits: Background, constraints, benefits and commercial opportunities. Hortic. Res. 2021, 8, 166. [Google Scholar] [CrossRef] [PubMed]
- Padmakar, B.; Dinesh, M.R.; Ravishankar, K.V. Marker-trait association for fruit characters in mango (Mangifera Indica L.) cultivars. J. Hortic. Sci. 2016, 11, 170–178. [Google Scholar]
- Azam, K.; Mir, H.; Prasad, B.D.; Ahmad, F. Identification of microsatellite markers associated with the horticultural traits in elite mango cultivars. J. Pharmac. Phytochem. 2018, 7, 2830–2834. [Google Scholar]
- Oak, P.; Deshpande, A.; Giri, A.; Gupta, V. Metabolomic dynamics reveals oxidative stress in spongy tissue disorder during ripening of Mangifera Indica L. fruit. Metabolites 2019, 9, 255. [Google Scholar] [CrossRef]
- Jonak, C.; Ökrész, L.; Bögre, L.; Hirt, H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002, 5, 415–424. [Google Scholar] [CrossRef]
- Asif, M.H.; Lakhwani, D.; Pathak, S.; Gupta, P.; Bag, S.K.; Nath, P.; Trivedi, P.K. Transcriptome analysis of ripe and unripe fruit tissue of banana identifies major metabolic networks involved in fruit ripening process. BMC Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, W.; Tie, W.; Ding, Z.; Ding, X.; Liu, Y.; Yan, Y. The MAPKKK and MAPKK gene families in banana: Identification, phylogeny and expression during development, ripening and abiotic stress. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Usha, K.; Jayaswal, P.K. In-silico analysis of WRKY transcription factors gene family in healthy and malformed stages of mango (Mangifera indica). Ind. J. Agric. Sci. 2021, 89, 111–116. [Google Scholar]
- Zheng, Y.; Liu, M.; Jia, C.; Wang, J.; Xu, B.; Jin, Z. Characteristics of banana B genome MADS box family demonstrate their roles in fruit development, ripening and stress. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, A.M.; Martinelli, F.; Reagan, R.L.; Uratsu, S.L.; Vo, A.; Tinoco, M.A.; Phu, M.L.; Chen, Y.; Rocke, D.M.; Dandekar, A.M. Transcriptome and metabolome analysis of citrus fruit to elucidate puffing disorder. Plant Sci. 2014, 217–218, 87–98. [Google Scholar] [CrossRef]
- Blas, A.L.; Yu, Q.; Veatch, O.J.; Paull, R.E.; Moore, P.H.; Ming, R. Genetic mapping of quantitative trait loci controlling fruit size and shape in papaya. Mol. Breed. 2012, 29, 457–466. [Google Scholar] [CrossRef]
- Mittal, A.; Yadav, I.S.; Arora, N.K.; Boora, R.S.; Mittal, M.; Kaur, P.; Erskine, W.; Chhuneja, P.; Indra, M.; Gill, S.; et al. RNA-Sequencing based gene expression landscape of guava cv Allahabad Safeda and comparative analysis to colored cultivars. BMC Genom. 2020, 21, 484. [Google Scholar]
- Usman, M.; Zaman, Q.; Fatima, B.; Rana, I.A.; Awan, F.S. Morpho-chemical diversity and RAPD fingerprinting in white flesh guava cultivars. J. Anim. Plant Sci. 2020, 30, 398–409. [Google Scholar]
- Shyamalamma, S.; Chandra, S.B.C.; Hegde, M.; Naryanswamy, P. Evaluation of genetic diversity in jackfruit (Artocarpus heterophyllus Lam.) based on amplified fragment length polymorphism markers. Genet. Mol. Res. 2008, 7, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Kavya, K.; Shyamalamma, S.; Gayatri, S. Morphological and molecular genetic diversity analysis using SSR markers in jackfruit (Artocarpus heterophyllus Lam.) Genotypes for Pulp Colour. Indian J. Agric. Res. 2019, 53, 8–16. [Google Scholar] [CrossRef]
- Goulao, L.F.; Oliveira, C.M. Cell wall modifications during fruit ripening: When a fruit is not the fruit. Trends Food Sci. Technol. 2008, 19, 4–25. [Google Scholar] [CrossRef]
- Ali, Z.M.; Chin, L.H.; Lazan, H. A comparative study on wall degrading enzymes, pectin modifications and softening during ripening of selected tropical fruits. Plant Sci. 2004, 167, 317–327. [Google Scholar] [CrossRef]
- Srivastava, S.; Singh, R.K.; Pathak, G.; Goel, R.; Asif, M.H.; Sane, A.P.; Sane, V.A. Comparative transcriptome analysis of unripe and mid-ripe fruit of Mangifera Indica (Var. “Dashehari”) unravels ripening associated genes. Sci. Rep. 2016, 6, 32557. [Google Scholar] [CrossRef]
- Fabi, J.P.; Broetto, S.G.; Silva, S.L.G.L.; Zhong, S.; Lajolo, F.M.; Nascimento, J.R.O.d. Analysis of papaya cell wall-related genes during fruit ripening indicates a central role of polygalacturonases during pulp softening. PLoS ONE 2014, 9, e105685. [Google Scholar] [CrossRef] [PubMed]
- Dautt-Castro, M.; López-Virgen, A.G.; Ochoa-Leyva, A.; Contreras-Vergara, C.A.; Sortillón-Sortillón, A.P.; Martínez-Téllez, M.A.; González-Aguilar, G.A.; Casas-Flores, J.S.; Sañudo-Barajas, A.; Kuhn, D.N.; et al. Genome-wide identification of mango (Mangifera indica L.) polygalacturonases: Expression analysis of family members and total enzyme activity during fruit ripening. Front. Plant Sci. 2019, 10, 969. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Awasthi, P.; Id, S.T. Comparative transcriptome analysis of unripe and ripe banana (cv. Nendran) unraveling genes involved in ripening and other related processes. PLoS ONE 2021, 16, e0254709. [Google Scholar] [CrossRef]
- Relacionados, A.; La, C.O.N.; Del, M.; Guayaba, F.D.E.; Silva, A.I.R.; Palenius, H.G.N.; Guzmán, G.H.; Solís, Á.G.A.; Garcidueñas, C.; Morales, J.F. Ripening-related cDNAs in guava fruit (Psidium guajava L.). characterization and expression analysis. Rev. Fitotec. Mex. 2013, 36, 117–125. [Google Scholar]
- Li, T.; Yun, Z.; Wu, Q.; Qu, H.; Duan, X.; Jiang, Y. Combination of transcriptomic, proteomic and metabolomic analysis reveals the ripening mechanism of banana pulp. Biomolecules 2019, 9, 523. [Google Scholar] [CrossRef]
- Yakir, E.; Zhangjun, F.; Sela, N.; Xu, Y.; Singh, V.; Dagar, A.; Joshi, J.R.; Müller, M.; Munné-bosch, S.; Giovannoni, J.J.; et al. MaMADS2 repression in banana fruits modifies hormone synthesis and signalling pathways prior to climacteric stage. BMC Plant Biol. 2018, 18, 267. [Google Scholar] [CrossRef]
- Song, C.; Shan, W.; Yang, Y.; Tan, X.; Fan, Z.; Chen, J.; Lu, W.; Kuang, J. BBA—Gene regulatory mechanisms heterodimerization of matcp proteins modulates the transcription of MaXTH10/11 genes during banana fruit ripening. BBA Gene Regul. Mech. 2018, 1861, 613–622. [Google Scholar]
- Shan, W.; Fan, Y.; Wei, G.; Jian, W.; Chen, Y.; Jin, W.; Bao, L.D.; Xin, Y.; Su, G.; Fei, J. Banana MaBZR1/2 associate with MaMPK14 to modulate cell wall modifying genes during fruit ripening. Plant Cell Rep. 2020, 39, 35–46. [Google Scholar] [CrossRef]
- Fan, Z.; Ba, L.; Shan, W.; Xiao, Y.; Lu, W.; Kuang, J.; Chen, J. A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. Plant J. 2018, 96, 1191–1205. [Google Scholar] [CrossRef]
- Wu, C.; Shan, W.; Liang, S.; Zhu, L.; Guo, Y.; Chen, J.; Lu, W.; Li, Q.; Page, S.D.S. MaMPK2 enhances MabZIP93 mediated transcriptional activation of cell wall modifying genes during banana fruit ripening. Plant Mol. Biol. 2019, 101, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.M.; Chen, S.C.; Liu, Z.l.; Shan, W.; Chen, J.Y.; Lu, W.J.; Lakshmanan, P.; Kuang, J.F. MabZIP74 interacts with MaMAPK11-3 to regulate the transcription of MaACO1/4 during banana fruit ripening. Postharvest Biol. Technol. 2020, 169, 111293. [Google Scholar] [CrossRef]
- Hu, W.; Wang, L.; Tie, W.; Yan, Y.; Ding, Z.; Liu, J.; Li, M.; Peng, M.; Xu, B.; Jin, Z. Genome-wide analyses of the bzip family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci. Rep. 2016, 6, 1–15. [Google Scholar]
- Feng, B.; Han, Y.; Xiao, Y.; Kuang, J.; Fan, Z.; Chen, J.; Lu, W. The banana fruit dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 2016, 67, 2263–2275. [Google Scholar] [CrossRef]
- Kuang, J.; Chen, J.; Liu, X.; Han, Y.; Xiao, Y.; Shan, W.; Tang, Y.; Wu, K.; He, J.; Lu, W.; et al. The transcriptional regulatory network mediated by banana (Musa acuminata) dehydration-responsive element binding (MaDREB) transcription factors in fruit ripening. New Phytol. 2017, 214, 762–781. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, Y.; Shan, W.; Cai, Y.; Chen, J.; Lu, W.; Kuang, J. Identification of two transcriptional activators MabZIP4/5 in controlling aroma biosynthetic genes during banana ripening. J. Agric. Food Chem. 2018, 66, 6142–6150. [Google Scholar] [CrossRef]
- Yan, H.; Wu, F.; Jiang, G.; Xiao, L.; Li, Z.; Duan, X. Postharvest biology and technology genome-wide identification, characterization and expression analysis of nf- y gene family in relation to fruit ripening in banana. Postharvest Biol. Technol. 2019, 151, 98–110. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, M.; Feng, J.; Wu, C.; Shan, W.; Kuang, J.; Chen, J.; Hu, Z.; Lu, W. Transcriptome analysis of low-temperature-affected ripening revealed myb transcription factors-mediated regulatory network in banana fruit. Food Res. Int. 2021, 148, 110616. [Google Scholar] [CrossRef]
- Id, B.P.; Pandey, A.; Id, B.W.; Id, R.S. The R2R3-MYB gene family in banana Musa. PLoS ONE 2020, 15, e0239275. [Google Scholar]
- Zhu, L.; Liang, S.; Chen, L.; Wu, C.; Wei, W.; Shan, W.; Chen, J.; Lu, W.; Su, X.; Kuang, J. Banana MaSPL16 modulates carotenoid biosynthesis during fruit ripening through activating the transcription of lycopene β cyclase genes. J. Agric. Food Chem. 2020, 68, 1286–1296. [Google Scholar] [CrossRef]
- Shan, W.E.I.; Chen, J.; Kuang, J.; Lu, W. Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis-related genes against Colletotrichum musae. Mol. Plant Pathol. 2015, 17, 330–338. [Google Scholar] [CrossRef]
- Lawson, T.; Lycett, G.W.; Mayes, S.; Ho, W.K.; Chin, C.F. Transcriptome-wide identification and characterization of the Rab GTPase family in mango. Mol. Biol. Rep. 2020, 47, 4183–4197. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Luo, C.; Huang, F.; Liu, Z.; An, Z.; Dong, L.; He, X. Identification and characterization of the mango EIF gene family reveals MieIF1A-a, which confers tolerance to salt stress in transgenic arabidopsis. Sci. Hortic. 2019, 248, 274–281. [Google Scholar] [CrossRef]
- Salih, H.; Tan, L.; Htet, N.N.W. Genome-Wide identification, characterization of bHLH transcription factors in mango. Trop. Plant Biol. 2021, 14, 72–81. [Google Scholar] [CrossRef]
- Tan, L.; Salih, H.; Htet, N.N.W.; Azeem, F.; Zhan, R. Genomic Analysis of WD40 protein family in the mango reveals a TTG1 protein enhances root growth and abiotic tolerance in arabidopsis. Sci. Rep. 2021, 11, 1–10. [Google Scholar]
- Figueroa-ya, L.; Pereira-santana, A.; Arroyo-herrera, A.; Rodriguez-corona, U.; Sanchez-teyer, F.; Espadas-alcocer, J.; Espadas-gil, F.; Barredo-pool, F.; Casta, E.; Rodriguez-zapata, C. RAP2. 4a is transported through the phloem to regulate cold and heat tolerance in papaya tree (Carica papaya cv. Maradol): Implications for protection against abiotic stress. PLoS ONE 2016, 11, e0165030. [Google Scholar] [CrossRef]
- Li, M.; Ren, L.; Zou, Z.; Hu, W.; Xiao, S.; Yang, X.; Ding, Z.; Yan, Y.; Tie, W.; Yang, J.; et al. Identification and Expression Analyses of the Special 14—3-3 Gene family in papaya and its involvement in fruit development, ripening, and abiotic stress responses. Biochem. Genet. 2021, 59, 1599–1616. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Chen, H.; Gao, H.; Wang, S.; Wang, N.; Jin, J.; Lu, Y.; Yu, Z.; Ma, Q.; Han, Y. Postharvest biology and technology papaya CpMADS4 and CpNAC3 co-operatively regulate ethylene signal genes CpERF9 and CpEIL5 during fruit ripening. Postharvest Biol. Technol. 2021, 175, 111485. [Google Scholar] [CrossRef]
- Fu, C.; Chen, H.; Gao, H.; Lu, Y. Two papaya MYB proteins function in fruit ripening by regulating some genes involved in cell-wall degradation and carotenoid biosynthesis. J. Sci. Food Agric. 2020, 100, 4442–4448. [Google Scholar] [CrossRef]
- Liu, K.; Yuan, C.; Li, H.; Lin, W.; Yang, Y.; Shen, C.; Zheng, X. Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica Papaya L.). BMC Genom. 2015, 16, 901. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, W.; Xie, R.; Xu, L.; Zhou, Y.; Li, H.; Yuan, C.; Zheng, X.; Xiao, L. CpARF2 and CpEIL1 interact to mediate auxin—Ethylene interaction and regulate fruit ripening in papaya. Plant J. 2020, 103, 1318–1337. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xu, H.; Wall, M.M.; Yang, J. Roles of transcription factor SQUAMOSA promoter binding protein-like gene family in papaya (Carica papaya) development and ripening. Genom. 2020, 112, 2734–2747. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Shen, Y.; Chen, L.; Ming, R. Papaya CpbHLH1/2 Regulate carotenoid biosynthesis-related genes during papaya fruit ripening. Hortic. Res. 2019, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Han, Y.; Fan, Z.; Chen, J.; Chen, W.; Lu, W. The papaya transcription factor cpnac1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during fruit ripening. J. Agric. Food Chem. 2016, 64, 5454–5463. [Google Scholar] [CrossRef]
- Fu, C.; Han, Y.; Kuang, J.; Chen, J.; Lu, W. Papaya CpEIN3a and CpNAC2 co-operatively regulate carotenoid biosynthesis-related genes cppds2/4, cplcy-e and CpCHY-b during fruit ripening. Plant Cell Physiol. 2017, 58, 2155–2165. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, C.; Yang, H.; Kuang, R.; Huang, B.; Wei, Y. Genome-wide analysis of basic helix-loop-helix transcription factors in papaya (Carica papaya L.). Peer J. 2020, 8, e9319. [Google Scholar] [CrossRef]
- Estrella-Maldonado, H.; Ramírez, A.G.; Ortiz, G.F.; Peraza-Echeverría, S.; Vega, O.M.; Góngora-Castillo, E.; Santamaría, J.M. Transcriptomic analysis reveals key transcription factors associated to drought tolerance in a wild papaya (Carica papaya) genotype. PLoS ONE 2021, 16, e0245855. [Google Scholar] [CrossRef] [PubMed]
- Moon, P.; Fu, Y.; Bai, J.; Plotto, A.; Crane, J.; Chambers, A. Assessment of fruit aroma for twenty-seven guava (Psidium guajava) accessions through three fruit developmental stages. Sci. Hortic. (Amsterdam) 2018, 238, 375–383. [Google Scholar] [CrossRef]
- Wang, L.; Hua, Q.; Ma, Y.; Hu, G.; Qin, Y. Comparative transcriptome analyses of a late-maturing mandarin mutant and its original cultivar reveals gene expression profiling associated with citrus fruit maturation. PeerJ 2017, 5, e3343. [Google Scholar] [CrossRef]
- Terol, J.; Nueda, M.J.; Ventimilla, D.; Tadeo, F.; Talon, M. transcriptomic analysis of Citrus clementina mandarin fruits maturation reveals a MADS- Box transcription factor that might be involved in the regulation of earliness. BMC Plant Biol. 2019, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Zeng, Y.; An, J.; Ye, J.; Xu, Q.; Deng, X. An integrative analysis of transcriptome and proteome provides new insights into carotenoid biosynthesis and regulation in sweet orange fruits. J. Proteom. 2012, 75, 2670–2684. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, Y.; Lin, Y.; Chen, Y.; Lu, X. Illumina® sequencing reveals candidate genes of carotenoid metabolism in three pummelo cultivars (Citrus maxima) with different pulp color. Int. J. Mol. Sci. 2019, 20, 2246. [Google Scholar] [CrossRef]
- Agusti, M.; Mesejo, C.; Mu, N.; Vera-sirera, F.; Lucas, M.D.; Reig, C.; Iglesias, D.J.; Primo-millo, E.; Miguel, A. Fruit-dependent epigenetic regulation of flowering in citrus. New Phytol. 2019, 225, 376–384. [Google Scholar] [CrossRef]
- Li, P.; Sun, L.; Zhou, H.; Zeng, R.; Ai, X.; Zhang, J.; Hu, C. HD-ZIP I transcription factor (pthb13) negatively regulates citrus flowering through binding to FLOWERING LOCUS C Promoter. Plants 2020, 9, 114. [Google Scholar]
- Wu, J.; Cao, J.; Su, M.; Feng, G.; Xu, Y.; Yi, H. Genome-wide comprehensive analysis of transcriptomes and small rnas offers insights into the molecular mechanism of alkaline stress tolerance in a citrus rootstock. Hortic. Res. 2019, 6, 1–19. [Google Scholar] [CrossRef]
- Xie, R.; Pan, X.; Zhang, J.; Ma, Y.; He, S.; Zheng, Y.; Ma, Y. effect of salt-stress on gene expression in citrus roots revealed by RNA-Seq. Funct. Integr. Genom. 2018, 18, 155–173. [Google Scholar] [CrossRef]
- Namita. Identification and Isolation of Genes Responsible for Increased Shelf Life in Guava (Psidium guajava L.). Master’s Degree, Punjab Agricultural University, Ludhiana, India, 2020.
- Chen, J.Y.; Xie, F.F.; Cui, Y.Z.; Chen, C.B.; Lu, W.J.; Hu, X.D.; Hua, Q.Z.; Zhao, J.; Wu, Z.J.; Gao, D.; et al. A chromosome-scale genome sequence of pitaya (Hylocereus Undatus) provides novel insights into the genome evolution and regulation of betalain biosynthesis. Hortic. Res. 2021, 8, 164. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Cheng, M.N.; Ba, L.J.; Zeng, R.X.; Luo, D.L.; Qin, Y.H.; Liu, Z.L.; Kuang, J.F.; Lu, W.J.; Chen, J.Y.; et al. Pitaya HpWRKY3 is associated with fruit sugar accumulation by transcriptionally modulating sucrose metabolic genes HpINV2 and HpSuSy1. Int. J. Mol. Sci. 2019, 20, 1890. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, J.; Han, X.; Qiao, G.; Yang, K.; Wen, Z.; Wen, X. Comparative transcriptome analysis combining SMRT and Illumina-based RNA-seq identifies potential candidate genes involved in betalain biosynthesis in pitaya fruit. Int. J. Mol. Sci. 2020, 21, 3288. [Google Scholar] [CrossRef]
- Xi, X.; Zong, Y.; Li, S.; Cao, D.; Sun, X.; Liu, B. Transcriptome analysis clarified genes involved in betalain biosynthesis in the fruit of red pitayas (Hylocereus costaricensis). Molecules. 2019, 24, 445. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Nong, Q.; Jian, S.; Lu, H.; Zhang, M.; Xia, K. An AP2/ERF Gene, HuERF1, from Pitaya (Hylocereus undatus) positively regulates salt tolerance. Int. J. Mol. Sci. 2020, 21, 4586. [Google Scholar] [CrossRef] [PubMed]
- Khaksar, G.; Sangchay, W.; Pinsorn, P.; Sangpong, L.; Sirikantaramas, S. Genome-wide analysis of the Dof gene family in durian reveals fruit ripening-associated and cultivar-dependent Dof transcription factors. Sci. Rep. 2019, 9, 12109. [Google Scholar] [CrossRef] [PubMed]
- Khaksar, G.; Sirikantaramas, S. Transcriptome-Wide Identification and expression profiling of the ERF gene family suggest roles as transcriptional activators and repressors of fruit ripening in durian. PLoS ONE 2021, 16, e0252367. [Google Scholar] [CrossRef]
- Khaksar, G.; Sirikantaramas, S. Auxin response factor 2A is part of the regulatory network mediating fruit ripening through auxin-ethylene crosstalk in durian. Front. Plant Sci. 2020, 11, 1393. [Google Scholar] [CrossRef]
- Dan, M.; Huang, M.; Liao, F.; Qin, R.; Liang, X.; Zhang, E.; Huang, M.; Huang, Z.; He, Q. Identification of ethylene responsive miRNAs and their targets from newly harvested banana fruits using high-throughput sequencing. J. Agric. Food Chem. 2018, 66, 10628–10639. [Google Scholar] [CrossRef]
- Sampangi-Ramaiah, M.H.; Ravishankar, K.V.; Ajitha, R. Long non-coding RNAs in banana: Prediction, mapping and their comparative studies using Musa balbisiana and Musa acuminata transcriptome. Trees 2019, 33, 359–369. [Google Scholar] [CrossRef]
- Lakhwani, D.; Pandey, S.A.; Sharma, D.; Asif, M.H.; Trivedi, P.K. Novel microRNAs regulating ripening associated processes in banana fruit. Plant Growth Regul. 2020, 90, 223–235. [Google Scholar] [CrossRef]
- Fu, C.; Han, Y.; Guo, Y.; Kuang, J.; Chen, J.; Shan, W.; Lu, W. Postharvest biology and technology differential expression of histone deacetylases during banana ripening and identification of MaHDA6 in regulating ripening-associated genes. Postharvest Biol. Technol. 2018, 141, 24–32. [Google Scholar] [CrossRef]
- Fu, C.; Chen, H.; Gao, H.; Han, Y. Histone deacetylase CpHDA3 is functionally associated with CpERF9 in suppression of CpPME1/2 and CpPG5 genes during papaya fruit ripening. J. Agric. Food Chem. 2019, 67, 8919–8925. [Google Scholar] [CrossRef]
- Liu, K.; Yuan, C.; Feng, S.; Zhong, S.; Li, H.; Zhong, J.; Shen, C. Genome-wide analysis and characterization of Aux / IAA family genes related to fruit ripening in papaya (Carica Papaya L.). BMC Genom. 2017, 18, 351. [Google Scholar] [CrossRef]
- Fabi, J.P.; Seymour, G.B.; Graham, N.S.; Broadley, M.R.; May, S.T.; Lajolo, F.M.; Cordenunsi, B.R.; Roberto, J. Analysis of ripening-related gene expression in papaya using an arabidopsis-based microarray. BMC Plant Biol. 2012, 12, 242. [Google Scholar] [CrossRef] [PubMed]
- Pinsorn, P.; Oikawa, A.; Watanabe, M.; Sasaki, R.; Ngamchuachit, P.; Hoefgen, R.; Saito, K.; Sirikantaramas, S. Metabolic Variation in the pulps of two durian cultivars: Unraveling the metabolites that contribute to the flavor. Food Chem. 2018, 268, 118–125. [Google Scholar] [CrossRef]
- Feng, G.; Wu, J.; Yi, H. Global tissue-specific transcriptome analysis of Citrus sinensis fruit across six developmental stages. Sci. Data 2019, 6, 153. [Google Scholar] [CrossRef]
- Wu, J.; Fu, L.; Yi, H. Genome-wide identification of the transcription factors involved in citrus fruit ripening from the transcriptomes of a late-ripening sweet orange mutant and its wild type. PLoS ONE 2016, 11, e0154330. [Google Scholar]
- Guo, D.L.; Xi, F.F.; Yu, Y.H.; Zhang, X.Y.; Zhang, G.H.; Zhong, G.Y. Comparative RNA-seq profiling of berry development between table grape “kyoho” and its early-ripening mutant “fengzao”. BMC Genom. 2016, 17, 795. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.-Y.; Liu, Z.-L.; Shu, Y.-S.; Wang, M.-L.; He, D.; Song, Z.-Q.; Zeng, H.-L.; Ning, Z.-C.; Lu, C.; Lu, A.-P.; et al. Chemotaxonomic classification applied to the identification of two closely-related citrus tcms using UPLC-Q-TOF-MS-based metabolomics. Molecules 2017, 22, 1721. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Pan, Z.; Wang, L.; Ding, Y.; Xu, Q.; Xiao, S.; Deng, X. Phosphoproteomic analysis of chromoplasts from sweet orange during fruit ripening. Physiol. Plant. 2014, 150, 252–270. [Google Scholar] [CrossRef]
- Arvind, K.; Sabana, A.A.; Rajesh, M.K. Computational identification of mirnas and their targets from mango (Magnifera indica L.) ests computational identification of mirnas and their targets from mango (Magnifera indica L.) ESTs. J. Appl. Biol. Biotechnol. 2018, 5, 53–58. [Google Scholar]
- Moh, N.M.M.; Zhang, P.; Chen, Y.; Chen, M. Computational identification of mirnas and temperature-responsive lncRNAs from mango (Mangifera indica L.). Front. Genet. 2021, 12, 1–25. [Google Scholar] [CrossRef]
- Li, W.; Zhu, X.G.; Zhang, Q.J.; Li, K.; Zhang, D.; Shi, C.; Gao, L.Z. SMRT sequencing generates the chromosome-scale reference genome of tropical fruit mango, Mangifera Indica. BioRxiv 2020. [Google Scholar] [CrossRef]
- Yadav, A.K.; Nigam, D.; Gautam, B.; Mishra, A.K. Computational approaches to decipher mirna-target association in mango (Mangifera indica L.). Plant Gene 2021, 27, 1–8. [Google Scholar] [CrossRef]
- Lee, W.S.; Gudimella, R.; Wong, G.R.; Tammi, M.T. Transcripts and micrornas responding to salt stress in Musa acuminata Colla (AAA Group) cv. berangan roots. PLoS ONE 2015, 10, e0127526. [Google Scholar] [CrossRef] [PubMed]
- Bi, F.; Meng, X.; Ma, C.; Yi, G. Identification of mirnas involved in fruit ripening in cavendish bananas by deep sequencing. BMC Genom. 2015, 16, 779. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cheng, C.; Lin, Y.; XuHan, X.; Lai, Z. Genome-wide identification and characterization of mrnas and lncrnas involved in cold stress in the wild banana (Musa itinerans). PLoS ONE 2018, 13, e0200002. [Google Scholar] [CrossRef]
- Cai, J.; Wu, Z.; Hao, Y.; Liu, Y.; Song, Z.; Chen, W.; Li, X.; Zhu, X. Small RNAs, degradome, and transcriptome sequencing provide insights into papaya fruit ripening regulated by 1-MCP. Foods. 2021, 10, 1643. [Google Scholar] [CrossRef]
- Sharma, A.; Ruiz-manriquez, L.M.; Serrano-cano, F.I.; Roxana, P.; Karina, C.; Alfaro, T.; Esmeralda, Y.; Karen, A.; Srivastava, A.; Paul, S. Identification of micrornas and their expression in leaf tissues of guava (Psidium guajava L.) under salinity stress. Agronomy 2020, 10, 1920. [Google Scholar] [CrossRef]
- Li, L.; Wu, H.; Ma, X.; Xu, W.; Liang, Q.; Zhan, R.; Wang, S. Genomics transcriptional mechanism of differential sugar accumulation in pulp of two contrasting mango (Mangifera Indica L.) cultivars. Genomics 2020, 112, 4505–4515. [Google Scholar] [CrossRef]
- Hu, L.; Wu, G.; Hao, C.; Yu, H.; Tan, L. Transcriptome and selected metabolite analyses reveal points of sugar metabolism in jackfruit (Artocarpus Heterophyllus Lam.). Plant Sci. 2016, 248, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Fuentealba, C.; Hernández, I.; Olaeta, J.A.; Defilippi, B.; Meneses, C.; Campos, R.; Lurie, S.; Carpentier, S.; Pedreschi, R. New insights into the heterogeneous ripening in hass avocado via LC–MS/MS proteomics. Postharvest Biol. Technol. 2017, 132, 51–61. [Google Scholar] [CrossRef]
- Gavicho Uarrota, V.; Fuentealba, C.; Hernández, I.; Defilippi-Bruzzone, B.; Meneses, C.; Campos-Vargas, R.; Lurie, S.; Hertog, M.; Carpentier, S.; Poblete-Echeverría, C.; et al. Integration of proteomics and metabolomics data of early and middle season hass avocados under heat treatment. Food Chem. 2019, 289, 512–521. [Google Scholar] [CrossRef]
- Charoensumran, P.; Pratumyot, K.; Vilaivan, T.; Praneenararat, T. Investigation of key chemical species from durian peduncles and their correlations with durian maturity. Sci. Rep. 2021, 11, 13301. [Google Scholar] [CrossRef]
- Deng, S.; Mai, Y.; Niu, J. Fruit characteristics, soluble sugar compositions and transcriptome analysis during the development of Citrus Maxima “Seedless”, and Identification of SUS and INV genes involved in sucrose degradation. Gene 2019, 689, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.B.; Anamika, K.; Jha, V.; Chidley, H.G.; Oak, P.S.; Kadoo, N.Y.; Pujari, K.H.; Giri, A.P.; Gupta, V.S. Transcriptional transitions in alphonso mango (Mangifera indica L.) during fruit development and ripening explain its distinct aroma and shelf life characteristics. Sci. Rep. 2017, 7, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Li, C.; Khoo, H.E.; Li, L.; He, X.; Yi, P.; Tang, Y.; Sun, J. Dynamic analyses of transcriptome and metabolic profiling: Revealing molecular insight of aroma synthesis of mango (Mangifera Indica L. Var. Tainong). Front. Plant Sci. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Cuevas-Glory, L.F.; Sauri-Duch, E.; Sosa-Moguel, O.; Pino, J.A. Characterization of odor-active compounds in mango ‘Ataulfo’ (Mangifera Indica L.) fruit. Chem. Pap. 2020, 74, 4025–4032. [Google Scholar] [CrossRef]
- Jaleel, W.; Li, Q.; Shi, Q.; Qi, G.; Latif, M.; Ali, S.; Yasin, N.A.; Lyu, L.; He, Y. Using GCMS to find out the volatile components in the aroma of three different commercial fruits in china. J. Anim. Plant Sci. 2021, 31, 166–174. [Google Scholar]
- Jiang, B.; Ou, S.; Xu, L.; Mai, W.; Ye, M.; Gu, H.; Zhang, T.; Yuan, C. Comparative proteomic analysis provides novel insights into the regulation mechanism underlying papaya (Carica papaya L.) exocarp during fruit ripening process. BMC Plant Biol. 2019, 19, 238. [Google Scholar] [CrossRef]
- Liu, R.; Du, Z.; Zhang, Y.; Shi, Y.; Chen, X.; Lin, L.; Xiong, Y.; Chen, M. Postharvest biology and technology volatile component quantification in combination with putative gene expression analysis reveal key players in aroma formation during fruit ripening in Carica papaya cv ‘Hong Fei’. Postharvest Biol. Technol. 2019, 158, 110987. [Google Scholar] [CrossRef]
- Zheng, J.; Meinhardt, L.W.; Goenaga, R.; Zhang, D.; Yin, Y. The Chromosome-level genome of dragon fruit reveals whole-genome duplication and chromosomal co-localization of betacyanin biosynthetic Genes. Hortic. Res. 2021, 8, 1–16. [Google Scholar] [CrossRef]
- Panpetch, P.; Sirikantaramas, S. Fruit ripening-associated leucylaminopeptidase with cysteinylglycine dipeptidase activity from durian suggests its involvement in glutathione recycling. BMC Plant Biol. 2021, 21, 69. [Google Scholar] [CrossRef] [PubMed]
- Sangpong, L.; Khaksar, G.; Pinsorn, P.; Oikawa, A.; Sasaki, R.; Erban, A.; Watanabe, M.; Wangpaiboon, K.; Tohge, T.; Kopka, J.; et al. Assessing dynamic changes of taste-related primary metabolism during ripening of durian pulp using metabolomic and transcriptomic analyses. Front. Plant Sci. 2021, 12, 687799. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.S.; Steinhaus, M. Identification of an important odorant precursor in durian: First evidence of ethionine in plants. J. Agric. Food Chem. 2020, 68, 10397–10402. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zheng, B.; Ma, Y.; Xu, W.; Wu, H.; Wang, S. Carotenoid accumulation and expression of carotenoid biosynthesis genes in mango flesh during fruit development and ripening. Sci. Hortic. (Amsterdam) 2018, 237, 201–206. [Google Scholar] [CrossRef]
- Yungyuen, W.; Thuong Vo, T.; Uthairatanakij, A.; Ma, G.; Zhang, L.; Tatmala, N.; Kaewsuksaeng, S.; Jitareerat, P.; Kato, M. Carotenoid accumulation and the expression of carotenoid metabolic genes in mango during fruit development and ripening. Appl. Sci. 2021, 11, 4249. [Google Scholar] [CrossRef]
- Palafox-Carlos, H.; Contreras-Vergara, C.A.; Muhlia-Almazán, A.; Islas-Osuna, M.A.; González-Aguilar, G.A. Expression and enzymatic activity of phenylalanine ammonia-lyase and p-Coumarate 3-Hydroxylase in mango (Mangifera Indica ’Ataulfo’) during Ripening. Genet. Mol. Res. 2014, 13, 3850–3858. [Google Scholar] [CrossRef]
- Karanjalker, G.R.; Ravishankar, K.V.; Shivashankara, K.S.; Dinesh, M.R.; Roy, T.K.; Sudhakar Rao, D.V. A study on the expression of genes involved in carotenoids and anthocyanins during ripening in fruit peel of green, yellow, and red colored mango cultivars. Appl. Biochem. Biotechnol. 2018, 184, 140–154. [Google Scholar] [CrossRef]
- Bajpai, A.; Khan, K.; Muthukumar, M.; Rajan, S.; Singh, N.K. Molecular analysis of anthocyanin biosynthesis pathway genes and their differential expression in mango peel. Genome 2018, 61, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Dars, A.G.; Liu, Q.; Xie, B.; Sun, Z. Phytochemical profiling of the ripening of chinese mango (Mangifera indica L.) cultivars by real-time monitoring using UPLC-ESI-QTOF-MS and its potential benefits as prebiotic ingredients. Food Chem. 2018, 256, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Marcillo-Parra, V.; Anaguano, M.; Molina, M.; Tupuna-Yerovi, D.S.; Ruales, J. Characterization and quantification of bioactive compounds and antioxidant activity in three different varieties of mango (Mangifera Indica L.) peel from the ecuadorian region using HPLC-UV/VIS and UPLC-PDA. NFS J. 2021, 23, 1–7. [Google Scholar] [CrossRef]
- Karanjalker, G.R.; Ravishankar, K.V.; Shivashankara, K.S.; Dinesh, M.R. Influence of bagging on color, anthocyanin and anthocyanin biosynthetic genes in peel of red colored mango cv. ‘Lily’. Erwerbs-Obstbau 2018, 60, 281–287. [Google Scholar] [CrossRef]
- Deng, S.; Cheng, C.; Liu, Z.; Chen, Y.; Zhang, Z.; Lin, Y.; Wang, T.; Lai, Z. Comparative transcriptome analysis reveals a role for anthocyanin biosynthesis genes in the formation of purple peel in minhou wild banana (Musa itinerans cheesman). J. Hortic. Sci. Biotechnol. 2018, 94, 184–200. [Google Scholar] [CrossRef]
- Zheng, X.; Zhu, K.; Sun, Q.; Zhang, W.; Wang, X.; Cao, H.; Tan, M.; Xie, Z.; Zeng, Y.; Ye, J.; et al. Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol. Plant 2019, 12, 1294–1307. [Google Scholar] [CrossRef]
- Guo, F.; Yu, H.; Xu, Q.; Deng, X. Transcriptomic analysis of differentially expressed genes in an orange-pericarp mutant and wild type in pummelo (Citrus grandis). BMC Plant Biol. 2015, 15, 44. [Google Scholar] [CrossRef]
- Ríos, G.; Naranjo, M.A.; Rodrigo, M.; Alós, E.; Zacarías, L.; Cercós, M.; Talón, M. Identification of a GCC transcription factor responding to fruit colour change events in citrus through the transcriptomic analyses of two mutants. BMC Plant Biol. 2010, 10, 276. [Google Scholar] [CrossRef]
- Shen, Y.H.; Yang, F.Y.; Lu, B.G.; Zhao, W.W.; Jiang, T.; Feng, L.; Chen, X.J.; Ming, R. Exploring the differential mechanisms of carotenoid biosynthesis in the yellow peel and red flesh of papaya. BMC Genom. 2019, 20, 1–11. [Google Scholar] [CrossRef]
- Gao, P.Y.X.Z.; Zhou, W.T.S.P. Cloning and expression analysis of phytoene desaturase and ζ-carotene desaturase genes in carica papaya. Mol. Biol. Rep. 2011, 38, 785–791. [Google Scholar]
- Chan-león, A.C.; Estrella-maldonado, H.; Dubé, P.; Fuentes, G.; Espadas-gil, F.; Talavera, C.; Ramírez, J.; Desjardins, Y.; Santamaría, J.M. The High content of β-carotene present in orange-pulp fruits of Carica Papaya L. is not correlated with a high expression of the CpLCY-β2 gene. Food Res. Int. 2017, 100, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Vetö, N.M.; Guzman, F.; Kulcheski, F.R.; Segatto, A.L.A.; Lacerda, M.E.G.; Margis, R.; Turchetto-Zolet, A.C. transcriptomics analysis of Psidium cattleyanum sabine (myrtaceae) unveil potential genes involved in fruit pigmentation. Genet. Mol. Biol. 2020, 43, e20190255. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.N.; Huang, Z.J.; Hua, Q.Z.; Shan, W.; Kuang, J.F.; Lu, W.J.; Qin, Y.H.; Chen, J.Y. The WRKY transcription factor HpWRKY44 regulates CytP450-like1 expression in red pitaya fruit (Hylocereus Polyrhizus). Hortic. Res. 2017, 4, 17039. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gao, H.; Ming, J.; Ding, Z.; Lin, X.; Zhan, R. Combined transcriptome and metabolome analysis of pitaya fruit unveiled the mechanisms underlying peel and pulp color formation. BMC Genom. 2020, 21, 734. [Google Scholar] [CrossRef] [PubMed]
- Qingzhu, H.; Chengjie, C.; Zhe, C.; Pengkun, C.; Yuewen, M.; Jingyu, W.; Jian, Z.; Guibing, H.; Jietang, Z.; Yonghua, Q. Transcriptomic analysis reveals key genes related to betalain biosynthesis in pulp coloration of Hylocereus polyrhizus. Front. Plant Sci. 2016, 6, 1179. [Google Scholar] [CrossRef] [PubMed]
- Al-Mekhlafi, N.A.; Mediani, A.; Ismail, N.H.; Abas, F.; Dymerski, T.; Lubinska-Szczygeł, M.; Vearasilp, S.; Gorinstein, S. Metabolomic and antioxidant properties of different varieties and origins of dragon fruit. Microchem. J. 2021, 160, 105687. [Google Scholar] [CrossRef]
- Lin, X.; Gao, H.; Ding, Z.; Zhan, R.; Zhou, Z.; Ming, J. Comparative metabolic profiling in pulp and peel of green and red pitayas (Hylocereus polyrhizus and Hylocereus undatus) reveals potential valorization in the pharmaceutical and food industries. Biomed Res. Int. 2021. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, J.; He, Y.; Shi, M.; Han, X.; Li, W.; Zhang, X.; Wen, X. Metabolic Profiling of Pitaya (Hylocereus polyrhizus) during Fruit Development and Maturation. Molecules 2019, 24, 1114. [Google Scholar] [CrossRef]
- Pucker, B.; Singh, H.B.; Kumari, M.; Khan, M.I.; Brockington, S.F. The report of anthocyanins in the betalain-pigmented genus hylocereus is not well evidenced and is not a strong basis to refute the mutual exclusion paradigm. BMC Plant Biol. 2021, 21, 4–9. [Google Scholar] [CrossRef]
- Wang, S.; Wu, H.; Xu, W.; Ma, X.; Li, L.; Zhan, R. Differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation in the pericarp of mango (Mangifera indica L.). IOP Conf. Ser. Mater. Sci. Eng. 2019, 569, 022036. [Google Scholar] [CrossRef]
- Fu, X.; Cheng, S.; Liao, Y.; Huang, B.; Du, B.; Zeng, W.; Jiang, Y.; Duan, X.; Yang, Z. Comparative analysis of pigments in red and yellow banana fruit. Food chem. 2017, 239, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Rahim, A.; Shazia, K.; Hee, A.; Jung, J.; Taek, H.; Jong, K.; Park, I.; Sup, I. Identification and characterization of carotenoid biosynthesis related genes in a novel dark skinned citrus mutant cultivar ‘Suneat’. Hortic. Environ. Biotechnol. 2020, 62, 99–111. [Google Scholar] [CrossRef]
- Youssef, M.; Ibrahim, R.A. Molecular markers associated with high vitamin-c content in guava. J. Agric. Chem. Biotechnol. 2016, 7, 49–55. [Google Scholar] [CrossRef][Green Version]
- Buah, S.; Mlalazi, B.; Khanna, H.; Dale, J.L.; Mortimer, C.L. The quest for golden bananas: Investigating carotenoid regulation in a fe’i group musa cultivar. J. Agric. Food Chem. 2016, 64, 3176–3185. [Google Scholar] [CrossRef] [PubMed]
- Wisutiamonkul, A.; Ampomah-Dwamena, C.; Allan, A.C.; Ketsa, S. Carotenoid Accumulation and Gene Expression during Durian (Durio zibethinus) Fruit Growth and Ripening. Sci. Hortic. 2017, 220, 233–242. [Google Scholar] [CrossRef]
- Huang, D.; Zhao, Y.; Cao, M.; Qiao, L.; Zheng, Z. Integrated systems biology analysis of transcriptomes reveals candidate genes for acidity control in developing fruits of sweet orange (Citrus Sinensis L. Osbeck). Front. Plant Sci. 2016, 7, 486. [Google Scholar] [CrossRef]
- Zinati, Z.; Sazegari, S.; Amin, H.; Tahmasebi, A. Mining transcriptome data to identify genes and pathways related to lemon taste using supervised and unsupervised data learning methods. Hortic. Environ. Biotechnol. 2021, 62, 593–603. [Google Scholar] [CrossRef]
- Mou, J.; Zhang, Z.; Qiu, H.; Lu, Y.; Zhu, X.; Fan, Z.; Zhang, Q.; Ye, J.; Fernie, A.R.; Cheng, Y.; et al. Multiomics-based dissection of citrus flavonoid metabolism using a Citrus reticulata × Poncirus trifoliata population. Hortic. Res. 2021, 8, 56. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef]
- Romero, P.; Romero, P. Abscisic acid deficiency alters epicuticular wax metabolism and morphology that leads to increased cuticle permeability during sweet orange (Citrus sinensis) Fruit Ripening. Front. Plant Sci. 2020, 11, 594184. [Google Scholar] [CrossRef]
- Wang, J.; Sun, L.; Xie, L.; He, Y.; Luo, T.; Sheng, L.; Luo, Y.; Zeng, Y.; Xu, J.; Deng, X.; et al. Plant science regulation of cuticle formation during fruit development and ripening in ‘Newhall’ navel orange (Citrus sinensis Osbeck) revealed by transcriptomic and metabolomic profiling. Plant Sci. 2016, 243, 131–144. [Google Scholar] [CrossRef]
- Ge, Y.; Cheng, Z.; Si, X.; Ma, W.; Tan, L.; Zang, X.; Wu, B.; Xu, Z.; Wang, N.; Zhou, Z.; et al. Transcriptome profiling provides insight into the genes in carotenoid biosynthesis during the mesocarp and seed developmental stages of avocado (Persea americana). Int. J. Mol. Sci. 2019, 20, 4117. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bhuj, B.D.; Singh, C.P. Alternate bearing in fruits trees: A review. Int. J. Curr. Microbiol. App. Sci. 2021, 10, 1218–1235. [Google Scholar] [CrossRef]
- Sharma, N.; Kumar, S.; Kumar, A.; Ravishankar, H.; Dubey, A.K.; Kumar, N. Physiological and molecular basis of alternate bearing in perennial fruit crops. Sci. Hortic. 2019, 243, 214–225. [Google Scholar] [CrossRef]
- Gottschalk, C.; Zhang, S.; Schwallier, P.; Rogers, S.; Bukovac, J.; Van Nocker, S. Genetic mechanisms associated with floral initiation and the repressive effect of fruit on flowering in apple (Malus x domestica Borkh). PLoS ONE 2021, 16, e0245487. [Google Scholar] [CrossRef]
- Fan, S.; Gao, X.; Gao, C.; Yang, Y.; Zhu, X.; Feng, W.; Li, R.; Tahir, M.M.; Zhang, D.; Han, M.; et al. Dynamic cytosine DNA methylation patterns associated with mRNA and siRNA expression profiles in alternate bearing apple trees. J. Agric. Food Chem. 2019, 67, 5250–5264. [Google Scholar] [CrossRef]
- Primo-millo, E. Fruit regulates seasonal expression of flowering genes in alternate-bearing ‘Moncada’ mandarin. Ann. Bot. 2011, 108, 511–519. [Google Scholar]
- Das, A.; Geetha, G.A.; Ravishankar, K.V.; Shivashankara, K.S.; Roy, T.K.; Dinesh, M.R. Interrelations of growth regulators, carbohydrates and expression of flowering genes (FT, LFY, AP1) in leaf and shoot apex of regular and alternate bearing mango (Mangifera Indica L.) cultivars during flowering. Sci. Hortic. 2019, 253, 263–269. [Google Scholar] [CrossRef]
- Sharma, N.; Singh, A.K.; Singh, S.K.; Mahato, A.K.; Srivastav, M.; Singh, N.K. Comparative RNA sequencing based transcriptome profiling of regular bearing and alternate bearing mango (Mangifera indica L.) varieties reveals novel insights into the regulatory mechanisms underlying alternate bearing. Biotechnol. Lett. 2020, 42, 1035–1050. [Google Scholar] [CrossRef]
- Choudhary, R.K.; Sahu, T.K.; Kumar, H. Computational identification of putative genes and vital amino acids involved in biennial rhythm in mango (Mangifera indica L.). J. Pharmacog. Phytochem. 2020, SP6, 267–272. [Google Scholar]
- Ding, Y.; Chang, J.; Ma, Q.; Chen, L.; Liu, S.; Jin, S.; Han, J. Network analysis of postharvest senescence process in citrus fruits revealed by transcriptomic and metabolomic profiling. Plant Physiol. 2015, 168, 357–376. [Google Scholar] [CrossRef] [PubMed]
- Luria, N.; Sela, N.; Yaari, M.; Feygenberg, O.; Kobiler, I.; Lers, A.; Prusky, D. De-novo assembly of mango fruit peel transcriptome reveals mechanisms of mango response to hot water treatment. BMC Genom. 2014, 15, 957. [Google Scholar] [CrossRef] [PubMed]
- Dautt-Castro, M.; Ochoa-Leyva, A.; Contreras-Vergara, C.A.; Muhlia-Almazán, A.; Rivera-Domínguez, M.; Casas-Flores, S.; Martinez-Tellez, M.A.; Sañudo-Barajas, A.; Osuna-Enciso, T.; Baez-Sañudo, M.A.; et al. Mesocarp RNA-Seq analysis of mango (Mangifera indica L.) identify quarantine postharvest treatment effects on gene expression. Sci. Hortic. 2018, 227, 146–153. [Google Scholar] [CrossRef]
- Hu, K.; Peng, D.; Wang, L.; Liu, H.; Xie, B.; Sun, Z. Effect of mild high hydrostatic pressure treatments on physiological and physicochemical characteristics and carotenoid biosynthesis in postharvest mango. Postharvest Biol. Technol. 2021, 172, 111381. [Google Scholar] [CrossRef]
- Brizzolara, S.; Manganaris, G.A.; Fotopoulos, V.; Watkins, C.B.; Tonutti, P. Primary metabolism in fresh fruits during storage. Front. Plant Sci. 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, Y.; Dong, C.; Terry, L.A.; Watkins, C.B.; Yu, Z.; Cheng, Z.-M. (Max). Meta-analysis of the effects of 1-Methylcyclopropene (1-MCP) treatment on climacteric fruit ripening. Hortic. Res. 2020, 7, 208. [Google Scholar]
- Zhu, X.; Song, Z.; Li, Q.; Li, J.; Chen, W.; Li, X. Physiological and transcriptomic analysis reveals the roles of 1-MCP in the ripening and fruit aroma quality of banana fruit (Fenjiao). Food Res. Int. 2020, 130, 108968. [Google Scholar] [CrossRef]
- Ma, Q.; Ding, Y.; Chang, J.; Sun, X.; Zhang, L.; Wei, Q.; Cheng, Y. Comprehensive insights on how 2,4-Dichlorophenoxyacetic acid retards senescence in post-harvest citrus fruits using transcriptomic and proteomic approaches. J. Exp. Bot. 2014, 65, 61–74. [Google Scholar] [CrossRef]
- Zhu, X.; Ye, L.; Ding, X.; Gao, Q.; Xiao, S.; Tan, Q.; Huang, J.; Chen, W.; Li, X. Transcriptomic analysis reveals key factors in fruit ripening and rubbery texture caused by 1-MCP in papaya. BMC Plant Biol. 2019, 19, 309. [Google Scholar] [CrossRef]
- Ding, X.; Zhu, X.; Ye, L.; Xiao, S.; Wu, Z.; Chen, W.; Li, X. The Interaction of CpEBF1 with CpMADSs is involved in cell wall degradation during papaya fruit ripening. Hortic. Res. 2019, 6, 13. [Google Scholar] [CrossRef]
- Ghedini, R.; Agopian, D.; Paulo, J.; Cordenunsi-lysenko, B.R. Metabolome and proteome of ethylene-treated papayas reveal different pathways to volatile compounds biosynthesis. Food Res. Int. 2020, 131, 108975. [Google Scholar]
- Chen, K.; Liu, T.; Liu, Y.; Wu, C. ‘Jen-Ju Bar’ Guava exhibited a non-climacteric ripening behavior resulting from a defect in the expression of system-2 ACC synthase PgACS1. ISHS Acta Hortic. 2017, 1166, 63–70. [Google Scholar] [CrossRef]
- Pang, X.; Zhao, S.; Zhang, M.; Cai, L.; Zhang, Y.; Li, X. Catechin gallate acts as a key metabolite induced by trypsin in Hylocereus undatus during storage indicated by omics. Plant Physiol. Biochem. 2021, 158, 497–507. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Yin, Y.; Yu, H.; Zhang, M.; Jing, H.; Ma, Y.; Xiong, X.; Pang, X. Transcriptomic analysis reveals key genes related to antioxidant mechanisms of Hylocereus undatus quality improvement by trypsin during storage. Food Funct. 2019, 10, 8116–8128. [Google Scholar] [CrossRef]
- Pang, X.; Li, X.; Liu, X.; Cai, L.; Li, B.; Li, X. Transcriptomic analysis reveals Cu/Zn SODs acting as hub genes of SODs in Hylocereus undatus induced by trypsin during storage. Antioxidants 2020, 9, 162. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Pang, X.; Yin, Y.; Yu, H.; Yuan, Y.; Li, B. Transcriptomic analysis reveals hub genes and subnetworks related to ros metabolism in Hylocereus undatus through novel superoxide scavenger trypsin treatment during storage. BMC Genom. 2020, 21, 437. [Google Scholar] [CrossRef]
- Li, X.; Li, B.; Guan, S.; Cai, L.; Xinyue, P. Hub genes and sub-networks of stoma-related genes in Hylocereus undatus through trypsin treatment during storage revealed by transcriptomic analysis. J. Food Biochem. 2021, 45, e13538. [Google Scholar] [CrossRef]
- Palapol, Y.; Kunyamee, S.; Thongkhum, M.; Ketsa, S.; Ferguson, I.B.; Van Doorn, W.G. Expression of expansin genes in the pulp and the dehiscence zone of ripening durian (Durio zibethinus) fruit. J. Plant Physiol. 2015, 182, 33–39. [Google Scholar] [CrossRef]
- Thongkum, M.; Imsabai, W.; Burns, P.; McAtee, P.A.; Schaffer, R.J.; Allan, A.C.; Ketsa, S. The effect of 1-Methylcyclopropene (1-MCP) on expression of ethylene receptor genes in durian pulp during ripening. Plant Physiol. Biochem. 2018, 125, 232–238. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhang, J.; Hou, X.-L.; Wang, F.; Xiong, A.-S. Transcriptomic, proteomic, metabolomic and functional genomic approaches for the study of abiotic stress in vegetable crops. Crit. Rev. Plant Sci. 2014, 33, 225–237. [Google Scholar] [CrossRef]
- Vasanthaiah, H.K.N.; Ravishankar, K.V.; Narayanaswamy, P.; Shivashankara, K.S. Influence of Temperature on spongy tissue formation in ‘Alphonso’ mango. Int. J. Fruit Sci. 2008, 8, 226–234. [Google Scholar] [CrossRef]
- Khanum, Z.; Tiznado-Hernández, M.E.; Ali, A.; Musharraf, S.G.; Shakeel, M.; Khan, I.A. Adaptation mechanism of mango fruit (Mangifera indica L. cv. Chaunsa White) to heat suggest modulation in several metabolic pathways. RSC Adv. 2020, 10, 35531–35544. [Google Scholar] [CrossRef]
- Sivankalyani, V.; Sela, N.; Feygenberg, O.; Zemach, H.; Maurer, D.; Alkan, N. Transcriptome dynamics in mango fruit peel reveals mechanisms of chilling stress. Front. Plant Sci. 2016, 7, 1–17. [Google Scholar] [CrossRef]
- Zhu, X.; Luo, J.; Li, Q.; Li, J.; Liu, T.; Wang, R.; Chen, W.; Li, X. Postharvest biology and technology low temperature storage reduces aroma-related volatiles production during shelf-life of banana fruit mainly by regulating key genes involved in volatile biosynthetic pathways. Postharvest Biol. Technol. 2018, 146, 68–78. [Google Scholar] [CrossRef]
- Yun, Z.; Jin, S.; Ding, Y.; Wang, Z.; Gao, H.; Pan, Z.; Xu, J.; Cheng, Y.; Deng, X. Comparative transcriptomics and proteomics analysis of citrus fruit, to improve understanding of the effect of low temperature on maintaining fruit quality during lengthy post-harvest storage. J. Exp. Bot. 2012, 63, 2873–2893. [Google Scholar] [CrossRef]
- Yungyuen, W.; Ma, G.; Zhang, L.; Futamura, M.; Tabuchi, M. Regulation of carotenoid metabolism in response to di ff erent temperatures in citrus juice sacs in vitro. Sci. Hortic. 2018, 238, 384–390. [Google Scholar] [CrossRef]
- Abouzaid, E.; Mohamed, E.A. Molecular analysis of drought tolerance in guava based on in vitro PEG evaluation. Trop. Plant Biol. 2016, 9, 73–81. [Google Scholar] [CrossRef]
- Gamboa-tuz, S.D.; Pereira-santana, A.; Zamora-, J.A.; Castano, E.; Espadas-gil, F.; Ayala-sumuano, J.T.; Keb-llanes, M.Á.; Sanchez-teyer, F.; Rodríguez-zapata, L.C. Transcriptomics and co-expression networks reveal tissue-specific responses and regulatory hubs under mild and severe drought in papaya (Carica papaya L.). Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Biradar, J. Molecular Characterisation of Root Specific Mapping Population of Mulberry by SSR Markers and Identification of QTLs Governing Drought Tolerance Traits. Master’s Thesis, University of Agricultural Sciences, Bengaluru, India, 2013. [Google Scholar]
- Fan, Q.J.; Yan, F.X.; Qiao, G.; Zhang, B.X.; Wen, X.P. Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus Undatus) by suppression subtractive hybridization and cDNA Microarray analysis. Gene 2014, 533, 322–331. [Google Scholar] [CrossRef]
- Nie, Q.; Gao, G.L.; Fan, Q.J.; Qiao, G.; Wen, X.P.; Liu, T.; Peng, Z.J.; Cai, Y.Q. Isolation and characterization of a catalase gene “HuCAT3” from Pitaya (Hylocereus Undatus) and its expression under abiotic stress. Gene 2015, 563, 63–71. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Z.; Mao, Y.; Wang, L.; Xiao, T.; Hu, Y.; Zhang, Y.; Ma, Y. Proteogenomic analysis of pitaya reveals cold stress-related molecular signature. Peer J. 2020, 2, 1–20. [Google Scholar] [CrossRef]
- Chaithanya, M.V.N.; Ramesh, S.; Dinesh, M.R.; Sailaja, D.; Aswath, C.; Chaithanya, M.V.N.; Ramesh, S.; Dinesh, M.R.; Sailaja, D.; Aswath, C. Developing mapping populations for identifying genomic regions controlling resistance to bark-eating caterpillar (indarbela tetraonis) in guava developing mapping populations for identifying genomic regions controlling resistance to bark-eating caterpillar. J. Crop Improv. 2016, 30, 371–377. [Google Scholar] [CrossRef]
- Santos, R.M.; Viana, A.P.; Santos, E.A.; Rodrigues, D.L.; Ricardo, P.; Santos, D.O.S. Genetic structuring of segregating populations of psidium spp resistant to the southern root-knot nematode by bayesian approach as basis for the guava breeding program. An. Acad. Bras. Cienc. 2020, 92, 1–18. [Google Scholar] [CrossRef]
- Noor Camellia, N.A.; Salma, I.; Mohd Norfaizal, G. Development of SCAR markers for rapid identification of resistance to phytophthora in durian using inter simple sequence repeat markers. Asian J. Adv. Basic Sci. 2019, 7, 30–34. [Google Scholar]
- Santoso, P.J.; Pancoro, A. Point mutation of ITS-NrDNA sequences as specific markers of three durian species: Durio zibethinus, Dkutejensis and D. lowianus. IOP Conf. Ser. Earth Environ. Sci. 2020, 482, 012020. [Google Scholar] [CrossRef]
- Kemal, R.A.; Sandjaja, E.B.L.; Santosa, A.P.; Ivan, J. Identification of Mildew Locus O (MLO) genes in Durio zibethinus genome corresponding with the powdery mildew disease. Biodiversitas 2018, 19, 2204–2212. [Google Scholar] [CrossRef]
- Zakaria, L. Diversity of colletotrichum species associated with anthracnose disease in tropical fruit crops—A review. Agriculture 2021, 11, 297. [Google Scholar] [CrossRef]
- Hong, K.; Gong, D.; Zhang, L.; Hu, H.; Jia, Z.; Gu, H.; Song, K. Transcriptome characterization and expression profiles of the related defense genes in postharvest mango fruit against Colletotrichum gloeosporioides. Gene 2016, 156, 275–283. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Straker, C.J. The isolation of high quality RNA from the fruit of avocado (Persea americana Mill.). S. Afr. J. Bot. 2012, 78, 44–46. [Google Scholar] [CrossRef]
- Xoca-Orozco, L.Á.; Cuellar-Torres, E.A.; González-Morales, S.; Gutiérrez-Martínez, P.; López-García, U.; Herrera-Estrella, L.; Vega-Arreguín, J.; Chacón-López, A. transcriptomic analysis of avocado hass (Persea americana Mill) in the interaction system fruit-chitosan-colletotrichum. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Wang, F.; Itai, R.N.; Nozoye, T.; Kobayashi, T.; Nishizawa, N.K.; Nakanishi, H. The bHLH protein OsIRO3 Is critical for plant survival and Iron (Fe) homeostasis in rice Oryza sativa L.) under Fe-deficient conditions. Soil Sci. Plant Nutr. 2020, 66, 579–592. [Google Scholar] [CrossRef]
- Miao, H.; Sun, P.; Liu, Q.; Miao, Y.; Liu, J.; Zhang, K.; Hu, W. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Tang, N.; Chen, N.; Hu, N.; Deng, W.; Chen, Z.; Li, Z. Postharvest biology and technology comparative metabolomics and transcriptomic pro fi ling reveal the mechanism of fruit quality deterioration and the resistance of citrus fruit against Penicillium digitatum. Postharvest Biol. Technol. 2018, 145, 61–73. [Google Scholar] [CrossRef]
- Li, Q.; Jia, R.; Dou, W.; Qi, J.; Qin, X.; Fu, Y.; He, Y.; Id, S.C. CsBZIP40, a bZIP transcription factor in sweet orange, plays a positive regulatory role in citrus bacterial canker response and tolerance. PLoS ONE 2019, 14, e0223498. [Google Scholar] [CrossRef]
- Deng, B.; Wang, W.; Deng, L.; Yao, S.; Ming, J.; Zeng, K. Comparative RNA-Seq analysis of citrus fruit in response to infection with three major postharvest fungi. Postharvest Biol. Technol. 2018, 146, 134–146. [Google Scholar] [CrossRef]
- Xu, M.; Liu, C.L.; Luo, J.; Qi, Z.; Yan, Z.; Fu, Y.; Wei, S.S.; Tang, H. Transcriptomic de Novo analysis of Pitaya (Hylocereus polyrhizus) canker disease caused by Neoscytalidium dimidiatum. BMC Genom. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Xu, M.; Liu, C.L.; Fu, Y.; Liao, Z.W.; Guo, P.Y.; Xiong, R.; Cheng, Y.; Wei, S.S.; Huang, J.Q.; Tang, H. Molecular characterization and expression analysis of pitaya (Hylocereus polyrhizus) HpLRR genes in response to Neoscytalidium dimidiatum infection. BMC Plant Biol. 2020, 20, 160. [Google Scholar] [CrossRef]
- Madronero, L.J.; Corredor-Rozo, Z.L.; Escobar-Perez, J.; Velandia-Romero, M.L. Next generation sequencing and proteomics in plant virology: How is colombia doing? Acta Biol. Colomb. 2019, 24, 423–438. [Google Scholar] [CrossRef]
- Fang, J.; Lin, A.; Qiu, W.; Cai, H.; Umar, M. Transcriptome profiling revealed stress-induced and disease resistance genes up-regulated in PRSV resistant transgenic papaya. Front. Plant Sci. 2016, 7, 1–20. [Google Scholar] [CrossRef]
- Jena, R.C.; Agarwal, K.; Chand, P.K. Fruit and leaf diversity of selected indian mangoes (Mangifera Indica L.). Sci. Hortic. 2021, 282, 109941. [Google Scholar] [CrossRef]
- Irish, B.M.; Cuevas, H.E.; Simpson, S.A.; Scheffler, B.E.; Sardos, J.; Ploetz, R.; Goenaga, R. Musa spp. germplasm management: Microsatellite fingerprinting of USDA-ARS national plant germplasm system collection. Crop Sci. 2014, 54, 2140–2151. [Google Scholar] [CrossRef]
- Razak, S.A.; Ariffin, M.A.T.; Mohamad, S.M.S.; Azman, N.H.E.N.; Hassan, M.A.; Sarip, J. Microsatellite markers for the molecular characterisation of potentially commercial mango (Mangifera Indica) progenies. Malays. Appl. Biol. 2020, 49, 81–85. [Google Scholar]
- Ravishankar, K.V.; Dinesh, M.R.; Nischita, P.; Sandya, B.S. Development and characterization of microsatellite markers in mango (Mangifera Indica) using next-generation sequencing technology and their transferability across species. Mol. Breed. 2015, 35, 1–13. [Google Scholar] [CrossRef]
- Srivastav, M.; Singh, S.K.; Prakash, J.; Singh, R.; Sharma, N.; Ramchandra, S.; Devi, R.; Gupta, A.; Mahto, A.K.; Jayaswal, P.K.; et al. New hyper-variable ssrs for diversity analysis in mango (Mangifera Indica L.). Indian J. Genet. Plant Breed. 2021, 81, 119–126. [Google Scholar] [CrossRef]
- Kuhn, D.N.; Dillon, N.; Bally, I.; Groh, A.; Rahaman, J.; Warschefsky, E.; Freeman, B.; Innes, D.; Chambers, A.H. Estimation of genetic diversity and relatedness in a mango germplasm collection using SNP markers and a simplified visual analysis Method. Sci. Hortic. 2019, 252, 156–168. [Google Scholar] [CrossRef]
- Rai, M.K.; Phulwaria, M.; Shekhawat, N.S. Transferability of simple sequence repeat (SSR) markers developed in guava (Psidium Guajava L.) to four myrtaceae species. Mol. Biol. Rep. 2013, 40, 5067–5071. [Google Scholar] [CrossRef]
- Kumar, C.; Kumar, R.; Singh, S.K.; Goswami, A.K.; Nagaraja, A.; Paliwal, R.; Singh, R. Development of novel g-SSR markers in guava (Psidium Guajava L.) cv. Allahabad Safeda and their application in genetic diversity, population structure and cross species transferability studies. PLoS ONE 2020, 15, e0237538. [Google Scholar] [CrossRef]
- Biswas, M.K.; Bagchi, M.; Biswas, D.; Harikrishna, J.A. Genome-wide novel genic microsatellite marker resource development and validation for genetic diversity and population structure analysis of banana. Genes 2020, 11, 1479. [Google Scholar] [CrossRef]
- Čížková, J.; Hřibová, E.; Christelová, P.; Houwe, I.v.d.; Häkkinen, M.; Roux, N.; Swennen, R.; Doležel, J. Molecular and cytogenetic characterization of wild musa species. PLoS ONE 2015, 10, e0134096. [Google Scholar]
- Amorim, E.P.; Vilarinhos, A.D.; Cohen, K.O.; Amorim, V.B.O.; Santos-Serejo, J.A.; Oliveira, S.; Pestana, K.N.; Santos, V.J.; Paes, N.S.; Monte, D.C.; et al. Genetic diversity of carotenoid-rich bananas evaluated by diversity arrays technology (DArT). Genet. Mol. Biol. 2009, 103, 96–103. [Google Scholar] [CrossRef]
- Devarajan, R.; Jayaraman, J.K.; Somasundaram, S.M.; Ragupathy, S.; Raman, P.; Sathiamoorthy, K.; Subbaraya, U. Genetic diversity in fresh fruit pulp mineral profile of 100 indian musa accessions. Food Chem. 2021, 361, 130080. [Google Scholar] [CrossRef]
- Vidal, N.M.; Grazziotin, A.L.; Christine, H.; Ramos, C.; Pereira, G.; Venancio, T.M. Development of a gene-centered SSR atlas as a resource for papaya (Carica papaya) marker-assisted selection and population genetic studies. PLoS ONE 2014, 9, e112654. [Google Scholar]
- Ahmed, B.; Ma, M.; Sa, H. Molecular characterization of guava (Psidium guajava L.) germplasm by RAPD Analysis. Int. J. Nat. Sci. 2011, 1, 62–67. [Google Scholar] [CrossRef]
- Sitther, V.; Zhang, D.; Harris, D.L.; Yadav, A.K.; Zee, F.T.; Meinhardt, L.W.; Dhekney, S.A. Genetic characterization of guava (Psidium guajava L.) germplasm in the united states using microsatellite markers. Genet. Resour. Crop Evol. 2014, 61, 829–839. [Google Scholar] [CrossRef]
- Latha, P.M.; Aswath, C.; Reddy, L.; Padmakar, B.; Dinesh, M.R. Cultivar identification and genetic fingerprinting of guava (Psidium guajava) using microsatellite markers. Int. J. Fruit Sci. 2011, 11, 184–196. [Google Scholar]
- Thaipong, K.; Promchot, S.; Auvuchanon, A. Genetic analysis of guava germplasm using AFLP Markers. Int. J. Agric. Technol. 2017, 13, 741–752. [Google Scholar]
- Feng, C.; Feng, C.; Lin, X.; Liu, S.; Li, Y.; Kang, M. A Chromosome-level genome assembly provides insights into ascorbic acid accumulation and fruit softening in guava (Psidium guajava). Hortic. Res. 2021, 19, 717–730. [Google Scholar] [CrossRef]
- Nakintu, J.; Albrecht, C.; Müller, C.M.; Kagoro-Rugunda, G.; Andama, M.; Olet, E.A.; Lejju, J.B.; Gemeinholzer, B. Exploring the genetic diversity of jackfruit (Artocarpus Heterophyllus Lam.) grown in uganda based on SSR Markers. Genet. Resour. Crop Evol. 2020, 67, 605–619. [Google Scholar] [CrossRef]
- Sahu, S.K.; Liu, M.; Yssel, A.; Kariba, R.; Muthemba, S.; Jiang, S.; Song, B.; Hendre, P.S.; Muchugi, A.; Jamnadass, R.; et al. Draft genomes of two artocarpus plants, jackfruit (A. heterophyllus) and breadfruit (A. altilis). Genes 2020, 11, 27. [Google Scholar] [CrossRef]
- Singh, N.K. Origin, Diversity and Genome Sequence of Mango (Mangifera Indica L.). Indian J. Hist. Sci. 2016, 51, 355–368. [Google Scholar] [CrossRef]
- Wang, P.; Luo, Y.; Huang, J.; Gao, S.; Zhu, G.; Dang, Z.; Gai, J.; Yang, M.; Zhu, M.; Zhang, H.; et al. The Genome evolution and domestication of tropical fruit mango. Genome Biol. 2020, 21, 1–17. [Google Scholar] [CrossRef]
- Bally, I.S.E.; Bombarely, A.; Chambers, A.H.; Cohen, Y.; Dillon, N.L.; Innes, D.J.; Islas-Osuna, M.A.; Kuhn, D.N.; Mueller, L.A.; Ophir, R.; et al. The ‘Tommy Atkins’ mango genome reveals candidate genes for fruit quality. BMC Plant Biol. 2021, 21, 108. [Google Scholar] [CrossRef]
- Hont, D.; Denoeud, F.; Aury, J.; Baurens, F.; Carreel, F.; Garsmeur, O.; Rouard, M.; Da Silva, C.; Jabbari, K.; Noel, B.; et al. The Banana (Musa acuminata) Genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar]
- Davey, M.W.; Gudimella, R.; Harikrishna, J.A.; Sin, L.W.; Khalid, N.; Keulemans, J. A Draft musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific musa hybrids. BMC Genom. 2013, 14, 683. [Google Scholar] [CrossRef]
- Wu, W.; Yang, Y.; He, W.; Rouard, M.; Li, W.; Xu, M. Whole genome sequencing of a banana wild relative musa itinerans provides insights into lineage- specific diversification of the musa genus. Sci. Rep. 2016, 6, 31586. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zheng, X.; Huang, Y.; Ye, J.; Chen, P.; Zhang, C.; Zhao, F.; Xie, Z. Genome sequencing and CRISPR/Cas9 gene editing of an early flowering mini-citrus (Fortunella hindsii). Plant Biotechnol. 2019, 17, 2199–2210. [Google Scholar] [CrossRef]
- Shimizu, T.; Tanizawa, Y.; Mochizuki, T.; Nagasaki, H. Draft sequencing of the heterozygous diploid genome of satsuma (Citrus unshiu Marc.) using a hybrid assembly approach. Front. Genet. 2017, 8, 180. [Google Scholar] [CrossRef]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, F.; Huang, Y.; He, J.; Yang, S.; Zeng, J.; Deng, C.; Jiang, X.; Fang, Y.; Wen, S.; et al. Genome of wild mandarin and domestication history of mandarin. Mol. Plant 2018, 11, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Zhang, S.; Cao, L.; Huang, Y.; Cheng, J.; Wu, G.; Tian, S.; Chen, C.; Liu, Y.; et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017, 49, 765–772. [Google Scholar] [CrossRef]
- Ming, R.; Hou, S.; Feng, Y.; Yu, Q.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.T.; et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya L.). Nature 2008, 452, 991–996. [Google Scholar] [CrossRef]
- Teh, B.T.; Lim, K.; Yong, C.H.; Ng, C.C.Y.; Rao, S.R.; Rajasegaran, V.; Lim, W.K.; Ong, C.K.; Chan, K.; Cheng, V.K.Y.; et al. The Draft genome of tropical fruit durian (Durio Zibethinus). Nat. Genet. 2017, 49, 1633–1641. [Google Scholar] [CrossRef]
- Abirami, K.; Swain, S.; Baskaran, V.; Venkatesan, K.; Sakthivel, K.; Bommayasamy, N. Distinguishing three dragon fruit (Hylocereus Spp.) species grown in andaman and nicobar islands of india using morphological, biochemical and molecular traits. Sci. Rep. 2021, 11, 2894. [Google Scholar] [CrossRef]
- Tao, J.; Qiao, G.; Wen, X.P.; Gao, G.L.; Liu, T.; Peng, Z.J.; Cai, Y.Q.; Chen, N.; Yan, F.X.; Zhang, B.X. Characterization of genetic relationship of dragon fruit accessions (Hylocereus spp.) by morphological traits and ISSR Markers. Sci. Hortic. 2014, 170, 82–88. [Google Scholar] [CrossRef]
- Pan, L.; Fu, J.; Zhang, R.; Qin, Y.; Lu, F.; Jia, L.; Hu, Q.; Liu, C.; Huang, L.; Liang, G. Genetic diversity among germplasms of pitaya based on SSR markers. Sci. Hortic. 2017, 225, 171–176. [Google Scholar] [CrossRef]
- Songnuan, W.; Pichakum, A.; Traiperm, P.; Rungjangsuwan, E.; Siriwattanakul, U. Diversity of durian (Durio zibethinus L.) from Nonthaburi, Thailand based on morpho-palatability characteristics and simple sequence repeat markers. Agric. Nat. Resour. 2019, 53, 218–227. [Google Scholar]
- Shearman, J.R.; Sonthirod, C.; Naktang, C.; Sangsrakru, D.; Yoocha, T.; Chatbanyong, R.; Vorakuldumrongchai, S.; Chusri, O.; Tangphatsornruang, S.; Pootakham, W. Assembly of the durian chloroplast genome using long PacBio Reads. Sci. Rep. 2020, 10, 15980. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.H.; Jo, S.; Kim, H.W.; Kim, Y.K.; Sohn, J.Y.; Kim, K.J. The complete plastome sequence of durian, Durio Zibethinus L. (Malvaceae). Mitochondrial DNA Part B Resour. 2017, 2, 763–764. [Google Scholar] [CrossRef] [PubMed]
- Blas, A.L.; Ming, R.; Liu, Z.; Veatch, O.J.; Paull, R.E.; Moore, P.H.; Yu, Q. Cloning of the Papaya Chromoplast-Specific Lycopene β-Cyclase, CpCYC-b, Controlling Fruit Flesh Color Reveals Conserved Microsynteny and a Recombination Hot Spot. Plant Physiol. 2010, 152, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- Gomes, V.M.; Ribeiro, R.M.; Viana, A.P. Inheritance of resistance to meloidogyne enterolobii and individual selection in segregating populations of Psidium spp. Eur. J. Plant Pathol. 2016, 148, 699–708. [Google Scholar] [CrossRef]
- Mulagund, J.; Dinesh, M.R.; Vasugi, C.; Rekha, A.; Ravishankar, K.V. Studies on sexual compatibility and or incompatibility among psidium species and their hybridity confirmation through SSR markers. Isr. J. Plant Sci. 2021, 1, 1–14. [Google Scholar] [CrossRef]
- Machado, R.M.; de Oliveira, F.A.; de Matos Alves, F.; de Souza, A.P.; Forni-Martins, E.R. Population genetics of polyploid complex psidium cattleyanum sabine (myrtaceae): Preliminary analyses based on new species-specific microsatellite loci and extension to other species of the genus. Biochem. Genet. 2020, 59, 219–234. [Google Scholar] [CrossRef]
- Grossi, L.L.; Fernandes, M.; Silva, M.A.; de Oliveira Bernardes, C.; Tuler, A.C.; dos Santos, P.H.D.; Ferreira, A.; da Silva Ferreira, M.F. DArTseq-derived SNPs for the genus psidium reveal the high diversity of native species. Tree Genet. Genomes 2021, 17, 1–13. [Google Scholar] [CrossRef]
- Rodríguez, N.; Becker, D.; Velázquez, B.; González, G.; Sourd, D. Characterization of guava accessions by ssr markers, extension of the molecular linkage map and mapping of qtls for vegetative and reproductive characters. Acta Hort. 2007, 735, 201–216. [Google Scholar] [CrossRef]
- Padmakar, B.; Kanupriya, C.; Latha, P.M.; Vasugi, C.; Dinesh, M.R.; Sailaja, D.; Aswath, C. Enrichment of genetic linkage maps and mapping qtls specific to seed strength—Hardness/softness—In guava (Psidium guajava L.). Sci. Hortic. 2016, 11, 13–20. [Google Scholar]
- ZhiGuo, D.; YeYuan, C. Construction of a genetic linkage map of mango based on SRAP, AFLP and ISSR markers. Agric. Biotechnol. 2017, 6, 9–16. [Google Scholar]
- Luo, C.; Shu, B.; Yao, Q.; Wu, H.; Xu, W.; Wang, S. Construction of a high-density genetic map based on large-scale marker development in mango using specific-locus amplified fragment sequencing (SLAF-Seq). Front. Plant Sci. 2016, 7, 1310. [Google Scholar] [CrossRef]
- Nantawan, U.; Kanchana-udomkan, C.; Bar, I.; Ford, R. Linkage mapping and quantitative trait loci analysis of sweetness and other fruit quality traits in papaya. BMC Plant Biol. 2019, 19, 449. [Google Scholar] [CrossRef] [PubMed]
- Bohry, D.; Christine, H.; Ramos, C.; Henrique, P.; Santana, M.; Boechat, B.; Abreu, F.; Arêdes, S. Discovery of SNPs and InDels in papaya genotypes and its potential for marker assisted selection of fruit quality traits. Sci. Rep. 2021, 11, 292. [Google Scholar] [CrossRef]
- Imai, A. Quantitative Trait Locus (QTL) Analysis of fruit-quality traits for mandarin breeding in Japan. PLoS ONE 2017, 14, e0221880. [Google Scholar] [CrossRef]
- Curtolo, M.; Cristofani-yaly, M.; Gazaffi, R.; Takita, M.A.; Figueira, A.; Machado, M.A. QTL Mapping for fruit quality in citrus using DArTseq markers. BMC Genom. 2017, 18, 1–16. [Google Scholar] [CrossRef]
- Fujii, H.; Nonaka, K.; Minamikawa, M.F.; Endo, T. Allelic composition of carotenoid metabolic genes in 13 founders influences carotenoid composition in juice sac tissues of fruits among japanese citrus breeding population. PLoS ONE 2021, 16, e0246468. [Google Scholar] [CrossRef]
- Rubinstein, M.; Eshed, R.; Rozen, A.; Zviran, T.; Kuhn, D.N.; Irihimovitch, V.; Sherman, A.; Ophir, R. Genetic diversity of avocado (Persea americana Mill.) germplasm using pooled sequencing. BMC Genom. 2019, 20, 1–19. [Google Scholar] [CrossRef]
- Kuhn, D.N.; Livingstone, D.S.; Richards, J.H.; Manosalva, P.; Van den Berg, N.; Chambers, A.H. Application of genomic tools to avocado (Persea americana) breeding: SNP discovery for genotyping and germplasm characterization. Sci. Hortic. 2019, 246, 1–11. [Google Scholar] [CrossRef]
- Minamikawa, M.F.; Takada, N.; Terakami, S.; Saito, T.; Onogi, A. Genome-wide association study and genomic prediction using parental and breeding populations of Japanese pear (Pyrus pyrifolia Nakai). Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Font, C.; Guajardo, V.; Chin-wo, S.R. Association mapping analysis for fruit quality traits in Prunus persica using SNP markers. Front. Plant Sci. 2019, 9, 1–12. [Google Scholar]
- Salazar, J.A.; Pacheco, I.; Shinya, P.; Zapata, P.; Silva, C.; Aradhya, M.; Velasco, D.; Ruiz, D.; Martínez-gómez, P. Genotyping by sequencing for SNP-based linkage analysis and identification of QTLs linked to fruit quality traits in japanese plum (Prunus salicina Lindl.). Front. Plant Sci. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias-pazaran, G.; Schlautman, B.; Diaz-garcia, L. Multivariate GBLUP improves accuracy of genomic selection for yield and fruit weight in biparental populations of vaccinium macrocarpon ait. Front. Plant Sci. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Connor, K.O. Prospects for increasing yield in macadamia using component traits and Genom. Tree Genet. Genomes 2018, 14, 1–14. [Google Scholar]
- Larsen, B.; Migicovsky, Z.; Jeppesen, A.A.; Gardner, K.M.; Toldam-, T.B.; Myles, S.; Orgaard, M.; Petersen, M.A.; Pedersen, C. Genome-wide association studies in apple reveal loci for aroma volatiles, sugar composition, and harvest Date. TPG 2019, 12, 180104. [Google Scholar] [CrossRef]
- Henry, R. Genome-wide association mapping of flowering and ripening periods in apple. Front. Plant Sci. 2017, 8, 1923. [Google Scholar]
- Campos, C.R. Marker Assisted Selection, Fine Mapping and Identification of Candidate Genes for Three Major Traits of Prunus persica L. (Batsh). Master’s Thesis, Universidade do Porto, Porto, Portugal, 2020. [Google Scholar]
- Zhang, Y.; López-Girona, E.; Aranzana, M.J. Region-wide association analysis and high-throughput resequencing strategies in peach to develop molecular markers for flat fruit marker-assisted selection. IV Int. Symp. Mol. Mark. Hortic. 2017, 1203, 79–84. [Google Scholar] [CrossRef]
- Biscarini, F.; Nazzicari, N.; Bink, M.; Arús, P.; Aranzana, M.J.; Verde, I.; Micali, S.; Pascal, T.; Quilot-Turion, B.; Lambert, P.; et al. Genome-enabled predictions for fruit weight and quality from repeated records in European peach progenies. BMC Genom. 2017, 18, 432. [Google Scholar] [CrossRef]
- Marimon de María, N. Towards an Integrated Control of Peach Powdery Mildew (Podosphaera pannosa) through the Application of Molecular Tools in Epidemiological and Genetic Resistance Studies. Master’s Thesis, Universitat de Lleida, Lleida, Spain, 2020. [Google Scholar]
- Lu, Z.; Pan, L.; Wei, B.; Niu, L.; Cui, G.; Wang, L.; Zeng, W.; Wang, Z. Fine mapping of the gene controlling the fruit skin hairiness of Prunus persica and its uses for MAS in progenies. Plants. 2021, 10, 1433. [Google Scholar] [CrossRef]
- Laurens, F.; Aranzana, M.J.; Arus, P.; Bassi, D.; Bink, M.; Bonany, J.; Caprera, A.; Corelli-Grappadelli, L.; Costes, E.; Durel, C.E.; et al. An integrated approach for increasing breeding efficiency in apple and peach in Europe. Hortic. Res. 2018, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Fujii, H.; Omura, M.; Shimada, T. Fast-track breeding system to introduce CTV resistance of trifoliate orange into citrus germplasm, by integrating early flowering transgenic plants with marker-assisted selection. BMC Plant Biol. 2020, 20, 224. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Norelli, J.L.; Howard, N.P.; Wisniewski, M.; Flachowsky, H.; Hanke, M.V.; Peace, C. Introgressing blue mold resistance into elite apple germplasm by rapid cycle breeding and foreground and background DNA-informed selection. Tree Genet. Genomes 2020, 16, 28. [Google Scholar] [CrossRef]
- Minamikawa, M.F.; Nonaka, K.; Kaminuma, E.; Kajiya-kanegae, H.; Goto, S.; Yoshioka, T.; Imai, A.; Hamada, H.; Hayashi, T. Genome-wide association study and genomic prediction in citrus: Potential of genomics-assisted breeding for fruit quality traits. Sci. Rep. 2017, 7, 4721. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Bai, J.; Chen, C.; Plotto, A.; Yu, Q.; Baldwin, E.A.; Gmitter, F.G. Identification of QTLs controlling aroma volatiles using a “Fortune” × “Murcott” (Citrus reticulata) population. BMC Genom. 2017, 18, 646. [Google Scholar] [CrossRef]
- Zhu, P.; Gu, B.; Li, P.; Shu, X.; Zhang, X.; Zhang, J. New cold-resistant, seedless grapes developed using embryo rescue and marker-assisted selection. Plant Cell Tissue Organ Culture 2020, 140, 551–562. [Google Scholar] [CrossRef]
- Sardos, J.; Perrier, X.; Dole, J. DArT Whole Genome Profiling Provides Insights on the Evolution and Taxonomy of Edible Banana (Musa spp.). Ann. Bot. 2016, 118, 1269–1278. [Google Scholar] [CrossRef]
- Pandey, A.; Alok, A.; Lakhwani, D.; Singh, J.; Asif, M.H. Genome-wide expression analysis and metabolite profiling elucidate transcriptional regulation of flavonoid biosynthesis and modulation under abiotic stresses in banana. Nat. Publ. Gr. 2016, 6, 31361. [Google Scholar] [CrossRef] [PubMed]
- Nyine, M.; Uwimana, B.; Akech, V.; Brown, A.; Ortiz, R.; Doležel, J.; Lorenzen, J.; Swennen, R. Association genetics of bunch weight and its component traits in east african highland banana (Musa spp. AAA Group). Theor. Appl. Genet. 2019, 132, 3295–3308. [Google Scholar] [CrossRef]
- Nyine, M.; Uwimana, B.; Blavet, N.; Hřibová, E.; Vanrespaille, H.; Batte, M.; Akech, V.; Brown, A.; Lorenzen, J.; Swennen, R.; et al. Genomic prediction in a multiploid crop: Genotype by environment interaction and allele dosage effects on predictive ability in banana. Plant Genome 2018, 11, 170090. [Google Scholar] [CrossRef] [PubMed]
- Wannemuehler, S.D.; Luby, J.J.; Yue, C.; Bedford, D.S.; Gallardo, R.K.; McCracken, V.A. A cost–benefit analysis of DNA informed apple breeding. Hort. Sci. 2019, 54, 1998–2004. [Google Scholar] [CrossRef]
- Wannemuehler, S.D.; Yue, C.; Shane, W.W.; Gallardo, R.K.; McCracken, V. Estimated implementation costs of DNA-informed breeding in a peach breeding program. Hort. Technol. 2020, 30, 356–364. [Google Scholar] [CrossRef]
- Coletta, R.D.; Qiu, Y.; Ou, S.; Hufford, M.B.; Hirsch, C.N. How the pan-genome is changing crop genomics and improvement. Genome Biol. 2021, 22, 3. [Google Scholar] [CrossRef]
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D. Super-Pangenome by integrating the wild side of a species for accelerated crop improvement trends in plant science. Trends Plant Sci. 2020, 25, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Tahir ul Qamar, M.; Zhu, X.; Khan, M.S.; Xing, F.; Chen, L.L. Pan-genome: A promising resource for noncoding RNA discovery in plants. Plant Genome 2020, 13, e20046. [Google Scholar] [CrossRef]
- Rijzaani, H.; Batley, J.; Edwards, D.; Rouard, M.; Doležel. The Pangenome of banana highlights differences between genera and genomes. Plant Genome 2021, e20100. [Google Scholar]
- Sun, X.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.; Duan, N.; Khan, A.; Ban, S.; Xu, K.; Cheng, L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef]
- Dale, J.; Paul, J.; Dugdale, B.; Harding, R. Modifying Bananas: From transgenics to organics? Sustainability 2017, 9, 333. [Google Scholar] [CrossRef]
- Kumar, G.B.S.; Srinivas, L.; Ganapathi, T.R. Iron fortification of banana by the expression of soybean ferritin. Biol. Trace Elem. Res. 2010, 142, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.; Khanna, H.; Kleidon, J.; Hoang, P.; Geijskes, J.; Daniells, J.; Zaplin, E.; Rosenberg, Y.; James, A.; Mlalazi, B.; et al. Golden bananas in the field: Elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol. J. 2017, 15, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Vishnevetsky, J.; White, T.L.; Palmateer, A.J.; Flaishman, M.; Cohen, Y.; Elad, Y.; Velcheva, M.; Hanania, U.; Sahar, N.; Dgani, O.; et al. Improved tolerance toward fungal diseases in transgenic cavendish banana (Musa spp. AAA Group) Cv. Grand Nain. Transgenic Res. 2011, 20, 61–72. [Google Scholar] [CrossRef]
- Dash, P.K.; Rai, R.; Wang, Y. Translating the “ Banana Genome ” to delineate stress resistance, dwarfing, parthenocarpy and mechanisms of fruit ripening. Front. Plant Sci. 2016, 7, 1543. [Google Scholar] [CrossRef]
- Castillo, X.A.O.; Fermin, G.; Tabima, J.; Rojas, Y.; Tennant, P.F.; Fuchs, M.; Sierra, R.; Bernal, A.J.; Restrepo, S. Phylogeography and molecular epidemiology of papaya ringspot virus. Virus Res. 2011, 159, 132–140. [Google Scholar] [CrossRef]
- Jia, R.; Zhao, H.; Huan, J.; Kong, H.; Zhang, Y.; Guo, J. Use of RNAi technology to develop a PRSV-resistant transgenic papaya. Sci. Rep. 2017, 7, 1–9. [Google Scholar]
- Gonsalves, D.; Suzuki, J.Y.; Tripathi, S.; Ferreira, S.A. Papaya Ringspot Virus (Potyviridae). Encycl. Virol. 2008, 4, 1–8. [Google Scholar]
- Yabor, L.; Pérez, L.; Gómez, D.; Villalobos-Olivera, A.; Mendoza, J.R.; Martínez, J.; Escalante, D.; Garro, G.; Hajari, E.; Lorenzo, J.C. Histological evaluation of pineapple transgenic plants following 8 years of field growth. Euphytica 2020, 216, 23. [Google Scholar] [CrossRef]
- Stowe, E.; Dhingra, A. Development of the Arctic® Apple. Plant Breed. Rev. 2021, 44, 273–296. [Google Scholar]
- Firoozabady, E.; Young, T.R. Pineapple plant named ‘Rosé’. Patent No.. USPP25763P3, 5 August 2015. [Google Scholar]
- Tricoli, D.M. Field evaluation of transgenic squash containing single or multiple virus coat protein gene constructs for resistance to cucumber mosaic virus, watermelon mosaic virus 2, and zucchini yellow mosaic virus. Nat. Biotechnol. 1995, 13, 1458–1465. [Google Scholar] [CrossRef]
- Elitzur, T.; Yakir, E.; Quansah, L.; Zhangjun, F.; Vrebalov, J.; Khayat, E.; Giovannoni, J.J.; Friedman, H. Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security. Plant Physiol. 2016, 171, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Sheng, O.; Deng, G.; He, W.; Dong, T.; Yang, Q.; Dou, T. CRISPR / Cas9-Mediated Genome Editing of MaACO1 the shelf life of banana fruit. Plant Biotechnol. J. 2021, 1, 654–656. [Google Scholar] [CrossRef]
- Kaur, N.; Alok, A.; Kumar, P.; Kaur, N.; Awasthi, P.; Chaturvedi, S.; Pandey, P.; Pandey, A.; Pandey, A.K.; Tiwari, S. CRISPR / Cas9 Directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana Fruit. Metab. Eng. 2020, 59, 76–86. [Google Scholar] [CrossRef]
- Green, J.C.; Hu, J.S. Editing plants for virus resistance using CRISPR-Cas. Acta Virol. 2017, 61, 138–142. [Google Scholar] [CrossRef][Green Version]
- Jia, H.; Xu, J.; Orbović, V.; Zhang, Y.; Wang, N. Editing citrus genome via SaCas9/SgRNA system. Front. Plant Sci. 2017, 8, 2135. [Google Scholar] [CrossRef]
- Pons, E. Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol. J. 2014, 12, 17–27. [Google Scholar] [CrossRef]
- Cabanos, C.S.; Sajise, A.G.; Garcia, R.N.; Siar, S.V.; Mae, E.; Mendoza, T. Compositional analysis of transgenic papaya with delayed ripening trait. Philipp. Agric. Sci. 2013, 96, 331–339. [Google Scholar]
- Richael, C.M. Development of the Genetically Modified Innate® Potato. Plant Breed. Rev. 2021, 44, 57–78. [Google Scholar]
- Li, L.-J.; Tan, W.-S.; Li, W.-J.; Zhu, Y.-B.; Cheng, Y.-S.; Ni, H. Citrus taste modification potentials by genetic engineering. Int. J. Mol. Sci. 2019, 20, 6194. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Naim, F.; Dugdale, B.; Kleidon, J.; Brinin, A.; Shand, K.; Waterhouse, P.; Dale, J. Gene editing the phytoene desaturase alleles of cavendish banana using CRISPR/Cas9. Transgenic Res. 2018, 27, 451–460. [Google Scholar] [CrossRef]
- Jia, H.; Wang, N. Xcc-Facilitated agroinfiltration of citrus leaves: A tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 2014, 33, 1993–2001. [Google Scholar] [CrossRef]
- Fan, D.; Liu, T.; Li, C.; Jiao, B.; Li, S.; Hou, Y.; Luo, K. Efficient CRISPR/Cas9-mediated targeted mutagenesis in populus in the first generation. Sci. Rep. 2015, 5, 12217. [Google Scholar] [CrossRef]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient genome editing in Apple using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef]
- Jia, H.; Orbović, V.; Wang, N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 2019, 17, 1928–1937. [Google Scholar] [CrossRef]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N.; Kumar, P.L. Application of CRISPR/Cas for diagnosis and management of viral diseases of banana. Front. Microbiol. 2021, 11, 609784. [Google Scholar] [CrossRef] [PubMed]
- Gumtow, R.; Wu, D.; Uchida, J.; Tian, M. A phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Mol. Plant-Microbe Interact. 2018, 31, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Ackiyarani, S.U.B.; Handrasekar, A.R.C.; Ma, S.U.U. MusatransSSRDB (a Transcriptome Derived SSR Database)—An advanced tool for banana improvement. J. Biosci. 2019, 44, 4. [Google Scholar] [CrossRef]
- Katz, E.; Fon, M.; Eigenheer, R.A.; Phinney, B.S.; Fass, J.N.; Lin, D.; Sadka, A.; Blumwald, E. A Label-free differential quantitative mass spectrometry method for the characterization and identification of protein changes during citrus fruit development. Proteome Sci. 2010, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Ruas, M.; Guignon, V.; Sempere, G.; Sardos, J.; Hueber, Y. Database tool MGIS: Managing Banana (Musa spp.) genetic resources information and high-throughput genotyping data. Database 2017, 2017, bax046. [Google Scholar] [CrossRef]
- Droc, G.; Larivière, D.; Guignon, V.; Yahiaoui, N.; This, D.; Garsmeur, O.; Dereeper, A.; Hamelin, C.; Argout, X.; Dufayard, J.F.; et al. The Banana Genome Hub. Database 2013, 2013, bat035. [Google Scholar] [CrossRef]
- Arora, V.; Kapoor, N.; Fatma, S.; Jaiswal, S.; Iquebal, M.A.; Rai, A.; Kumar, D. BanSatDB, a whole-genome-based database of putative and experimentally validated microsatellite markers of three musa species. Crop J. 2018, 6, 642–650. [Google Scholar] [CrossRef]
- Qamar-ul-Islam, T.; Khan, M.A.; Faizan, R.; Mahmood, U. MGDb: An analyzed database and a genomic resource of mango (Mangifera indica L.) cultivars for mango research. bioRxiv 2018, 301358. [Google Scholar] [CrossRef]
- Mango Bienniality Gene Database IASRI, New Delhi, India. Available online: http://webapp.cabgrid.res.in/mangodb/index.php?a=reset (accessed on 10 August 2021).
- Iquebal, M.A.; Jaiswal, S.; Mahato, A.K.; Jayaswal, P.K.; Angadi, U.B.; Kumar, N.; Sharma, N.; Singh, A.K.; Srivastav, M.; Prakash, J.; et al. MiSNPDb: A web-based genomic resources of tropical ecology fruit mango (Mangifera indica L.) for phylogeography and varietal differentiation. Sci. Rep. 2017, 7, 14968. [Google Scholar] [CrossRef]
- Li, Q.; Qi, J.; Qin, X.; Dou, W.; Lei, T.; Hu, A.; Jia, R.; Jiang, G.; Zou, X.; Long, Q.; et al. CitGVD: A comprehensive database of citrus genomic variations. Hortic. Res. 2020, 7, 12. [Google Scholar] [CrossRef]
- Dong, Q.; Schlueter, S.D.; Brendel, V. PlantGDB, Plant Genome Database and Analysis Tools. Nucleic Acids Res. 2004, 32, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fan, W.; Guo, X.; Wu, K.; Zhou, S.; Chen, Z.; Li, D.; Wang, K.; Zhu, Y.; Zhou, Y. MaGenDB: A functional genomics hub for malvaceae plants. Nucleic Acids Res. 2020, 48, 1076–1084. [Google Scholar] [CrossRef]
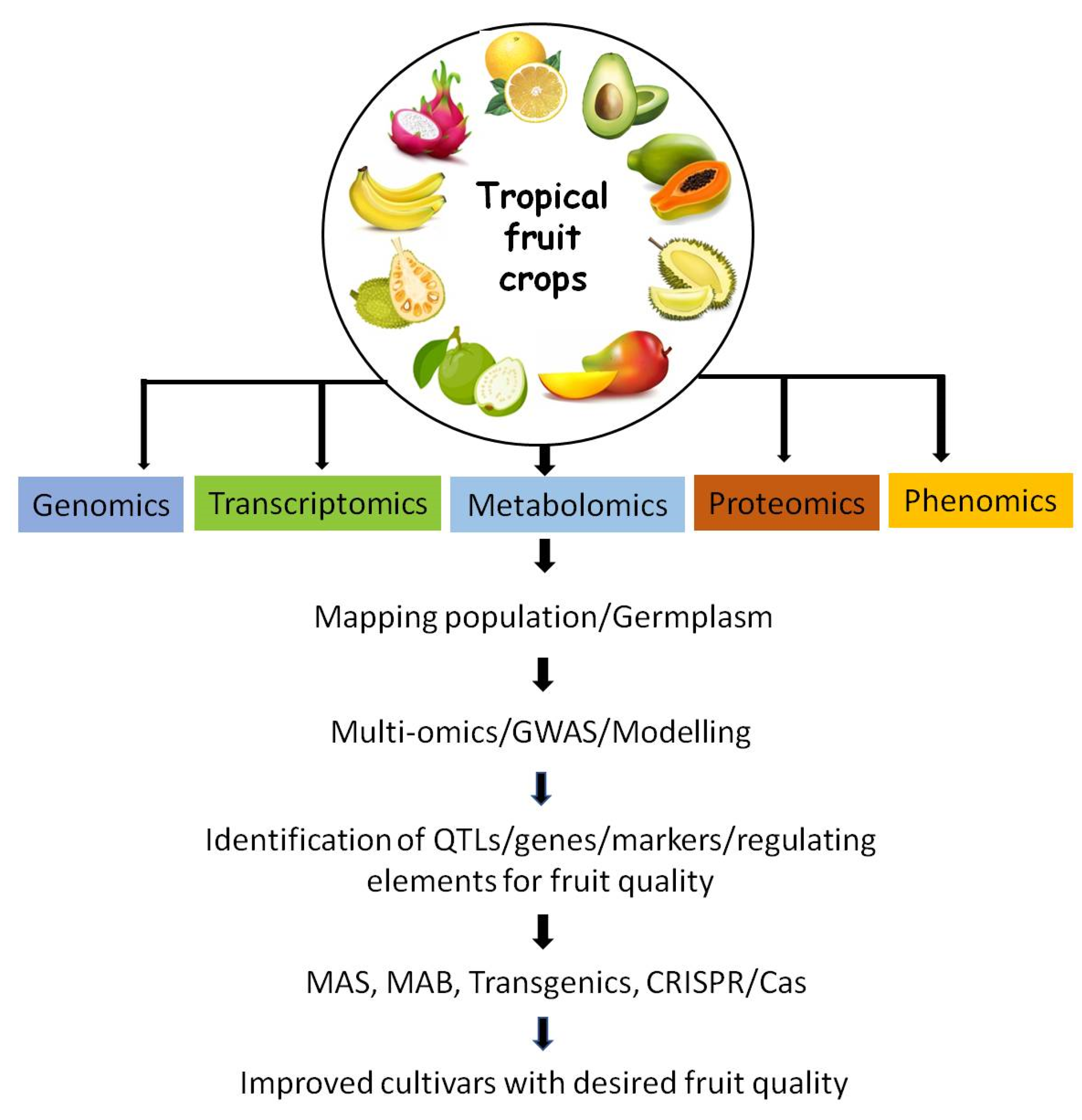
| Crop | Gene Family | Number of Transcription Factors | Fruit Traits | Reference |
|---|---|---|---|---|
| Banana | basic leucine zipper (bZIP) TF—MabZIP93 | 1 | Fruit ripening | [37] |
| MabZIP | 121 | Fruit development, ripening and abiotic stress | [39] | |
| MabZIP74 | 1 | Fruit ripening | [38] | |
| MaMADS2-box | 1 | Fruit ripening | [33] | |
| MaDof | 4 | Fruit ripening | [40] | |
| MaDREB1-MaDREB4 | 4 | Fruit ripening | [41] | |
| MabZIP4 & MabZIP5 | 2 | Fruit aroma | [42] | |
| serine/threonine protein kinases—MAPKK and MAPKKK genes | 10 77 | Fruit development, ripening and responses to abiotic stress | [16] | |
| Basic helix–loop–helix (bHLH)—MaTCP5, MaTCP19 and MaTCP20 | 3 | fruit softening | [34] | |
| MaNF-Ys | 13 | Fruit ripening | [43] | |
| MYB MYB3 R2R3- MYB | 9 1 285 | Fruit softening | [36,44,45] | |
| MaBZR1/2 | 1 | Fruit softening | [36] | |
| MaSPL16 | 1 | Carotenoid biosynthesis | [46] | |
| MaNAC5, MaWRKY1 and MaWRKY2 | 1 | Biotic stress | [47] | |
| Mango | MADS-box EIN3 factors APETALA2-like TFs Ethylene response element factors | 39 4 4 1 | Fruit softening | [29] |
| RabGTpase | - | Fruit softening | [48] | |
| Eukaryotic translation initiation factors (eIFs) | 18 | Abiotic stress | [49] | |
| WRKY | 10 | Mango malformation | [17] | |
| MibHLH | 212 | Hormone responses, abiotic stress | [50] | |
| WD40 family | 315 | Protein–protien interaction, abiotic stress response | [51] | |
| Papaya | CpAP2/ERF—RAP2.4 | 1 | Tolerance to cold and heat stress | [52] |
| CpGRF1–8 | 8 | Fruit ripening and abiotic stress | [53] | |
| CpMADS4 and CpNAC3 | 1 | Fruit ripening | [54] | |
| CpMYB1, CpMYB2 | 1 | Fruit softening | [55] | |
| CpARFs CpEIL1 | 11 1 | Fruit ripening | [56,57] | |
| CpSPLs | 14 | Fruit ripening | [58] | |
| cpNAC1, cpMYB1, cpMYB2, CpbHLH1, CpbHLH2 and CpEIN3a | 1 | Carotenoid biosynthesis in fruits | [55,59,60,61] | |
| CpbHLH | 73 | Abiotic stress | [62] | |
| CpHSF, CpMYB, CpNAC, CpNFY-A, CpERF and CpWRKY | 1 | Drought stress | [63] | |
| Citrus | C2H2, Dof, bHLH, ERF1, MYB, NAC, LBD, ARF1 and TGA9 | 16 | Late fruit ripening | [64] |
| ERF1, ARF1 and TGA9 | 1 | Late fruit ripening | [65] | |
| MADS | 1 | Early fruit ripening | [66] | |
| CBF/DREB, MYB, AP2, Zinc finger, NAC, WRKY, bHLH and ERF | - | Carotenoid biosynthesis | [67,68] | |
| CcMADS19 and homeodomain leucine zipper I (HD-ZIP I) | 1 | Alternate bearing | [69,70] | |
| ERF bHLH genes C2H2 genes WRKY46 HRA1 and Chitin-inducible gibberellinresponsive protein1 | 6 2 2 1 1 1 | Alkalinity stress | [71] | |
| WRKY, NAC, MYB, AP2/ERF, bZIP, GATA, bHLH, ZFP, SPL, CBF and CAMTA | - | Salinity stress | [72] | |
| Guava | WRKY, AP2, bHLH, PHOR1, MYB and C2C2.CO | - | Fruit ripening | [21] |
| TDR4/FUL1, MYB, MADS-RIN, ERF6 and TAG1 | - | Delayed fruit ripening | [73] | |
| Dragon fruit | MYB, bHLH WRKY | 185 168 80 | - | [74] |
| HpWRKY44 and HpWRKY3 | 1 | Betalain biosynthesis | [75] | |
| HpNAC, HpGSTs and MYB | 1 | Betalain biosynthesis | [76,77] | |
| HuERF1 | 1 | Salt stress | [78] | |
| Durian | DNA binding with one finger (Dof) | 24 | Fruit ripening | [79] |
| Ethylene response factor (ERF) transcription factor | 63 | Fruit ripening | [80] | |
| Auxin response factors (ARFs) | 15 | Fruit ripening | [81] |
| Crop | Peel Colour | Genes Involved | Metabolites | Reference | |
|---|---|---|---|---|---|
| Mango | Green |  | UDP-glucose:flavonoid-O-glycosyl-transferase (UFGT), dihydroflavonol 4reductase, anthocyanin synthase (ANS), chalcone synthase and basic helix loop helix (BHLHX) | All trans-violaxanthin butyrate | [123,124] |
| Yellow |  | Carotenoid biosynthesis viz. lycopene-β-cyclase and violaxanthin-de-epoxidase | β-carotene and violaxanthin | [123] | |
| Red |  | Anthocyanin biosynthesis genes viz. phenylalanine ammonia lyase (PAL) and p-coumarate 3-hydroxylase (C3H), flavanone 3-hydroxylase, anthocyanin synthase, MiC4H, Mi4CL2, anthocyanin synthase (MiANS) and UDP-glucose:flavonoid-O-glycosyl-transferase (MiUFGT2), flavonoid 3′hydroxylase and transcription factors MYB and basic helix loop | cyanidin-3-O-monoglucosides and peonidin-3-O-glucosides | [123,124,143] | |
| Banana | Yellow |  | Carotenoid biosynthesis genes | Lutein, D-carotene, and E-carotene | [144] |
| Red |  | Anthocyanin biosynthesis genes | Rutinoside derivatives of cyanidin, peonidin, petunidin, and malvidin | [144] | |
| Purple Ex: M. itinerans |  | Anthocyanin biosynthesis genes and transcription factorssuch as MYB, bHLH, WD40 gene and R2R3-MYB | — | [128] | |
| Papaya | Yellow (ArkaPrabhat) |  | Carotenoid biosynthesis pathway genes such as PSY1, PDS1, ZDS, LCYB1, CHYB, LUT1, ZEP and VDE | lutein and β-carotene | [132] |
| Guava | Yellow (Allahabad Safeda) |  | anthocyanin biosynthesis genes such as phytoene synthase and Aminocyclopropane-1-carboxylate oxidase 1-like | — | [21] |
| Red (Apple colour) |  | Phenypropanoid and lignin biosynthesis pathway genes and anthocyanin biosynthesis genes such as phenylalanine ammonia lyase (PAL), reticulin o-methyltransferase, glycerol-3-phosphate acyltransferase 5 (GPAT 5), peamaclein, CTP synthase-like, chloroplastic monodehydroascorbate (MDA), probable 2-oxoglutarate dependent dioxygenase AOP1 (2OG-AOP1) and methionine synthase (MS) | Reticulin o-methyltransferase | [21] | |
| Citus | Yellow |  | Carotenoid biosynthesis genes and TFCcGCC1 (Garp and coiled-coil) | β-carotene | [131] |
| Red/dark orange |  | Carotenoid biosynthesis genes such as CsPSY, CsZDS, CsZ-ISO, CsBCH1 and carotenoid cleavage dioxygenases genes and TFs such as CsFUL2, CsTAGL1, CsRIN1, CsRIN2 and CsHY5 | β –citraurinene, β -citraurin, phytoene and phytofluene | [129,145] | |
| Dragon fruit | Green |  | Carotenoid biosynthesis gene such as ZEP and Betalain biosynthesis pathway genes such as CYP76Ads | Gomphrenin-I, Cyanidin 3- O-malonylhexoside, Cyanidin chloride, Betanin, malvidin 3-o-galactoside and oenin chloride | [137] |
| Yellow |  | Betalain biosynthesis pathway genes such as TYDC and 2-aminoindan 2-phosphonic acid gene | Cyanidin 3- O-malonylhexoside, cyanidin O-syringic acid, 6-C-Hexosyl-hesperetin O-hexoside, citric acid, isochlorogenic acid A, verbascoside and luteolin-7,3′-Di-O-β-D-Glucoside | [137] | |
| Red |  | Betalain biosynthesis pathway genes such as CYP76ADs, Carotenoid biosynthesis gene such as phytoene synthases (PSY) and PLIS and WRKY transcription factors | Gomphrenin-I, Phyllocactin-II, Isophyllocactin, L-tyrosine, Cyanidin 3-O-galactoside, Cyanidin -O-glucoside-O-rhamnoside, amaranthine, Petunidin, Betanin, malvidin 3-o- galactoside and oenin chloride | [137] | |
| Crop | Reference | Sequencing Technology Used | Percent of Assembly in Pseudo Chromosomes | Assembly Level | Estimated Genome Assembly size | Public Availability (NCBI ID) |
|---|---|---|---|---|---|---|
| Mango (Cultivar ‘Amrapalli’) | [234] | SMRT sequencing | 73.2 | Chromosome | 323 Mb | Not available |
| Mango (Cultivar ‘Alphonso’) | [235] | SMRT sequencing | 91.1 | Chromosome | 392.9 Mb | CATAS_Mindica_2.1 GenBank accession: GCA_011075055.1 |
| Mango (Cultivar ‘Hong Xiang Ya’) | [97] | SMRT sequencing | 98.7 | Chromosome | 389 Mb | Not available |
| Mango (Cultivar ‘Tommy Atkins’) | [236] | Illumina HiSeq2500 | 89 | Chromosome | 377 Mb | ASM1674641v1 GenBank accession: GCA_016746415.1 |
| Banana- (Musa acuminata) | [237] | Sanger and Roche/454 (GSFLX pyrosequencing) | 87.8 | Contig | 523 Mb | DDBJ/EMBL/ GenBank accession numbers CAIC01000001–CAIC01024424 (contigs) |
| Banana (Musa balbisiana) | [238] | Illumina HiSeq 2000 II | 78.9 | Contig | 402.5 Mb. | GenBank SAMN02333823 |
| Banana (Musa itinerans) | [239] | Illumina Hiseq2000 | 75.2 | Contigs | 462.1 Mb | PRJNA312694 |
| Citrus (Fortunella hindsii) | [240] | PacBio Sequel, 10×Genomics Chromium | 96.9 | Contig | 373.6 Mb | ASM480246v1 PRJNA487160 |
| Citrus (Citrus unshiu) | [241] | Illumina HiSeq 2000 | 94.2 | Contig | 359.2 Mb | CUMW_v1.0 PRJDB5882 |
| Citrus (Citrus clementina) | [242] | Long-read 454 and Sanger expressed-sequence-tags (ESTs) | 97 | Scaffold | 301.3 Mb | PRJNA232045, PRJNA223006 |
| Citrus (Citrus reticulata) | [243] | Illumina shotgun sequencing | 90 | Scaffold | 344.2 Mb | PRJNA388397, INSDC: NIHA00000000.1 |
| Citrus (Citrus medica) | [244] | PacBio RS II platform via a shotgun approach | 87 | Scaffold | 406 Mb | PRJNA320023 |
| Papaya (Cultivar ‘SunUp’) | [245] | Whole-genome shotgun (WGS) sequencing | 74.2 | contigs | 372 Mb | GenBank under accession number ABIM00000000. |
| Guava (Cultivar ‘New Age’) | [231] | SMRT sequencing | 95.7 | Chromosome | 463.8 Mb | guava_v11.23 GenBank assembly accession: GCA_016432845.1 |
| Jack fruit (A. heterophyllus) | [233] | IIlumina HiSeq 2000 | 98.98 | Chromosome | 1.2 Gb | Not available |
| Dragon fruit (Cultivar ‘Guanhuabai’) | [74] | PacBio, Illumina, 10× Genomics, and Hi-C, | 97.67 | Chromosome | 1.41 Gb | Not available |
| Dragon fruit (Cultivar ‘David Bowie’) | [116] | 10×chromium sequencing, Hi-C | 88.7 | Chromosome | 1.33 Gb | ASM1758966v1 GenBank assembly accession: GCA_017589665.1 |
| Durian (Durio ziberthinus) | [246] | PacBio, Hi-C | 96.88 | Chromosome | 738 Mb | Duzib1.0 GenBank assembly accession: GCA_002303985.1 |
| Fruit Crop | Targeted Genes/Pathways | Genetic Engineering Approach | Modified Traits | Reference |
|---|---|---|---|---|
| Apple: Arctic®Golden Delicious, Arctic® Granny Smith, and Arctic® Fuji | Polyphenoloxidases- PPO2, GPO3, APO5 and pSR7 | RNAi technology | Non-browning | [307] |
| Pineapple (Pinkglow™) | Carotenoid pathway- tangerine (Citrus reticulata) PSY gene | Transgenic | Pink flesh | [308] |
| Squash | Coat protein gene transfer | Transgenic | Resistant to potyviruses | [309] |
| Banana | PSY and Ferritin gene | Transgenic | Biofortifed pro-vitamin A and Iron | [299,300] |
| Grand Naine banana | Endochitinase gene ThEn-42 (Trichoderma harzianum), grape stilbene synthase (StSy), and superoxide dismutase Cu, Zn-SOD was taken from tomato | Transgenic | Improved tolerance toward fungal disease | [301] |
| Banana | MaMADS, MaMADS36 | RNAi technology | Delayed ripening | [310] |
| Cavendish banana | Banana cisgenes | Cisgenic | Enhanced level of provitamin-A and to increase the resistance to Panama disease | [298] |
| Banana | MaACO1 (amino-cyclopropane-1-carboxylate oxidase 1) | CRISPR/Cas9 | Reduced ethylene synthesis and extended shelf life | [311] |
| Cavendish banana | Lycopene epsilon-cyclase (LCY ε) gene | CRISPR/Cas9 | β-carotene enrichment | [312] |
| Papaya | Coat protein (CP) mediated | RNAi technology | PRSV-resistant | [304] |
| Papaya | S-genes | CRISPR/Cas9 | Resistance to PRSV | [313] |
| Duncan grapefruit | CsLOB1 | CRISPR/Cas9-mediated gene knockouts | Duncan grapefruit canker | [314] |
| Sweet orange | Silencing of β-carotene hydroxylase | RNAi technology | Accumulation of carotenoids in fruit pulp | [315] |
| Crop | Database | Developed Using | Purpose | Reference | Weblink (Accessed on 10 September 2021) |
|---|---|---|---|---|---|
| Banana | MusatransSSRDB | Transcriptome | selection of SSR primers for a specific objective | [328] | http://nrcb.res.in/nrcbbio/about.html |
| Musa Germplasm Information System (MGIS) | Accession-based data and genotyping studies | global ex situ-held banana genetic resources | [330] | https://www.crop-diversity.org/mgis/ | |
| Banana Genome Hub | Genomic data on banana | integration between various systems (Jbrowse, Galaxy, Gigwa | [331] | banana-genome-hub. southgreen.fr | |
| BanSatDB | Whole genome-based | (>341,000) of putative STR markers from Musa genera and 580 validated STR markers from the published literature | [332] | http://webtom.cabgrid.res.in/bansatdb/ | |
| Mango | MGdb | Transcriptome | Genomic resource | [333] | — |
| Mango Bienniality Gene Database (MBGDB) | NCBI | Genes related to bienniality rhythm of mango | [334] | http://webapp.cabgrid.res.in/mangodb | |
| MiSNPDb | Sequence based | Phylogenetic and evolutionary studies using SNPs | [335] | http://webtom.cabgrid.res.in/mangosnps/ | |
| Citrus | CitGVD | Published resources | Citrus genomic variation database | [336] | http://citgvd.cric.cn/home |
| Citrus genome database (CGD) | Genomics, genetics and breeding | Genomics, genetics and breeding and disease resistance | — | https://www.citrusgenomedb.org/ | |
| iCitrus | NCBI | Citrus protein identification | [329] | — | |
| Papaya | Carica papaya Genome DB—PlantGDB | Whole genome | Comparative plant genomics | [337] | http://www.plantgdb.org/CpGDB/ |
| Durian | MaGenDB | Genome information from public databases | Genomic resource for Malvaceae family | [338] | http://magen.whu.edu.cn |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathiazhagan, M.; Chidambara, B.; Hunashikatti, L.R.; Ravishankar, K.V. Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content. Genes 2021, 12, 1881. https://doi.org/10.3390/genes12121881
Mathiazhagan M, Chidambara B, Hunashikatti LR, Ravishankar KV. Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content. Genes. 2021; 12(12):1881. https://doi.org/10.3390/genes12121881
Chicago/Turabian StyleMathiazhagan, Malarvizhi, Bhavya Chidambara, Laxman R. Hunashikatti, and Kundapura V. Ravishankar. 2021. "Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content" Genes 12, no. 12: 1881. https://doi.org/10.3390/genes12121881
APA StyleMathiazhagan, M., Chidambara, B., Hunashikatti, L. R., & Ravishankar, K. V. (2021). Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content. Genes, 12(12), 1881. https://doi.org/10.3390/genes12121881






