DNA Damage Responses during the Cell Cycle: Insights from Model Organisms and Beyond
Abstract
1. DNA Damage Responses: A Brief Overview
2. The Role of Genetic Screens in Model Organisms to Reveal DDR Genes
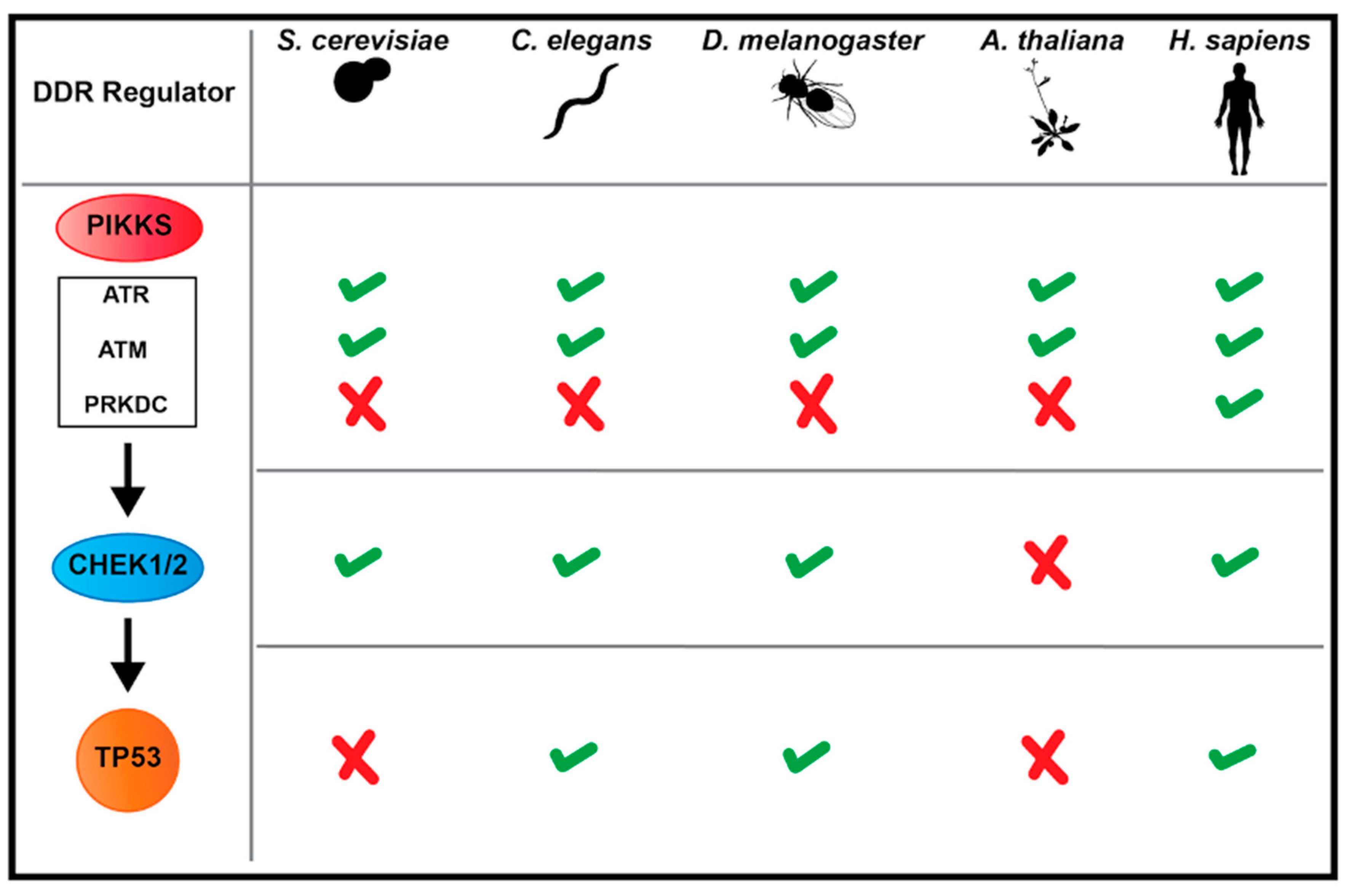
3. Major Players in DDR Mechanisms: Conservation and Differences across Model Systems
3.1. Kinase Signaling in Response to DSBs
3.2. DSB Repair Pathways
4. DDR Responses During the Mitotic Cell Cycle Across Model Systems
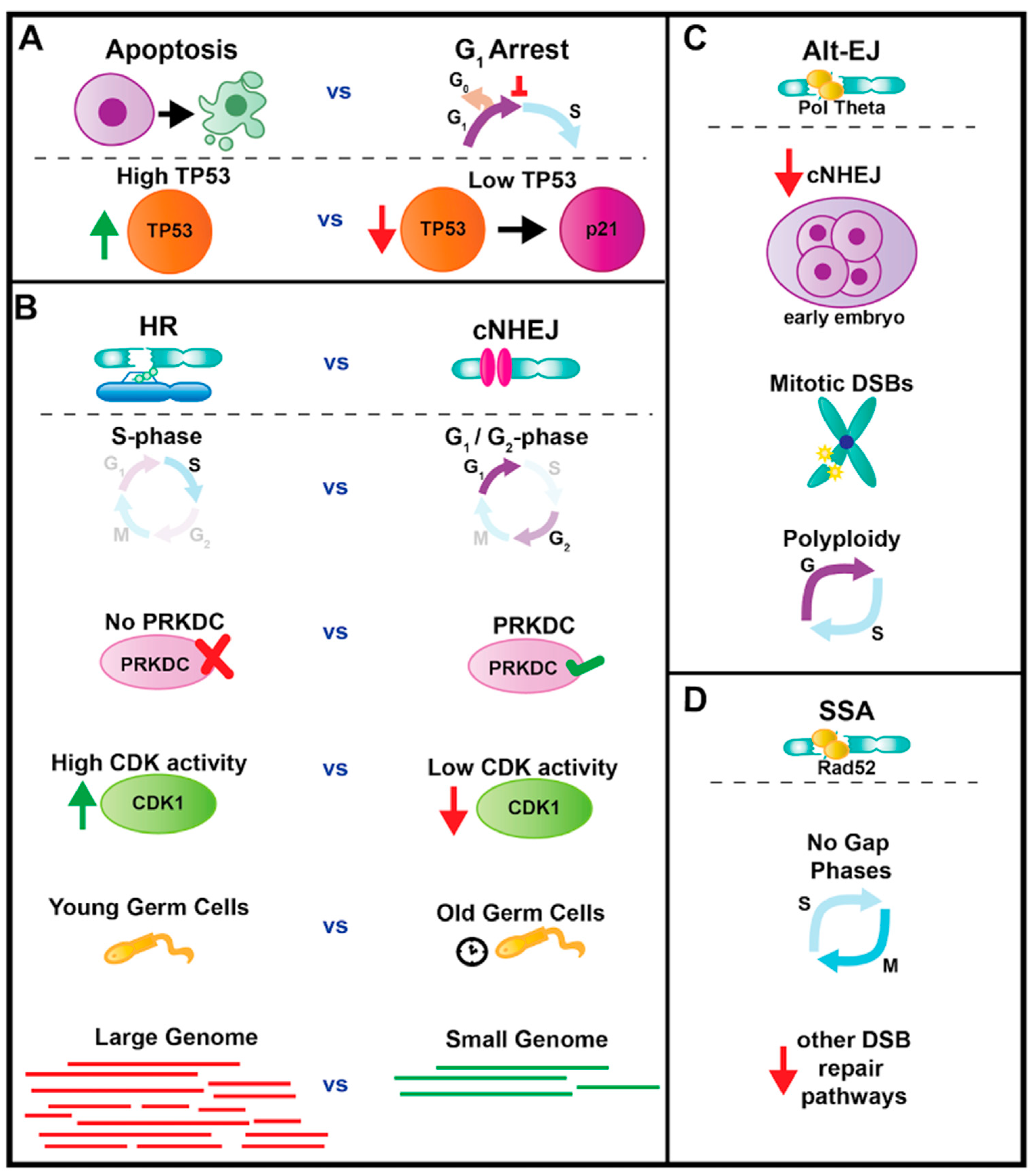
4.1. Interphase: Cell Cycle Arrest or Apoptosis
4.2. Interphase Repair: cNHEJ or HR
4.3. Roles for Alt-EJ and SSA Repair
4.4. The DDR During Mitosis
5. DDR Responses during Other Cell Cycles in Model Systems
5.1. Meiotic DDR Regulation
5.2. DDRs during and after Endoreplication Cycles
6. Emerging Model DDR Systems
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Harper, J.W.; Elledge, S.J. The DNA Damage Response: Ten Years After. Mol. Cell 2007, 28, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making it safe to play with knives. Mol. Cell 2010, 40, 179. [Google Scholar] [CrossRef] [PubMed]
- Finn, K.; Lowndes, N.F.; Grenon, M. Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell. Mol. Life Sci. 2012, 69, 1447–1473. [Google Scholar] [CrossRef]
- Su, T. Cellular responses to DNA damage: One signal, multiple choices. Annu. Rev. Genet. 2006, 40, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Gelling, C. Hermann Muller on Measuring Mutation Rates. Genetics 2016, 202, 369. [Google Scholar] [CrossRef][Green Version]
- Hughes, D.M.; Bovie, W.T. The Effects of Fluorite Ultra-Violet Light on the Rate of division of Paramecium Caudatum. J. Med. Res. 1918, 39, 233–238. [Google Scholar]
- Blum, H.F. Carcinogenesis by Ultraviolet Light; Princeton University Press: Princeton, NJ, USA, 1959. [Google Scholar]
- Sax, K. An Analysis of X-Ray Induced Chromosomal Aberrations in Tradescantia. Genetics 1940, 25, 41–68. [Google Scholar] [CrossRef]
- McClintock, B. The fusion of broken ends of sister half-chromatids following chromatid breakage at meiotic anaphases. Missouri Agric. Exp. Stn. Res. Bull. 1938, 290, 1–48. [Google Scholar]
- Nakai, S.; Matsumoto, S. Two types of radiation-sensitive mutant in yeast. Mutat. Res. 1967, 4, 129–136. [Google Scholar] [CrossRef]
- Cox, B.; Parry, J. The isolation, genetics and survival characteristics of ultraviolet light-sensitive mutants in yeast. Mutat. Res. 1968, 6, 37–55. [Google Scholar] [CrossRef]
- Weinert, T.; Hartwell, L. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 1988, 241, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Shinoura, Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet. 1977, 156, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hannan, M.A.; Duck, P.; Nasim, A. UV-induced lethal sectoring and pure mutant clones in yeast. Mutat. Res. Mol. Mech. Mutagen. 1976, 36, 171–175. [Google Scholar] [CrossRef]
- Prakash, L.; Prakash, S. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 1977, 86, 33–35. [Google Scholar] [CrossRef]
- Birrell, G.W.; Giaever, G.; Chu, A.M.; Davis, R.W.; Brown, J.M. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. USA 2001, 98, 12608–12613. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.; Lewis, L.; Karthikeyan, G.; Lobachev, K.; Jin, Y.; Sterling, J.; Snipe, J.; Resnick, M. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 2001, 29, 426–434. [Google Scholar] [CrossRef]
- Ooi, S.L.; Shoemaker, D.D.; Boeke, J.D. A DNA Microarray-Based Genetic Screen for Nonhomologous End-Joining Mutants in Saccharomyces cerevisiae. Science 2001, 294, 2552–2556. [Google Scholar] [CrossRef]
- Alvaro, D.; Lisby, M.; Rothstein, R. Genome-Wide Analysis of Rad52 Foci Reveals Diverse Mechanisms Impacting Recombination. PLOS Genet. 2007, 3, e228. [Google Scholar] [CrossRef]
- Andersen, M.P.; Nelson, Z.W.; Hetrick, E.D.; Gottschling, D.E. A Genetic Screen for Increased Loss of Heterozygosity in Saccharomyces cerevisiae. Genetics 2008, 179, 1179–1195. [Google Scholar] [CrossRef]
- Cheng, E.; Vaisica, J.A.; Ou, J.; Baryshnikova, A.; Lu, Y.; Roth, F.P.; Brown, G.W. Genome Rearrangements Caused by Depletion of Essential DNA Replication Proteins in Saccharomyces cerevisiae. Genetics 2012, 192, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Hendry, J.A.; Tan, G.; Ou, J.; Boone, C.; Brown, G.W. Leveraging DNA Damage Response Signaling to Identify Yeast Genes Controlling Genome Stability. G3 Genes Genomes Genet. 2015, 5, 997–1006. [Google Scholar] [CrossRef]
- Puddu, F.; Herzog, M.; Selivanova, A.; Wang, S.; Zhu, J.; Klein-Lavi, S.; Gordon, M.; Meirman, R.; Millan-Zambrano, G.; Ayestaran, I.; et al. Genome architecture and stability in the Saccharomyces cerevisiae knockout collection. Nature 2019, 573, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Hartman, P.S.; Herman, R.K. Radiation-Sensitive Mutants of Caenorhabditis Elegans. Genetics 1982, 102, 159. [Google Scholar] [CrossRef] [PubMed]
- Pothof, J.; van Haaften, G.; Thijssen, K.; Kamath, R.S.; Fraser, A.G.; Ahringer, J.; Plasterk, R.H.A.; Tijsterman, M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003, 17, 443–448. [Google Scholar] [CrossRef]
- Lemmens, B.; Tijsterman, M. DNA double-strand break repair in Caenorhabditis elegans. Chromosoma 2011, 120, 1–21. [Google Scholar] [CrossRef]
- Craig, A.L.; Moser, S.C.; Bailly, A.P.; Gartner, A. Methods for Studying the DNA Damage Response in the Caenorhabdatis elegans Germ Line. Methods Cell Biol. 2012, 107, 321–352. [Google Scholar] [PubMed]
- Rieckher, M.; Bujarrabal, A.; Doll, M.; Soltanmohammadi, N.; Schumacher, B. A simple answer to complex questions: Caenorhabditis elegans as an experimental model for examining the DNA damage response and disease genes. J. Cell. Physiol. 2018, 233, 2781–2790. [Google Scholar] [CrossRef]
- Tang, L.; Machacek, T.; Mamnun, Y.; Penkner, A.; Gloggnitzer, J.; Wegrostek, C.; Konrat, R.; Jantsch, M.; Loidl, J.; Jantsch, V. Mutations in Caenorhabditis elegans him-19 show meiotic defects that worsen with age. Mol. Biol. Cell 2010, 21, 885–896. [Google Scholar] [CrossRef]
- Smith, P.D. Mutagen sensitivity of Drosophila melanogaster—III. X-linked loci governing sensitivity to methyl methanesulfonate. MGG Mol. Gen. Genet. 1976, 149, 73–85. [Google Scholar] [CrossRef]
- Boyd, J.B.; Golino, M.D.; NGUYENL, T.D.; Green, M.M. Isolation And Characterization Of X-Linked Mutants Of Drosophila Melanogaster Which Are Sensitive To Mutagens. Genetics 1976, 84, 485–506. [Google Scholar] [CrossRef]
- Graf, U.; Green, M.; Würgler, F. Mutagen-sensitive mutants in Drosophila melanogaster: Effects on premutational damage. Mutat. Res. 1979, 63, 101–112. [Google Scholar] [CrossRef]
- Laurencon, A.; Orme, C.M.; Peters, H.K.; Boulton, C.L.; Vladar, E.K.; Langley, S.A.; Bakis, E.P.; Harris, D.T.; Harris, N.J.; Wayson, S.M.; et al. A large-scale screen for mutagen-sensitive loci in Drosophila. Genetics 2004, 167, 217–231. [Google Scholar] [CrossRef]
- Sekelsky, J. DNA Repair in Drosophila: Mutagens, Models, and Missing Genes. Genetics 2017, 205, 471–490. [Google Scholar] [CrossRef]
- Rong, Y.S.; Titen, S.W.; Xie, H.B.; Golic, M.M.; Bastiani, M.; Bandyopadhyay, P.; Olivera, B.M.; Brodsky, M.; Rubin, G.M.; Golic, K.G. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002, 16, 1568–1581. [Google Scholar] [CrossRef]
- Carvajal-Garcia, J.; Crown, K.N.; Ramsden, D.A.; Sekelsky, J. DNA polymerase theta suppresses mitotic crossing over. PLOS Genet. 2021, 17, e1009267. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, K.P.; Montague, R.A.; Paramore, S.V.; Lennox, A.L.; Mahowald, A.P.; Fox, D.T. Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development 2014, 141, 3551–3560. [Google Scholar] [CrossRef] [PubMed]
- Clay, D.E.; Bretscher, H.S.; Jezuit, E.A.; Bush, K.B.; Fox, D.T. Persistent DNA damage signaling and DNA polymerase theta promote broken chromosome segregation. J. Cell Biol. 2021, 220, e202106116. [Google Scholar] [CrossRef] [PubMed]
- Bretscher, H.S.; Fox, D.T. Proliferation of Double-Strand Break-Resistant Polyploid Cells Requires Drosophila FANCD2. Dev. Cell 2016, 37, 444–457. [Google Scholar] [CrossRef]
- Harlow, G.; Jenkins, M.; Pittalwala, T.; Mount, D. Isolation of uvh1, an Arabidopsis mutant hypersensitive to ultraviolet light and ionizing radiation. Plant Cell 1994, 6, 227–235. [Google Scholar]
- Preuss, S.; Britt, A. A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics 2003, 164, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Shima, N.; Hartford, S.A.; Duffy, T.; Wilson, L.A.; Schimenti, K.J.; Schimenti, J.C. Phenotype-Based Identification of Mouse Chromosome Instability Mutants. Genetics 2003, 163, 1031–1040. [Google Scholar] [CrossRef]
- Hoogenboom, W.S.; Klein Douwel, D.; Knipscheer, P. Xenopus egg extract: A powerful tool to study genome maintenance mechanisms. Dev. Biol. 2017, 428, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Cannan, W.; Pederson, D. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.; Jackson, S. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Cussiol, J.R.R.; Soares, B.L.; Oliveira, F.M.B. From yeast to humans: Understanding the biology of DNA DamageResponse (DDR) kinases. Genet. Mol. Biol. 2020, 43, e20190071. [Google Scholar] [CrossRef]
- Lees-Miller, J.P.; Cobban, A.; Katsonis, P.; Bacolla, A.; Tsutakawa, S.E.; Hammel, M.; Meek, K.; Anderson, D.W.; Lichtarge, O.; Tainer, J.A.; et al. Uncovering DNA-PKcs ancient phylogeny, unique sequence motifs and insights for human disease. Prog. Biophys. Mol. Biol. 2021, 163, 87–108. [Google Scholar] [CrossRef]
- Vitale, I.; Galluzzi, L.; Castedo, M.; Kroemer, G. Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 2011, 12, 385–392. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529. [Google Scholar] [CrossRef]
- Shima, N.; Munroe, R.J.; Schimenti, J.C. The Mouse Genomic Instability Mutation chaos1 Is an Allele of Polq That Exhibits Genetic Interaction with Atm. Mol. Cell. Biol. 2004, 24, 10381. [Google Scholar] [CrossRef]
- Boyd, J.B.; Sakaguchi, K.; Harris, P.V. Mus308 Mutants of Drosophila Exhibit Hypersensitivity to DNA Cross-Linking Agents and Are Defective in a Deoxyribonuclease. Genetics 1990, 125, 813. [Google Scholar] [CrossRef] [PubMed]
- Muzzini, D.; Plevani, P.; Boulton, S.; Cassata, G.; Marini, F. Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair 2008, 7, 941–950. [Google Scholar] [CrossRef]
- Wyatt, D.; Feng, W.; Conlin, M.; Yousefzadeh, M.; Roberts, S.; Mieczkowski, P.; Wood, R.; Gupta, G.; Ramsden, D. Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks. Mol. Cell 2016, 63, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Simpson, D.A.; Carvajal-Garcia, J.; Price, B.A.; Kumar, R.J.; Mose, L.E.; Wood, R.D.; Rashid, N.; Purvis, J.E.; Parker, J.S.; et al. Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun. 2019, 10, 4286. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Liu, C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.; O’Connor, K.; Konstantinopoulos, P.; Elledge, S.; Boulton, S.; et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 2015, 518, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.H.; Yu, A.M.; McVey, M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010, 6, e1001005. [Google Scholar] [CrossRef] [PubMed]
- Beagan, K.; Armstrong, R.L.; Witsell, A.; Roy, U.; Renedo, N.; Baker, A.E.; Scharer, O.; McVey, M. Drosophila DNA polymerase theta utilizes both helicase-like and polymerase domains during microhomology-mediated end joining and interstrand crosslink repair. PLoS Genet. 2017, 13, e1006813. [Google Scholar] [CrossRef]
- Mara, K.; Charlot, F.; Guyon-Debast, A.; Schaefer, D.G.; Collonnier, C.; Grelon, M.; Nogué, F. POLQ plays a key role in the repair of CRISPR/Cas9-induced double-stranded breaks in the moss Physcomitrella patens. New Phytol. 2019, 222, 1380–1391. [Google Scholar] [CrossRef]
- Decottignies, A. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics 2007, 176, 1403–1415. [Google Scholar] [CrossRef]
- Lee, K.; Lee, S.E. Saccharomyces cerevisiae Sae2- and Tel1-Dependent Single-Strand DNA Formation at DNA Break Promotes Microhomology-Mediated End Joining. Genetics 2007, 176, 2003. [Google Scholar] [CrossRef]
- Chen, Y.; Dui, W.; Yu, Z.; Li, C.; Ma, J.; Jiao, R. Drosophila RecQ5 is required for efficient SSA repair and suppression of LOH in vivo. Protein Cell 2010, 1, 478–490. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Holsclaw, J.; Sekelsky, J. Annealing of Complementary DNA Sequences During Double-Strand Break Repair in Drosophila Is Mediated by the Ortholog of SMARCAL1. Genetics 2017, 206, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Brodsky, M.H.; Weinert, B.T.; Tsang, G.; Rong, Y.S.; McGinnis, N.M.; Golic, K.G.; Rio, D.C.; Rubin, G.M. Drosophila melanogaster MNK/Chk2 and p53 Regulate Multiple DNA Repair and Apoptotic Pathways following DNA Damage. Mol. Cell. Biol. 2004, 24, 1219–1231. [Google Scholar] [CrossRef]
- Derry, W.; Putzke, A.; Rothman, J. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science 2001, 294, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.E.; Brehm, A. Of flies and men; p53, a tumour suppressor. FEBS Lett. 2004, 567, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ko, L.; Jayaraman, L.; Prives, C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996, 10, 2438–2451. [Google Scholar] [CrossRef]
- Purvis, J.; Karhohs, K.; Mock, C.; Batchelor, E.; Loewer, A.; Lahav, G. p53 dynamics control cell fate. Science 2012, 336, 1440–1444. [Google Scholar] [CrossRef]
- Zhang, X.-P.; Liu, F.; Cheng, Z.; Wang, W. Cell fate decision mediated by p53 pulses. Proc. Natl. Acad. Sci. USA 2009, 106, 12245–12250. [Google Scholar] [CrossRef]
- Morgan, D.O. The Cell Cycle: Principles of Control (Primers in Biology Series); New Science Press Ltd.: London, UK, 2007. [Google Scholar]
- Song, Y.-H. Drosophila melanogaster: A Model for the Study of DNA Damage Checkpoint Response. Mol. Cells 2005, 19, 167–179. [Google Scholar]
- Wahl, G.; Carr, A. The evolution of diverse biological responses to DNA damage: Insights from yeast and p53. Nat. Cell Biol. 2001, 3, E277–E286. [Google Scholar] [CrossRef]
- Abegglen, L.; Caulin, A.; Chan, A.; Lee, K.; Robinson, R.; Campbell, M.; Kiso, W.; Schmitt, D.; Waddell, P.; Bhaskara, S.; et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 2015, 314, 1850–1860. [Google Scholar] [CrossRef]
- Sulak, M.; Fong, L.; Mika, K.; Chigurupati, S.; Yon, L.; Mongan, N.; Emes, R.; Lynch, V. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 2016, 5, e11994. [Google Scholar] [CrossRef]
- Hafner, A.; Bulyk, M.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef]
- Barnum, K.; O’Connell, M. Cell cycle regulation by checkpoints. Methods Mol. Biol. 2014, 1170, 29–40. [Google Scholar] [PubMed]
- Yoshiyama, K.; Conklin, P.A.; Huefner, N.D.; Britt, A.B. Suppressor of γ response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 2009, 106, 12843–12848. [Google Scholar] [CrossRef] [PubMed]
- Adachi, S.; Minamisawa, K.; Okushima, Y.; Inagaki, S.; Yoshiyama, K.; Kondou, Y.; Kaminuma, E.; Kawashima, M.; Toyoda, T.; Matsui, M.; et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 10004–10009. [Google Scholar] [CrossRef] [PubMed]
- Ogita, N.; Okushima, Y.; Tokizawa, M.; Yamamoto, Y.Y.; Tanaka, M.; Seki, M.; Makita, Y.; Matsui, M.; Okamoto-Yoshiyama, K.; Sakamoto, T.; et al. Identifying the target genes of SUPPRESSOR OF γ RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018, 94, 439–453. [Google Scholar] [CrossRef]
- Delacôte, F.; Lopez, B.S. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: The trans-S double-strand break repair model. Cell Cycle 2008, 7, 33–38. [Google Scholar] [CrossRef] [PubMed]
- McArt, D.; McKerr, G.; Saetzler, K.; Howard, C.; Downes, C.; Wasson, G. Comet sensitivity in assessing DNA damage and repair in different cell cycle stages. Mutagenesis 2010, 25, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Xie, A. In my end is my beginning: Control of end resection and DSBR pathway ‘choice’ by cyclin-dependent kinases. Oncogene 2005, 24, 2871–2876. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008, 7, 2902–2906. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks. Int. J. Mol. Sci. 2020, 21, 6461. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.F.; Meyenberg, M.; Loizou, J.I. Tissue specificity of DNA repair: The CRISPR compass. Trends Genet. 2021, 37, 958–962. [Google Scholar] [CrossRef]
- Jeggo, P. Studies on mammalian mutants defective in rejoining double-strand breaks in DNA. Mutat. Res. 1990, 239, 1–16. [Google Scholar] [CrossRef]
- Kakarougkas, A.; Jeggo, P.A. DNA DSB repair pathway choice: An orchestrated handover mechanism. Br. J. Radiol. 2014, 87, 20130685. [Google Scholar] [CrossRef]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2007, 18, 134–147. [Google Scholar] [CrossRef]
- Li, J.; Xu, X. DNA double-strand break repair: A tale of pathway choices. Acta Biochim. Biophys. Sin. 2016, 48, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.; Bloomer, H.; Kellam, N.; LaRocque, J.R. Chromosome Preference During Homologous Recombination Repair of DNA Double-Strand Breaks in Drosophila melanogaster. G3 Genes|Genomes|Genet. 2019, 9, 3773. [Google Scholar] [CrossRef]
- Grabarz, A.; Barascu, A.; Guirouilh-Barbat, J.; Lopez, B.S. Initiation of DNA double strand break repair: Signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am. J. Cancer Res. 2012, 2, 249. [Google Scholar] [PubMed]
- Ferretti, L.; Lafranchi, L.; Sartori, A. Controlling DNA-end resection: A new task for CDKs. Front. Genet. 2013, 4, 99. [Google Scholar] [CrossRef] [PubMed]
- Delabaere, L.; Ertl, H.A.; Massey, D.J.; Hofley, C.M.; Sohail, F.; Bienenstock, E.J.; Sebastian, H.; Chiolo, I.; LaRocque, J.R. Aging impairs double-strand break repair by homologous recombination in Drosophila germ cells. Aging Cell 2017, 16, 320. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, J.; Karg, T.; Golic, K. Homolog-Dependent Repair Following Dicentric Chromosome Breakage in Drosophila melanogaster. Genetics 2019, 212, 615–630. [Google Scholar] [CrossRef]
- Malkova, A.; Ivanov, E.L.; Haber, J.E. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 1996, 93, 7131–7136. [Google Scholar] [CrossRef] [PubMed]
- Thyme, S.; Schier, A. Polq-Mediated End Joining Is Essential for Surviving DNA Double-Strand Breaks during Early Zebrafish Development. Cell Rep. 2016, 15, 707–714. [Google Scholar] [CrossRef]
- Bhargava, R.; Onyango, D.; Stark, J. Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet. 2016, 32, 566–575. [Google Scholar] [CrossRef]
- Preston, C.R.; Flores, C.; Engels, W.R. Age-Dependent Usage of Double-Strand-Break Repair Pathways. Curr. Biol. 2006, 16, 2009–2015. [Google Scholar] [CrossRef]
- Ahuja, A.K.; Jodkowska, K.; Teloni, F.; Bizard, A.H.; Zellweger, R.; Herrador, R.; Ortega, S.; Hickson, I.D.; Altmeyer, M.; Mendez, J.; et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 2016, 7, 10660. [Google Scholar] [CrossRef]
- Bakhoum, S.; Kabeche, L.; Murnane, J.; Zaki, B.; Compton, D. DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov. 2014, 4, 1281–1289. [Google Scholar] [CrossRef]
- Giunta, S.; Belotserkovskaya, R.; Jackson, S.P. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 2010, 190, 197–207. [Google Scholar] [CrossRef]
- Gomez-Godinez, V.; Wu, T.; Sherman, A.J.; Lee, C.S.; Liaw, L.H.; Zhongsheng, Y.; Yokomori, K.; Berns, M.W. Analysis of DNA double-strand break response and chromatin structure in mitosis using laser microirradiation. Nucleic Acids Res. 2010, 38, e202. [Google Scholar] [CrossRef] [PubMed]
- Orthwein, A.; Fradet-Turcotte, A.; Noordermeer, S.; Canny, M.; Brun, C.; Strecker, J.; Escribano-Diaz, C.; Durocher, D.; Orthwein, A.; Fradet-Turcotte, A.; et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science 2014, 344, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Kabeche, L.; Wood, M.D.; Laucius, C.D.; Qu, D.; Laughney, A.M.; Reynolds, G.E.; Louie, R.J.; Phillips, J.; Chan, D.A.; et al. Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat. Commun. 2015, 6, 5990. [Google Scholar] [CrossRef] [PubMed]
- Benada, J.; Burdová, K.; Lidak, T.; von Morgen, P.; Macurek, L. Polo-like kinase 1 inhibits DNA damage response during mitosis. Cell Cycle 2015, 14, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Kabeche, L.; Compton, D.A.; Powell, S.N.; Bastians, H. Mitotic DNA Damage Response: At the Crossroads of Structural and Numerical Cancer Chromosome Instabilities. Trends Cancer 2017, 3, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Heijink, A.; Krajewska, M.; van Vugt, M. The DNA damage response during mitosis. Mutat. Res. 2013, 750, 45–55. [Google Scholar] [CrossRef]
- Leimbacher, P.A.; Jones, S.E.; Shorrocks, A.M.; de Marco Zompit, M.; Day, M.; Blaauwendraad, J.; Bundschuh, D.; Bonham, S.; Fischer, R.; Fink, D.; et al. MDC1 Interacts with TOPBP1 to Maintain Chromosomal Stability during Mitosis. Mol. Cell 2019, 74, 571–583.e8. [Google Scholar] [CrossRef]
- Ishii, K.; Ogiyama, Y.; Chikashige, Y.; Soejima, S.; Masuda, F.; Kakuma, T.; Hiraoka, Y.; Takahashi, K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 2008, 321, 1088–1091. [Google Scholar] [CrossRef]
- Ohno, Y.; Ogiyama, Y.; Kubota, Y.; Kubo, T.; Ishii, K. Acentric chromosome ends are prone to fusion with functional chromosome ends through a homology-directed rearrangement. Nucleic Acids Res. 2016, 44, 232–244. [Google Scholar] [CrossRef]
- Landmann, C.; Pierre-Elies, P.; Goutte-Gattat, D.; Montembault, E.; Claverie, M.-C.; Royou, A. The Mre11-Rad50-Nbs1 complex mediates the robust recruitment of Polo to DNA lesions during mitosis in Drosophila. J. Cell Sci. 2020, 133, jcs244442. [Google Scholar] [CrossRef] [PubMed]
- Royou, A.; Gagou, M.E.; Karess, R.; Sullivan, W. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell 2010, 140, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Karg, T.; Warecki, B.; Sullivan, W. Aurora B-mediated localized delays in nuclear envelope formation facilitate inclusion of late-segregating chromosome fragments. Mol. Biol. Cell 2015, 26, 2227–2241. [Google Scholar] [CrossRef]
- Karg, T.; Elting, M.W.; Vicars, H.; Dumont, S.; Sullivan, W. The chromokinesin Klp3a and microtubules facilitate acentric chromosome segregation. J. Cell Biol. 2017, 216, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Derive, N.; Landmann, C.; Montembault, E.; Claverie, M.-C.C.; Pierre-Elies, P.; Goutte-Gattat, D.; Founounou, N.; McCusker, D.; Royou, A. Bub3-BubR1-dependent sequestration of Cdc20Fizzy at DNA breaks facilitates the correct segregation of broken chromosomes. J. Cell Biol. 2015, 211, 517–532. [Google Scholar] [CrossRef]
- Minocherhomji, S.; Ying, S.; Bjerregaard, V.A.; Bursomanno, S.; Aleliunaite, A.; Wu, W.; Mankouri, H.W.; Shen, H.; Liu, Y.; Hickson, I.D. Replication stress activates DNA repair synthesis in mitosis. Nature 2015, 528, 286–290. [Google Scholar] [CrossRef]
- Chan, K.L.; Palmai-Pallag, T.; Ying, S.; Hickson, I.D. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009, 11, 753–760. [Google Scholar] [CrossRef]
- Llorens-Agost, M.; Ensminger, M.; Le, H.P.; Gawai, A.; Liu, J.; Cruz-García, A.; Bhetawal, S.; Wood, R.D.; Heyer, W.-D.; Löbrich, M. POLθ-mediated end joining is restricted by RAD52 and BRCA2 until the onset of mitosis. Nat. Cell Biol. 2021. [Google Scholar] [CrossRef]
- Schwacha, A.; Kleckner, N. Interhomolog Bias during Meiotic Recombination: Meiotic Functions Promote a Highly Differentiated Interhomolog-Only Pathway. Cell 1997, 90, 1123–1135. [Google Scholar] [CrossRef]
- Da Ines, O.; Degroote, F.; Goubely, C.; Amiard, S.; Gallego, M.; White, C. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 2013, 9, e1003787. [Google Scholar] [CrossRef]
- Joyce, E.; Paul, A.; Chen, K.; Tanneti, N.; McKim, K. Multiple barriers to nonhomologous DNA end joining during meiosis in Drosophila. Genetics 2012, 191, 739–746. [Google Scholar] [CrossRef]
- Li, W.; Yanowitz, J. ATM and ATR Influence Meiotic Crossover Formation Through Antagonistic and Overlapping Functions in Caenorhabditis elegans. Genetics 2019, 212, 431–443. [Google Scholar] [CrossRef]
- Macaisne, N.; Kessler, Z.; Yanowitz, J. Meiotic Double-Strand Break Proteins Influence Repair Pathway Utilization. Genetics 2018, 210, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Mateo, A.-R.F.; Kessler, Z.; Jolliffe, A.K.; McGovern, O.; Yu, B.; Nicolucci, A.; Yanowitz, J.L.; Derry, W.B. The p53-like Protein CEP-1 Is Required for Meiotic Fidelity in C. elegans. Curr. Biol. 2016, 26, 1148. [Google Scholar] [CrossRef]
- Dumont, J.; Oegema, K.; Desai, A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 2010, 12, 894–901. [Google Scholar] [CrossRef]
- Yadav, V.; Claeys Bouuaert, C. Mechanism and Control of Meiotic DNA Double-Strand Break Formation in S. cerevisiae. Front. Cell Dev. Biol. 2021, 9, 642737. [Google Scholar] [CrossRef]
- Rubin, T.; Macaisne, N.; Huynh, J. Mixing and Matching Chromosomes during Female Meiosis. Cells 2020, 9, 696. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez Pinzón, Y.; González Kise, J.; Rueda, P.; Ronceret, A. The Formation of Bivalents and the Control of Plant Meiotic Recombination. Front. Plant Sci. 2021, 12, 717423. [Google Scholar] [CrossRef]
- Peterson, N.G.; Fox, D.T. Communal living: The role of polyploidy and syncytia in tissue biology. Chromosome Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Edgar, B.A.; Zielke, N.; Gutierrez, C. Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 2014, 15, 197–210. [Google Scholar] [CrossRef]
- Frawley, L.E.; Orr-Weaver, T.L. Polyploidy. Curr. Biol. 2015, 25, R353–R358. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.; Cragg, M.S.; Erenpreisa, J.; Emzinsh, D.; Lukman, H.; Illidge, T.M. Endopolyploid cells produced after severe genotoxic damage have the potential to repair DNA double strand breaks. J. Cell Sci. 2003, 116, 4095–4106. [Google Scholar] [CrossRef]
- Chitnis, M.M.; Lodhia, K.A.; Aleksic, T.; Gao, S.; Protheroe, A.S.; Macaulay, V.M. IGF-1R inhibition enhances radiosensitivity and delays double-strand break repair by both non-homologous end-joining and homologous recombination. Oncogene 2013, 33, 5262–5273. [Google Scholar] [CrossRef] [PubMed]
- Colin, D.J.; Limagne, E.; Ragot, K.; Lizard, G.; Ghiringhelli, F.; Solary, É.; Chauffert, B.; Latruffe, N.; Delmas, D. The role of reactive oxygen species and subsequent DNA-damage response in the emergence of resistance towards resveratrol in colon cancer models. Cell Death Dis. 2014, 5, e1533. [Google Scholar] [CrossRef]
- Shang, Z.-F.; Huang, B.; Xu, Q.-Z.; Zhang, S.-M.; Fan, R.; Liu, X.-D.; Wang, Y.; Zhou, P.-K. Inactivation of DNA-Dependent Protein Kinase Leads to Spindle Disruption and Mitotic Catastrophe with Attenuated Checkpoint Protein 2 Phosphorylation in Response to DNA Damage. Cancer Res. 2010, 70, 3657–3666. [Google Scholar] [CrossRef]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Fox, D.T.; Duronio, R.J. Endoreplication and polyploidy: Insights into development and disease. Development 2013, 140, 3–12. [Google Scholar] [CrossRef]
- Schoenfelder, K.P.; Fox, D.T. The expanding implications of polyploidy. J. Cell Biol. 2015, 209, 485–491. [Google Scholar] [CrossRef]
- Mahapatra, K.; Roy, S. An insight into the mechanism of DNA damage response in plants- role of SUPPRESSOR OF γ RESPONSE 1: An overview. Mutat. Res. Mol. Mech. Mutagen. 2020, 819–820, 111689. [Google Scholar] [CrossRef]
- Cohen, E.; Allen, S.R.; Sawyer, J.K.; Fox, D.T. Fizzy-related dictates a cell cycle switch during organ repair and tissue growth responses in the drosophila hindgut. Elife 2018, 7, e38327. [Google Scholar] [CrossRef]
- Grendler, J.; Lowgren, S.; Mills, M.; Losick, V.P. Wound-induced polyploidization is driven by Myc and supports tissue repair in the presence of DNA damage. Development 2019, 146, 173005. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.; Peterson, N.G.; Sawyer, J.K.; Fox, D.T. Accelerated cell cycles enable organ regeneration under developmental time constraints in the Drosophila hindgut. Dev. Cell 2021, 56, 2059–2072. [Google Scholar] [CrossRef]
- Stormo, B.M.; Fox, D.T. Polyteny: Still a giant player in chromosome research. Chromosom. Res. 2017, 25, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Yarosh, W.; Spradling, A.C. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev. 2014, 28, 1840–1855. [Google Scholar] [CrossRef]
- Alexander, J.L.; Beagan, K.; Orr-Weaver, T.L.; McVey, M. Multiple mechanisms contribute to double-strand break repair at rereplication forks in Drosophila follicle cells. Proc. Natl. Acad. Sci. USA 2016, 113, 13809–13814. [Google Scholar] [CrossRef]
- Mehrotra, S.; Maqbool, S.B.; Kolpakas, A.; Murnen, K.; Calvi, B.R. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008, 22, 3158–3171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Mehrotra, S.; Ng, W.L.; Calvi, B.R. Low levels of p53 protein and chromatin silencing of p53 target genes repress apoptosis in Drosophila endocycling cells. PLoS Genet. 2014, 10, e1004581. [Google Scholar] [CrossRef]
- Guarner, A.; Morris, R.; Korenjak, M.; Boukhali, M.; Zappia, M.P.; Van Rechem, C.; Whetstine, J.R.; Ramaswamy, S.; Zou, L.; Frolov, M.V.; et al. E2F/DP Prevents Cell-Cycle Progression in Endocycling Fat Body Cells by Suppressing dATM Expression. Dev. Cell 2017, 43, 689–703.e5. [Google Scholar] [CrossRef]
- Vazquez-Martin, A.; Anatskaya, O.V.; Giuliani, A.; Erenpreisa, J.; Huang, S.; Salmina, K.; Inashkina, I.; Huna, A.; Nikolsky, N.N.; Vinogradov, A.E. Somatic polyploidy is associated with the upregulation of c-MYC interacting genes and EMT-like signature. Oncotarget 2016, 7, 75235. [Google Scholar] [CrossRef]
- Nano, M.; Gemble, S.; Simon, A.; Pennetier, C.; Fraisier, V.; Marthiens, V.; Basto, R. Cell-Cycle Asynchrony Generates DNA Damage at Mitotic Entry in Polyploid Cells. Curr. Biol. 2019, 29, 3937–3945.e7. [Google Scholar] [CrossRef]
- Chitikova, Z.V.; Gordeev, S.A.; Bykova, T.V.; Zubova, S.G.; Pospelov, V.A.; Pospelova, T.V. Sustained activation of DNA damage response in irradiated apoptosis-resistant cells induces reversible senescence associated with mTOR downregulation and expression of stem cell markers. Cell Cycle 2014, 13, 1424–1439. [Google Scholar] [CrossRef]
- Horikawa, D.D.; Cumbers, J.; Sakakibara, I.; Rogoff, D.; Leuko, S.; Harnoto, R.; Arakawa, K.; Katayama, T.; Kunieda, T.; Toyoda, A.; et al. Analysis of DNA repair and protection in the Tardigrade Ramazzottius varieornatus and Hypsibius dujardini after exposure to UVC radiation. PLoS ONE 2013, 8, e64793. [Google Scholar] [CrossRef]
- Horikawa, D.D.; Sakashita, T.; Katagiri, C.; Watanabe, M.; Kikawada, T.; Nakahara, Y.; Hamada, N.; Wada, S.; Funayama, T.; Higashi, S.; et al. Radiation tolerance in the tardigrade Milnesium tardigradum. Int. J. Radiat. Biol. 2006, 82, 843–848. [Google Scholar] [CrossRef]
- Jönsson, K.I.; Harms-Ringdahl, M.; Torudd, J. Radiation tolerance in the eutardigrade Richtersius coronifer. Int. J. Radiat. Biol. 2009, 81, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-Pardo, E.; Jönsson, K.I.; Harms-Ringdahl, M.; Haghdoost, S.; Wojcik, A. Tolerance to γ Radiation in the Tardigrade Hypsibius dujardini from Embryo to Adult Correlate Inversely with Cellular Proliferation. PLoS ONE 2015, 10, e0133658. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, K.I.; Rabbow, E.; Schill, R.O.; Harms-Ringdahl, M.; Rettberg, P. Tardigrades survive exposure to space in low Earth orbit. Curr. Biol. 2008, 18, R729–R731. [Google Scholar] [CrossRef]
- Jönsson, K.I.; Wojcik, A. Tolerance to X-rays and Heavy Ions (Fe, He) in the Tardigrade Richtersius coronifer and the Bdelloid Rotifer Mniobia russeola. Astrobiology 2017, 17, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Horikawa, D.D.; Saito, Y.; Kuwahara, H.; Kozuka-Hata, H.; Shin, I.T.; Minakuchi, Y.; Ohishi, K.; Motoyama, A.; Aizu, T.; et al. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat. Commun. 2016, 7, 12808. [Google Scholar] [CrossRef]
- Chavez, C.; Cruz-Becerra, G.; Fei, J.; Kassavetis, G.A.; Kadonaga, J.T. The tardigrade damage suppressor protein binds to nucleosomes and protects DNA from hydroxyl radicals. Elife 2019, 8, e47682. [Google Scholar] [CrossRef]
- Delmas, S.; Shunburne, L.; Ngo, H.-P.; Allers, T. Mre11-Rad50 Promotes Rapid Repair of DNA Damage in the Polyploid Archaeon Haloferax volcanii by Restraining Homologous Recombination. PLOS Genet. 2009, 5, e1000552. [Google Scholar] [CrossRef]
- Bentchikou, E.; Servant, P.; Coste, G.; Sommer, S. Additive Effects of SbcCD and PolX Deficiencies in the In Vivo Repair of DNA Double-Strand Breaks in Deinococcus radiodurans. J. Bacteriol. 2007, 189, 4784. [Google Scholar] [CrossRef]
- Doré, A.S.; Drake, A.C.B.; Brewerton, S.C.; Blundell, T.L. Identification of DNA-PK in the arthropods: Evidence for the ancient ancestry of vertebrate non-homologous end-joining. DNA Repair 2004, 3, 33–41. [Google Scholar] [CrossRef]
- Block, W.; Lees-Miller, S. Putative homologues of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and other components of the non-homologous end joining machinery in Dictyostelium discoideum. DNA Repair 2005, 4, 1061–1065. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, L.; Pachebat, J.A.; Glöckner, G.; Rajandream, M.A.; Sucgang, R.; Berriman, M.; Song, J.; Olsen, R.; Szafranski, K.; Xu, Q.; et al. The genome of the social amoeba Dictyostelium discoideum. Nature 2005, 435, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Smith-Stocking, H. Genetic Studies on Selective Segregation of Chromosomes in Sciara Coprophila Lintner. Genetics 1936, 21, 421–443. [Google Scholar] [CrossRef]
- Metz, C.W.; Boche, R.D. Observations on the Mechanism of Induced Chromosome Rearrangements in Sciara. Proc. Natl. Acad. Sci. USA 1939, 25, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Crouse, H.V. The resistance of Sciara (Diptera) to the mutagenic effects of irradiation. Biol. Bull. 1949, 97, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Bozeman, M.; Metz, C. Further studies on sensitivity of chromosomes to irradiation at different meiotic stages in oöcytes of Sciara. Genetics 1949, 34, 285–314. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.; Foulk, M.; Bliss, J.; Coleman, C.; Lu, N.; Mazloom, R.; Brown, S.; Spradling, A.; Gerbi, S. High contiguity de novo genome assembly and DNA modification analyses for the fungus fly, Sciara coprophila, using single-molecule sequencing. BMC Genomics 2021, 22, 643. [Google Scholar] [CrossRef]
- Latif, C.; Harvey, S.H.; O’Connell, M.J. Ensuring the stability of the genome: DNA damage checkpoints. Sci. World J. 2001, 1, 684–702. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clay, D.E.; Fox, D.T. DNA Damage Responses during the Cell Cycle: Insights from Model Organisms and Beyond. Genes 2021, 12, 1882. https://doi.org/10.3390/genes12121882
Clay DE, Fox DT. DNA Damage Responses during the Cell Cycle: Insights from Model Organisms and Beyond. Genes. 2021; 12(12):1882. https://doi.org/10.3390/genes12121882
Chicago/Turabian StyleClay, Delisa E., and Donald T. Fox. 2021. "DNA Damage Responses during the Cell Cycle: Insights from Model Organisms and Beyond" Genes 12, no. 12: 1882. https://doi.org/10.3390/genes12121882
APA StyleClay, D. E., & Fox, D. T. (2021). DNA Damage Responses during the Cell Cycle: Insights from Model Organisms and Beyond. Genes, 12(12), 1882. https://doi.org/10.3390/genes12121882





