Evaluation of Reference Genes for Transcriptional Profiling in Two Cockroach Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cockroach Rearing and Sample Preparation
2.2. Candidate Reference Genes and Primer Design
2.3. RNA Isolation, cDNA Synthesis, and qPCR
2.4. Juvenile Hormone (JH) Treatment and RNAi Experiment in B. germanica
2.5. Inhibitor and Antibiotic Treatment in P. americana
2.6. Data Analysis and Statistics
3. Results
3.1. Validation of Primer Sets
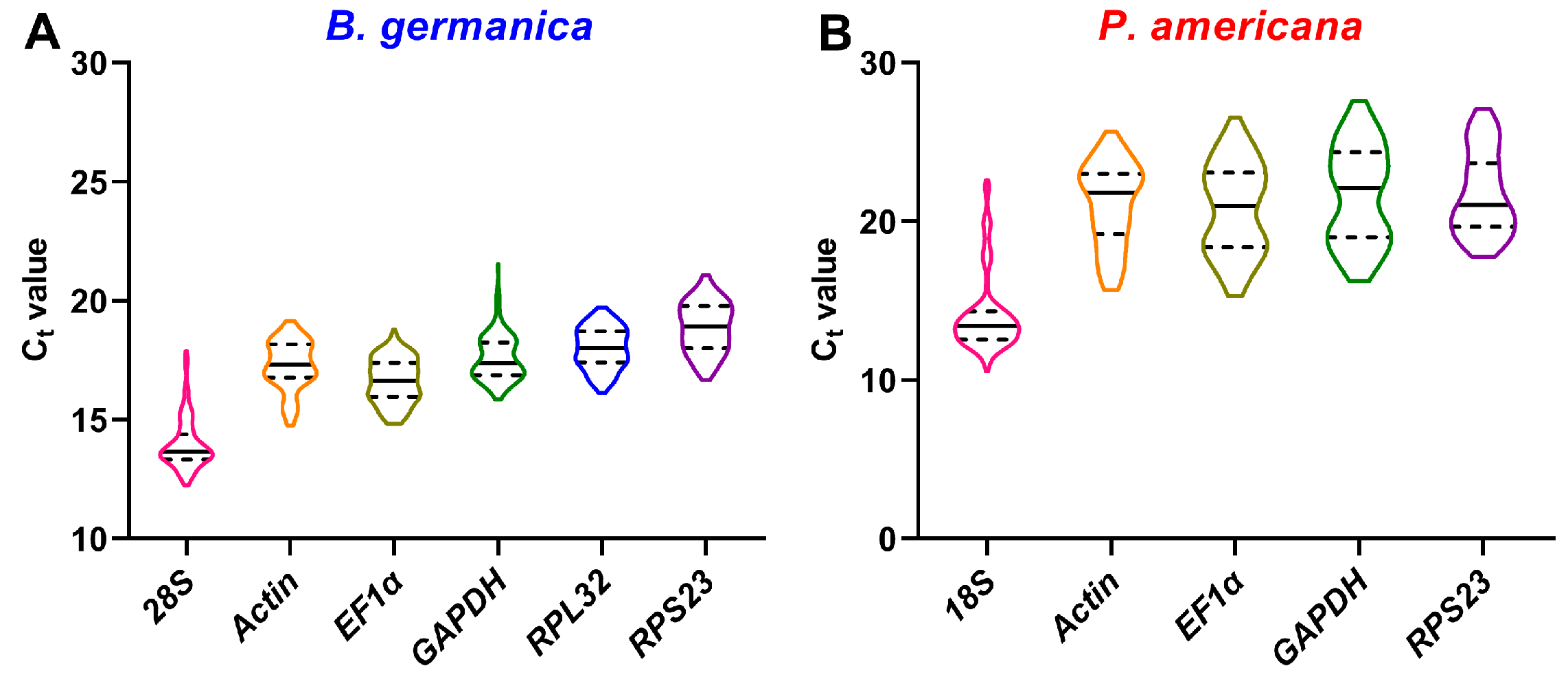
3.2. Transcriptional Profiles of Candidate Reference Genes
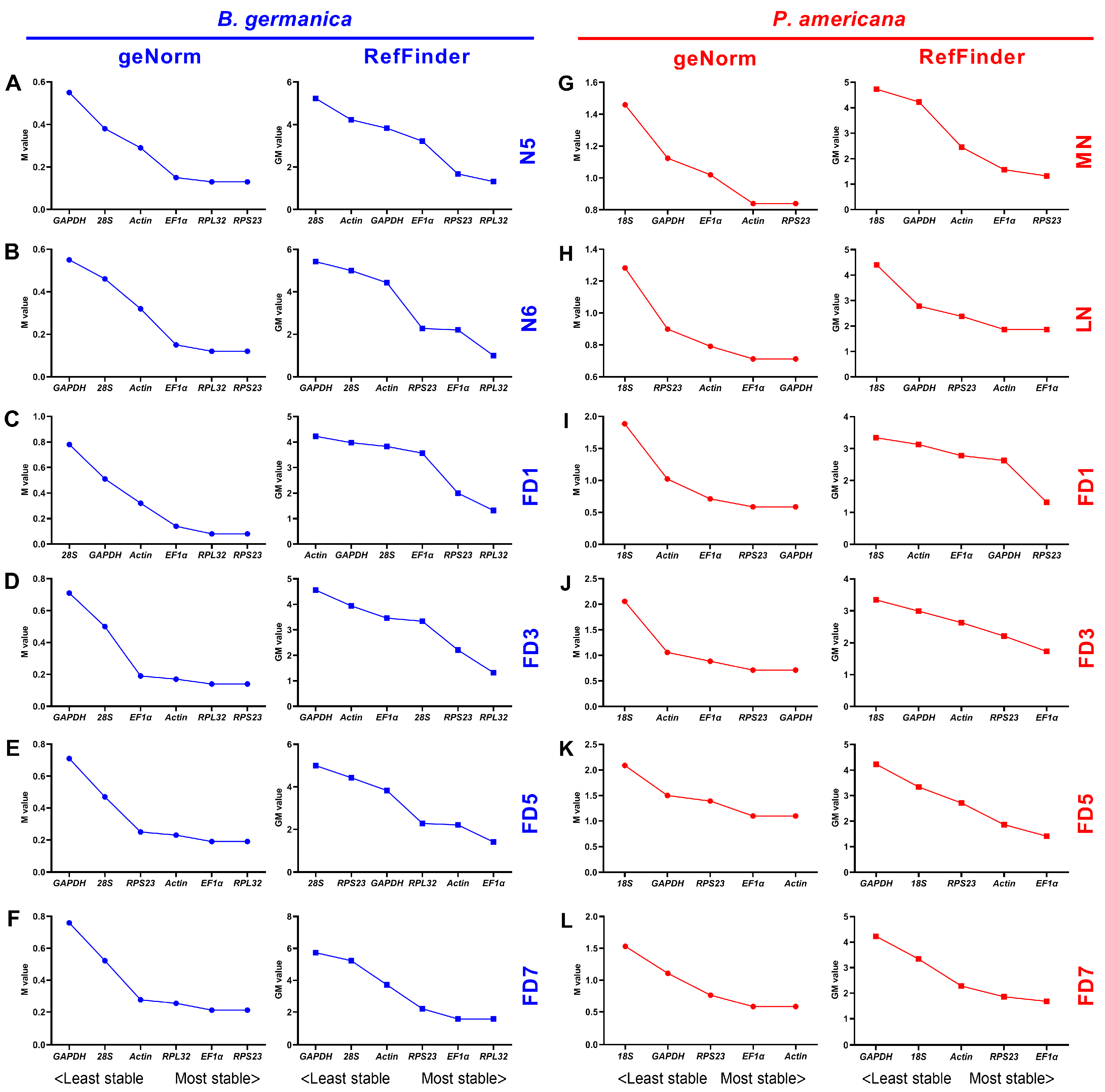
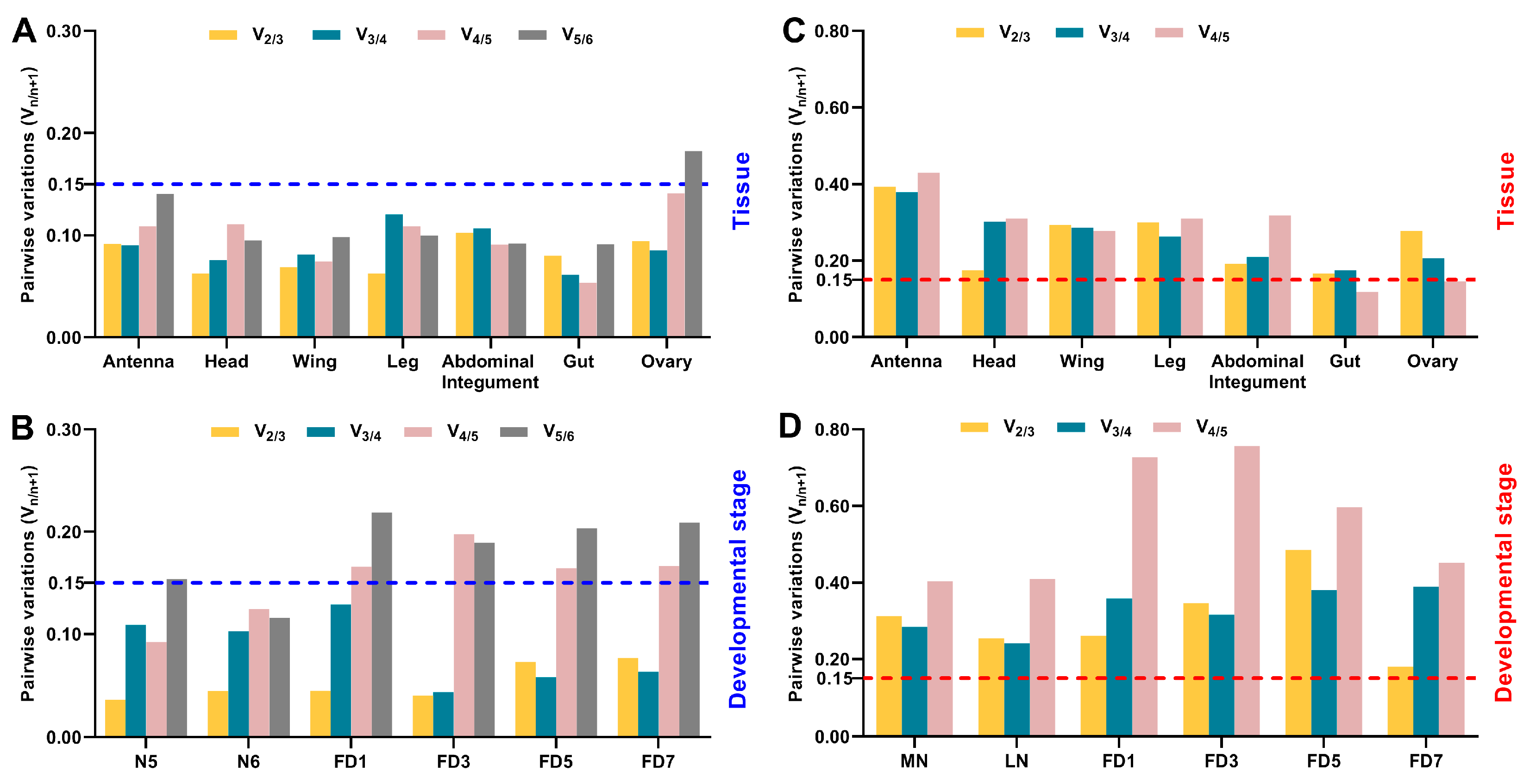
3.3. Expression Stability of Candidate Reference Genes throughout Various Stages in Specific Tissues
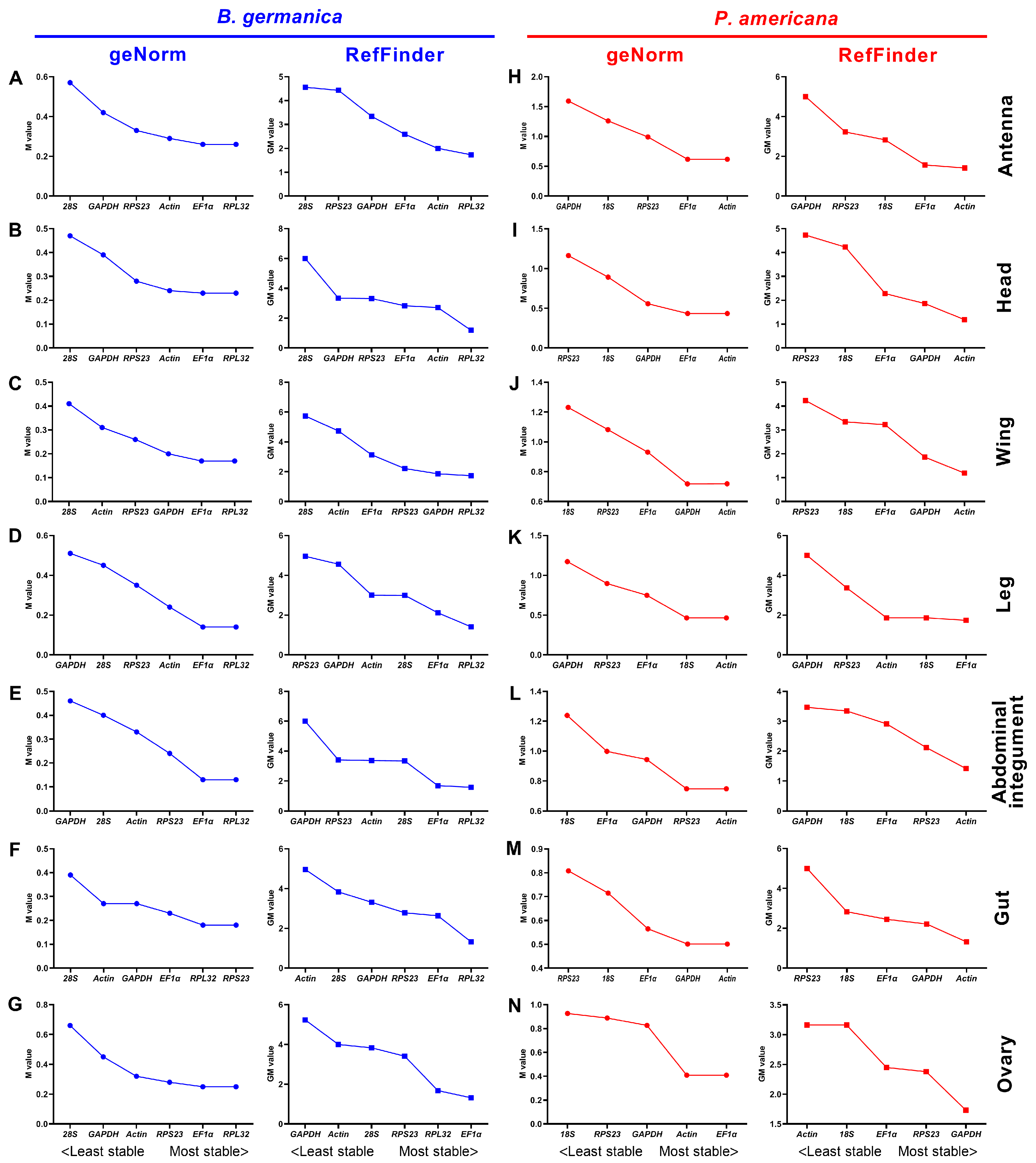
3.4. Expression Stability of Candidate Reference Genes across Various Tissues at Given Stages
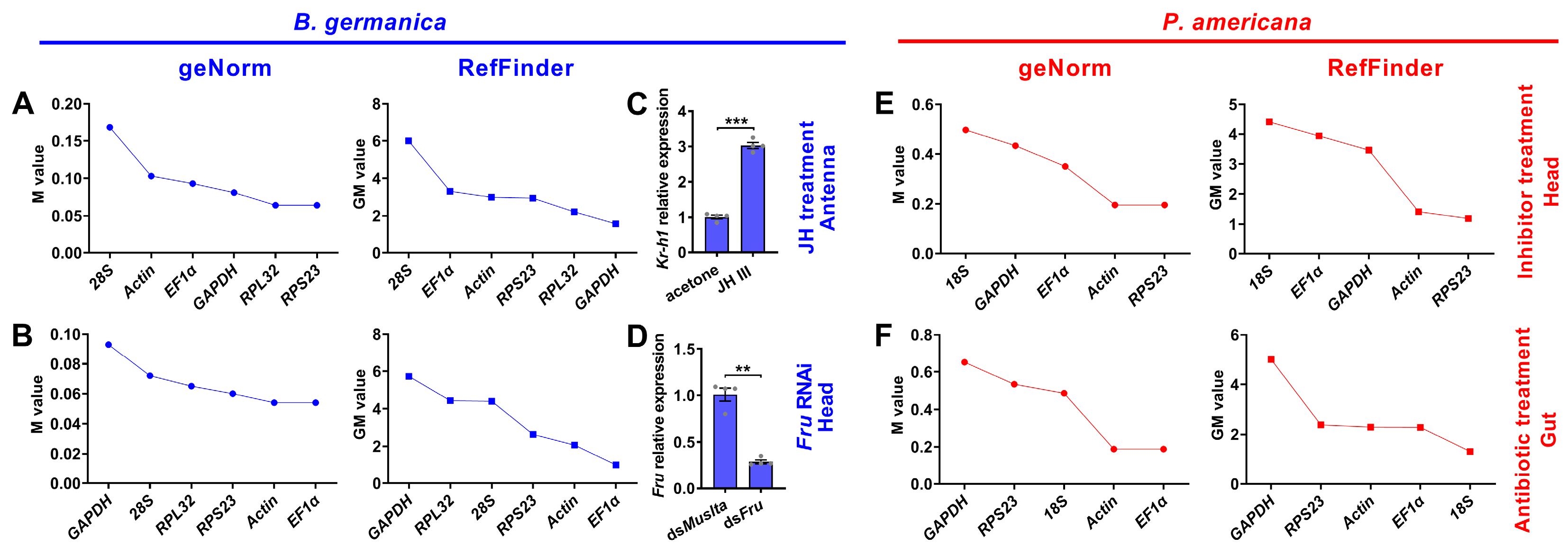
3.5. Expression Stability of Candidate Reference Genes under Experimental Treatment Conditions
3.6. Optimization of Gene Numbers Needed for qPCR Normalization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bell, W.J.; Roth, L.M.; Nalepa, C.A. Cockroaches: Ecology, Behavior, and Natural History; The Johns Hopkins University Press: Baltimore, MD, USA, 2007; pp. 1–6. [Google Scholar]
- Gore, J.C.; Schal, C. Cockroach Allergen Biology and Mitigation in the Indoor Environment. Annu. Rev. Entomol. 2007, 52, 439–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, P.J. The Cockroach as a Vector of Pathogenic Agents. Bol. Ofcina. Sanit. Panam. 1989, 107, 41–53. [Google Scholar]
- Treiblmayr, K.; Pascual, N.; Piulachs, M.-D.; Keller, T.; Belles, X. Juvenile Hormone Titer versus Juvenile Hormone Synthesis in Female Nymphs and Adults of the German Cockroach, Blattella germanica. J. Insect Sci. 2006, 6, 43. [Google Scholar] [CrossRef] [Green Version]
- Mané-Padrós, D.; Cruz, J.; Vilaplana, L.; Pascual, N.; Belles, X.; Martín, D. The Nuclear Hormone Receptor BgE75 Links Molting and Developmental Progression in the Direct-Developing Insect Blattella germanica. Dev. Biol. 2008, 315, 147–160. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Orte, E.; Belles, X. MicroRNA-Dependent Metamorphosis in Hemimetabolan Insects. Proc. Natl. Acad. Sci. USA 2009, 106, 21678–21682. [Google Scholar] [CrossRef] [Green Version]
- Lozano, J.; Belles, X. Conserved Repressive Function of Krüppel Homolog 1 on Insect Metamorphosis in Hemimetabolous and Holometabolous Species. Sci. Rep. 2011, 1, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belles, X.; Santos, C. The MEKRE93 (Methoprene Tolerant-Krüppel Homolog 1-E93) Pathway in the Regulation of Insect Metamorphosis, and the Homology of the Pupal Stage. Insect Biochem. Mol. Biol. 2014, 52, 60–68. [Google Scholar] [CrossRef]
- Li, S.; Zhu, S.; Jia, Q.; Yuan, D.; Ren, C.; Li, K.; Liu, S.; Cui, Y.; Zhao, H.; Cao, Y.; et al. The Genomic and Functional Landscapes of Developmental Plasticity in the American Cockroach. Nat. Commun. 2018, 9, 1008. [Google Scholar] [CrossRef]
- Wexler, J.; Delaney, E.K.; Belles, X.; Shcal, C.; Wada-Katsumata, A.; Amicucci, K.; Kopp, A. Hemimetabolous Insects Elucidate the Origin of Sexual Development via Alternative Splicing. eLife 2019, 8, e47490. [Google Scholar] [CrossRef]
- Maestro, J.; Cobo, J.; Belles, X. Target of Rapamycin (TOR) Mediates the Transduction of Nutritional Signals into Juvenile Hormone Production. J. Biol. Chem. 2009, 284, 5506–5513. [Google Scholar] [CrossRef] [Green Version]
- Süren-Castillo, S.; Abrisqueta, M.; Maestro, J. FoxO Inhibits Juvenile Hormone Biosynthesis and Vitellogenin Production in the German Cockroach. Insect Biochem. Mol. Biol. 2012, 42, 491–498. [Google Scholar] [CrossRef] [Green Version]
- Abrisqueta, M.; Süren-Castillo, S.; Maestro, J. Insulin Receptor-Mediated Nutritional Signalling Regulates Juvenile Hormone Biosynthesis and Vitellogenin Production in the German Cockroach. Insect Biochem. Mol. Biol. 2014, 49, 14–23. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Liu, F.; Zeng, H.; Li, N.; Ren, C.; Su, Y.; Zhou, S.; Wang, G.; Palli, S.R.; Wang, J.; et al. Insulin/IGF Signaling and TORC1 Promote Vitellogenesis via Inducing Juvenile Hormone Biosynthesis in the American Cockroach. Development 2020, 147, dev188805. [Google Scholar] [CrossRef]
- Okada, K.; Mori, M.; Shimazaki, K.; Chuman, T. Behavioral Responses of Male Periplaneta americana L. to Female Sex Pheromone Components, Periplanone-A and Periplanone-B. J. Chem. Ecol. 1990, 16, 2605–2614. [Google Scholar] [CrossRef]
- Schal, C.; Burns, E.; Gadot, M.; Chase, J.; Blomquist, G.J. Biochemistry and Regulation of Pheromone Production in Blattella germanica (L.) (Dictyoptera, Blattellidae). Insect Biochem. 1991, 21, 73–79. [Google Scholar] [CrossRef]
- Chase, J.; Touhara, K.; Prestwich, G.D.; Schal, C.; Blomquist, G.J. Biosynthesis and Endocrine Control of the Production of the German Cockroach Sex Pheromone 3,11-Dimethylnonacosan-2-One. Proc. Natl. Acad. Sci. USA 1992, 89, 6050–6054. [Google Scholar] [CrossRef] [Green Version]
- Nojima, S.; Schal, C.; Webster, F.X.; Santangelo, R.G.; Roelofs, W.L. Identification of the Sex Pheromone of the German Cockroach, Blattella germanica. Science 2005, 307, 1104–1106. [Google Scholar] [CrossRef]
- Eliyahu, D.; Nojima, S.; Mori, K.; Schal, C. New Contact Sex Pheromone Components of the German Cockroach, Blattella Germanica, Predicted from the Proposed Biosynthetic Pathway. J. Chem. Ecol. 2008, 34, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Wada-Katsumata, A.; Schal, C. Antennal Grooming Facilitates Courtship Performance in a Group-Living Insect, the German Cockroach Blattella germanica. Sci. Rep. 2019, 9, 2942. [Google Scholar] [CrossRef]
- Tinker, K.A.; Ottesen, E.A. The Core Gut Microbiome of the American Cockroach, Periplaneta americana, is Stable and Resilient to Dietary Shifts. Appl. Environ. Microbiol. 2016, 82, 6603–6610. [Google Scholar] [CrossRef] [Green Version]
- Jahnes, B.C.; Herrmann, M.; Sabree, Z.L. Conspecific Coprophagy Stimulates Normal Development in a Germ-Free Model Invertebrate. PeerJ 2019, 7, e6914. [Google Scholar] [CrossRef]
- Guzman, J.; Vilcinskas, A. Bacteria Associated with Cockroaches: Health Risk or Biotechnological Opportunity? Appl. Microbiol. Biotechnol. 2020, 104, 10369–10387. [Google Scholar] [CrossRef]
- Vera-Ponce de Leon, A.; Jahnes, B.C.; Otero-Bravo, A.; Sabree, Z.L. Microbiota Perturbation or Elimination Can Inhibit Normal Development and Elicit a Starvation-like Response in an Omnivorous Model Invertebrate. mSystems 2021, 6, e00802–e00821. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA Using Real-Time Reverse Transcription PCR (RT-PCR): Trends and Problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Zhou, X.; Qian, K.; Tong, Y.; Zhu, J.; Qiu, X.; Zeng, X. De Novo Transcriptome of the Hemimetabolous German Cockroach (Blattella germanica). PLoS ONE 2014, 9, e106932. [Google Scholar] [CrossRef]
- Niu, D.-J.; Liu, Y.; Dong, X.-T.; Dong, S.-L. Transcriptome Based Identification and Tissue Expression Profiles of Chemosensory Genes in Blattella germanica (Blattaria: Blattidae). Comp. Biochem. Physiol. D 2016, 18, 30–43. [Google Scholar] [CrossRef]
- Chen, N.; Pei, X.; Li, S.; Fan, Y.-L.; Liu, T.-X. Involvement of integument-rich CYP4G19 in hydrocarbon biosynthesis and cuticular penetration resistance in Blattella germanica (L.). Pest. Manag. Sci. 2019, 76, 215–226. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Harrison, M.C.; Jongepier, E.; Robertson, H.M.; Arning, M.; Bitard-Feildel, T.; Chao, H.; Childers, C.P.; Dinh, H.; Doddapaneni, H.; Dugan, S.; et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2018, 2, 557–566. [Google Scholar] [CrossRef] [Green Version]
- Clynen, E.; Ciudad, L.; Bellés, X.; Piulachs, M.-D. Conservation of fruitless’ role as master regulator of male courtship behaviour from cockroaches to flies. Dev. Genes Evol. 2011, 221, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ylla, G.; Piulachs, M.-D.; Belles, X. Comparative Transcriptomics in Two Extreme Neopterans Reveal General Trends in the Evolution of Modern Insects. Iscience 2018, 4, 164–179. [Google Scholar] [CrossRef] [Green Version]
- Pei, X.-J.; Fan, Y.-L.; Bai, Y.; Bai, T.-T.; Schal, C.; Zhang, Z.-F.; Chen, N.; Li, S.; Liu, T.-X. Modulation of Fatty Acid Elongation in Cockroaches Sustains Sexually Dimorphic Hydrocarbons and Female Attractiveness. PLoS Biol. 2021, 19, e3001330. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper—Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant. Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Jacobsen, A.V.; Yemaneab, B.T.; Jass, J.; Scherbak, N. Reference Gene Selection for qPCR Is Dependent on Cell Type Rather Than Treatment in Colonic and Vaginal Human Epithelial Cell Lines. PLoS ONE 2014, 9, e115592. [Google Scholar] [CrossRef]
- Rocha-Martins, M.; Njaine, B.; Silveira, M.S. Avoiding Pitfalls of Internal Controls: Validation of Reference Genes for Analysis by qRT-PCR and Western Blot throughout Rat Retinal Development. PLoS ONE 2012, 7, e43028. [Google Scholar] [CrossRef]
- Uddin, M.J.; Cinar, M.U.; Tesfaye, D.; Looft, C.; Tholen, E.; Schellander, K. Age-Related Changes in Relative Expression Stability of Commonly Used Housekeeping Genes in Selected porciNE Tissues. BMC Res. Notes 2011, 4, 441. [Google Scholar] [CrossRef] [Green Version]
| Rank | Delta Ct | geNorm | BestKeeper | NormFinder | RefFinder | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Average Ct | SD | Gene Name | M | Gene Name | CV | Gene Name | SV | Gene Name | GM | |
| 1 | RPL32 | 18.00 | 0.57 | RPL32 | 0.24 | RPL32 | 4.03 | RPL32 | 0.12 | RPL32 | 1.00 |
| 2 | EF1α | 16.62 | 0.60 | EF1α | 0.24 | GAPDH | 4.39 | EF1α | 0.22 | EF1α | 2.00 |
| 3 | RPS23 | 18.87 | 0.64 | RPS23 | 0.31 | RPS23 | 4.69 | RPS23 | 0.31 | RPS23 | 3.57 |
| 4 | Actin | 17.28 | 0.67 | Actin | 0.36 | EF1α | 4.73 | Actin | 0.37 | Actin | 4.23 |
| 5 | 28S | 13.94 | 1.06 | 28S | 0.61 | Actin | 4.74 | 28S | 0.94 | 28S | 4.40 |
| 6 | GAPDH | 17.58 | 1.10 | GAPDH | 0.77 | 28S | 5.55 | GAPDH | 0.98 | GAPDH | 4.56 |
| Rank | Delta Ct | geNorm | BestKeeper | NormFinder | RefFinder | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Average Ct | SD | Gene Name | M | Gene Name | CV | Gene Name | SV | Gene Name | GM | |
| 1 | EF1α | 20.79 | 1.54 | EF1α | 0.98 | RPS23 | 10.38 | EF1α | 0.44 | EF1α | 1.41 |
| 2 | Actin | 21.10 | 1.64 | Actin | 0.98 | Actin | 10.58 | Actin | 0.74 | Actin | 1.68 |
| 3 | RPS23 | 21.76 | 1.77 | RPS23 | 1.25 | 18S | 11.74 | RPS23 | 1.10 | RPS23 | 3.00 |
| 4 | GAPDH | 21.64 | 1.84 | GAPDH | 1.34 | EF1α | 12.00 | GAPDH | 1.32 | 18S | 3.34 |
| 5 | 18S | 14.07 | 2.75 | 18S | 1.91 | GAPDH | 12.62 | 18S | 2.59 | GAPDH | 4.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, S.; Liu, Y.; Liao, M.; Yang, Y.; Bai, Y.; Li, N.; Li, S.; Luan, Y.; Chen, N. Evaluation of Reference Genes for Transcriptional Profiling in Two Cockroach Models. Genes 2021, 12, 1880. https://doi.org/10.3390/genes12121880
Zhu S, Liu Y, Liao M, Yang Y, Bai Y, Li N, Li S, Luan Y, Chen N. Evaluation of Reference Genes for Transcriptional Profiling in Two Cockroach Models. Genes. 2021; 12(12):1880. https://doi.org/10.3390/genes12121880
Chicago/Turabian StyleZhu, Shen, Yongjun Liu, Mingtao Liao, Yang Yang, Yu Bai, Na Li, Sheng Li, Yunxia Luan, and Nan Chen. 2021. "Evaluation of Reference Genes for Transcriptional Profiling in Two Cockroach Models" Genes 12, no. 12: 1880. https://doi.org/10.3390/genes12121880
APA StyleZhu, S., Liu, Y., Liao, M., Yang, Y., Bai, Y., Li, N., Li, S., Luan, Y., & Chen, N. (2021). Evaluation of Reference Genes for Transcriptional Profiling in Two Cockroach Models. Genes, 12(12), 1880. https://doi.org/10.3390/genes12121880







