Propagation of a De Novo Gene under Natural Selection: Antifreeze Glycoprotein Genes and Their Evolutionary History in Codfishes
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen and Sample Collection
2.2. PCR Amplification of mt COI and Sequencing
2.3. Gadid Phylogenetic Analyses
2.4. AFGP Trait Characterization and Mapping to Phylogeny
2.5. Characterization and Comparison of AFGP Loci and Neighboring Genomic Regions
2.6. AFGP Gene Feature and Phylogenetic Analyses
2.7. Codon Usage Analyses of AFGP Tripeptide Repeats
3. Results and Discussion
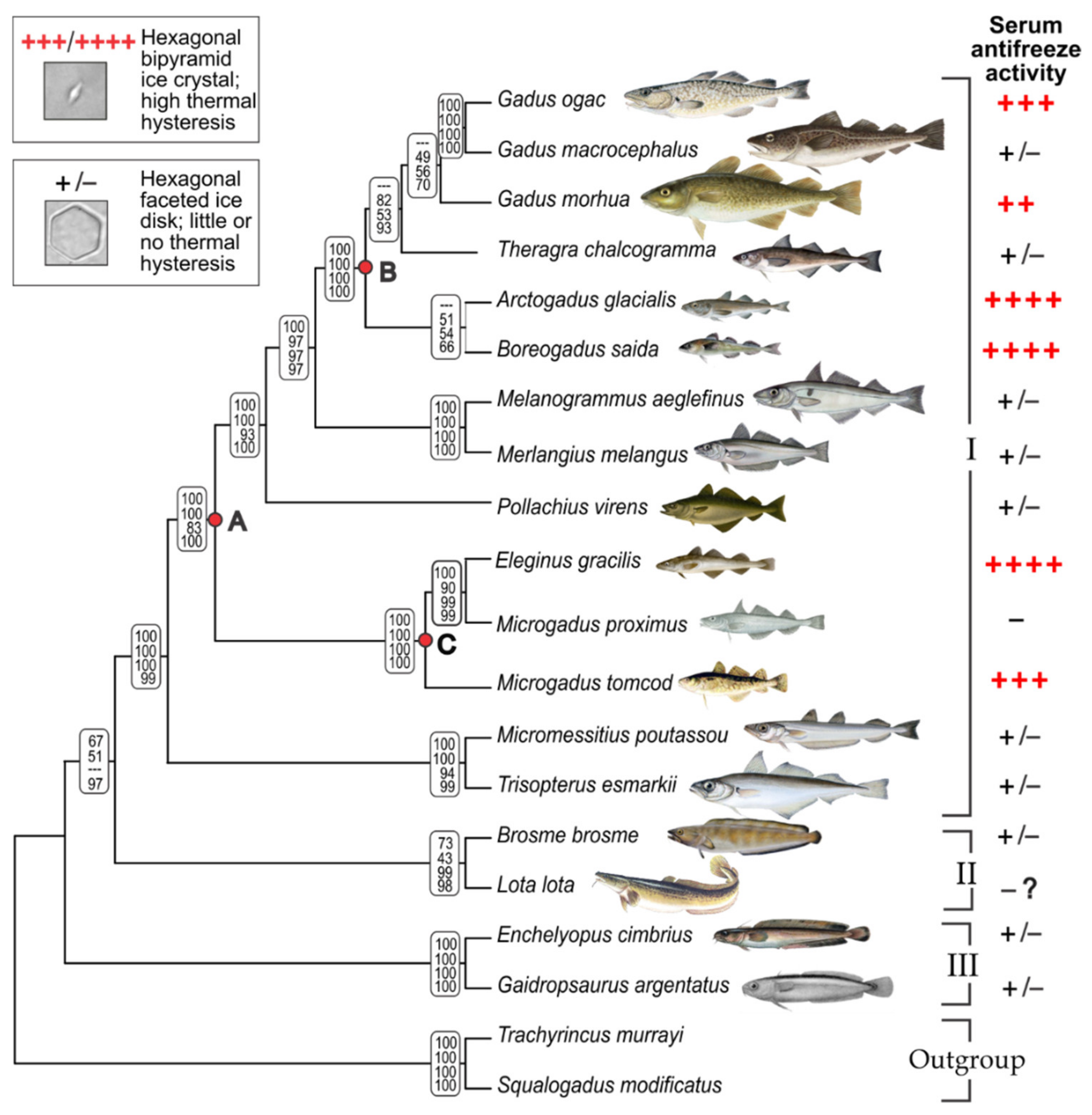
3.1. Gadid Phylogeny and Mapping AFGP Phenotype
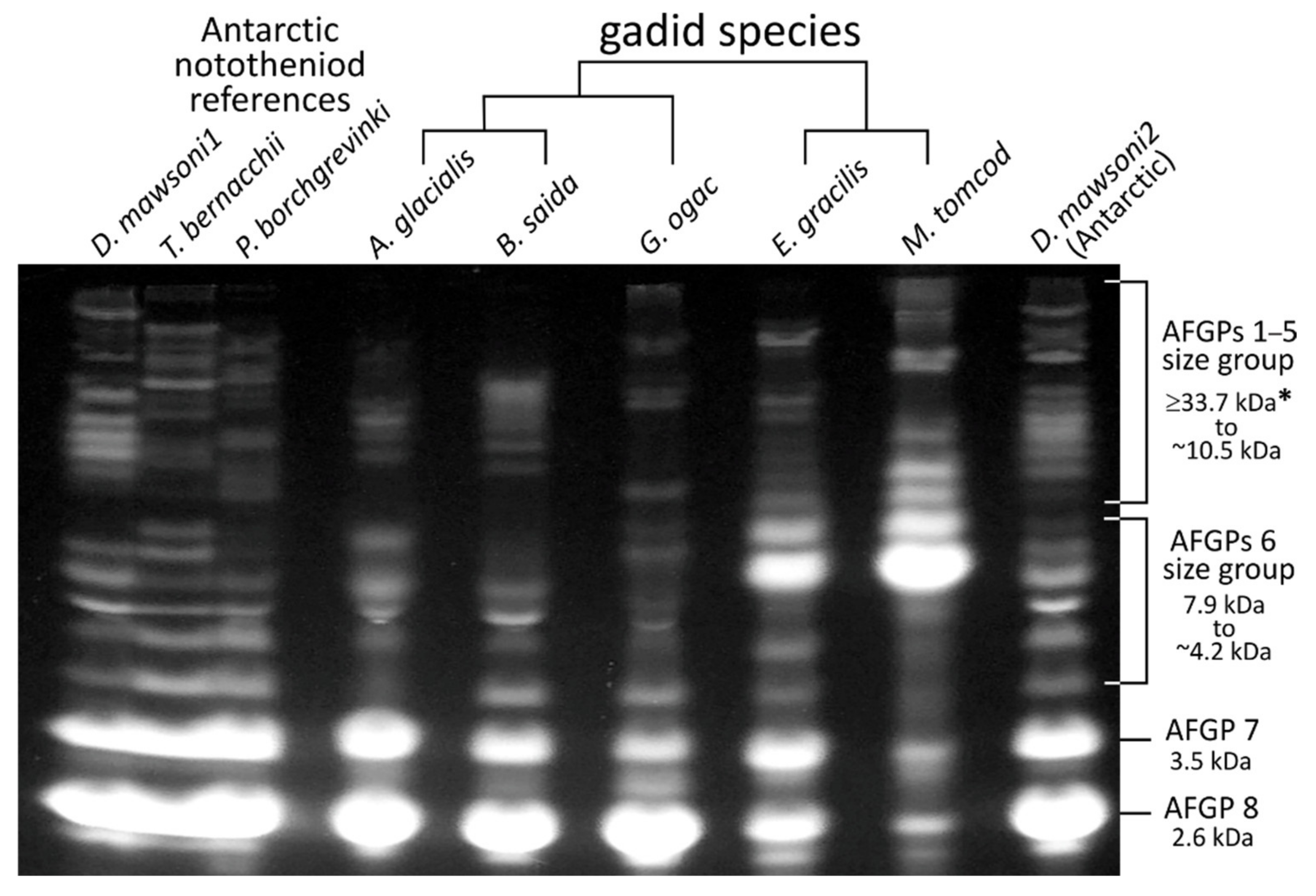
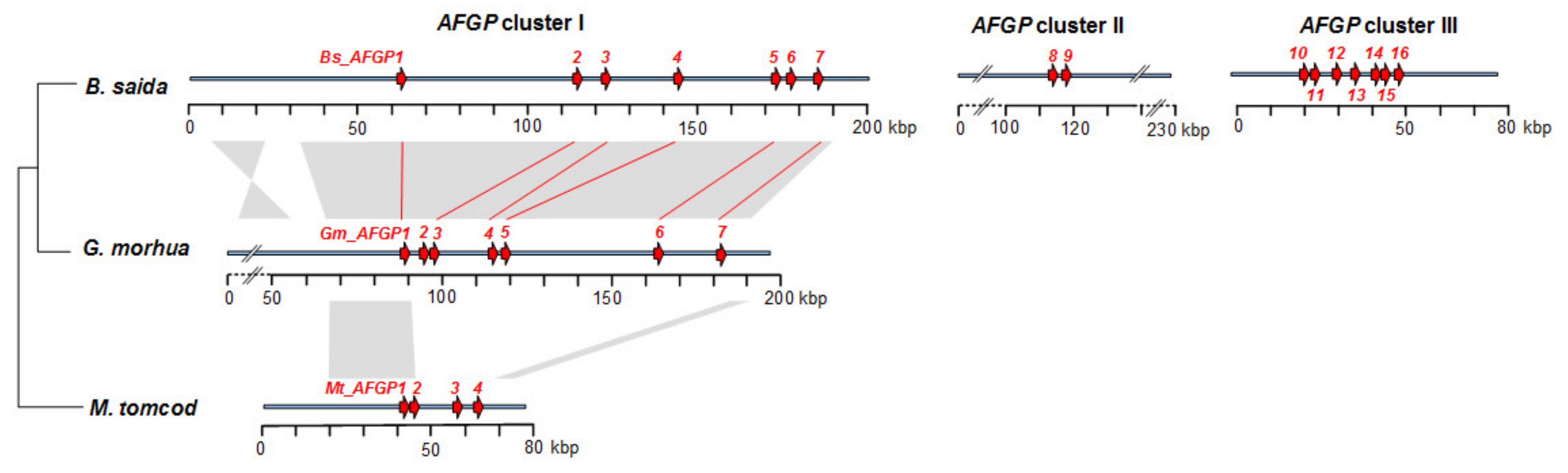
3.2. AFGP Gene Family Genomic Locus
3.3. AFGP Polypeptide Cleavage Sites and Size Isoforms
3.4. Codon Usage of AFGP Tripeptide Repeats
3.5. Molecular Mechanisms of the Duplication and Stabilization of the Tripeptide Repeats
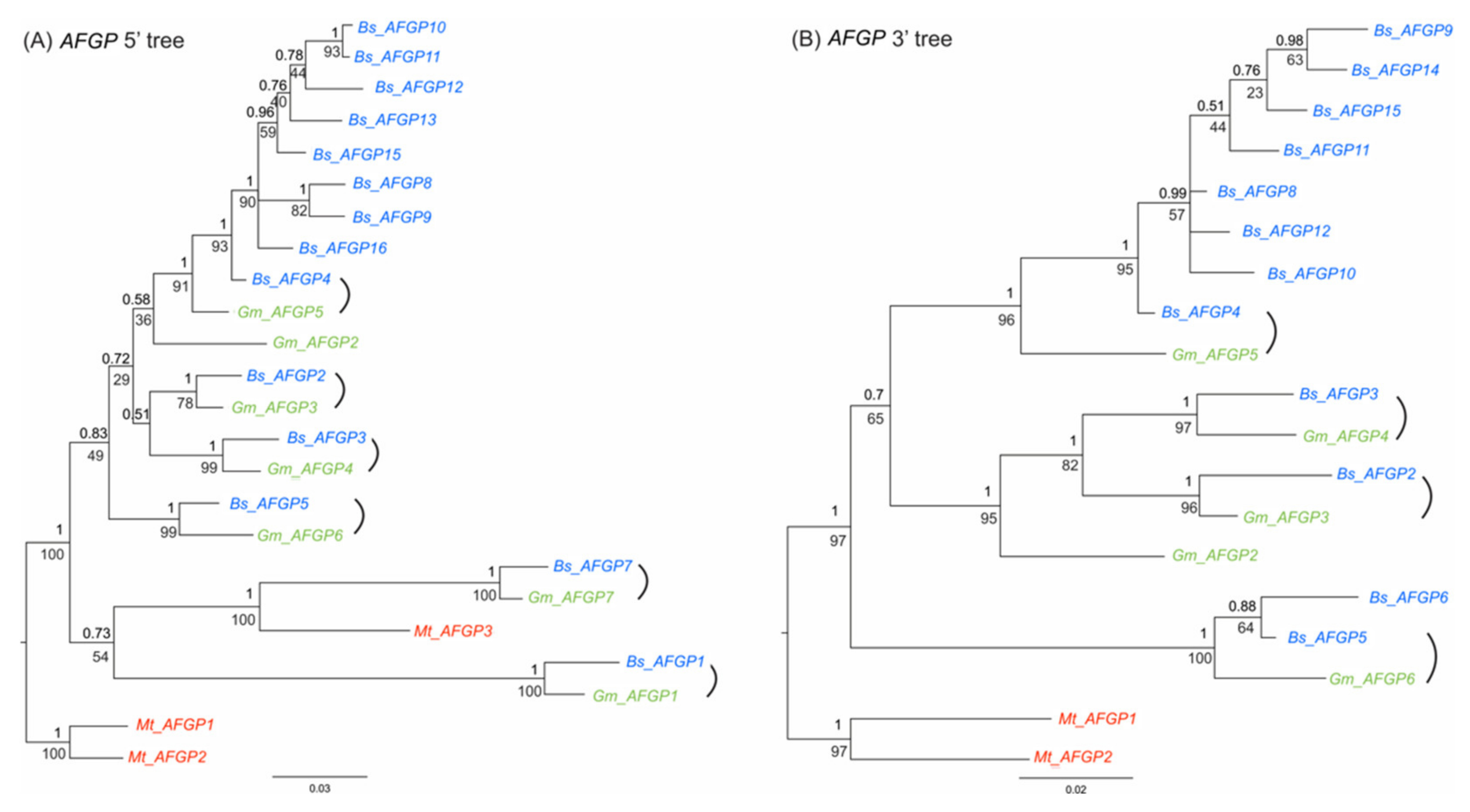
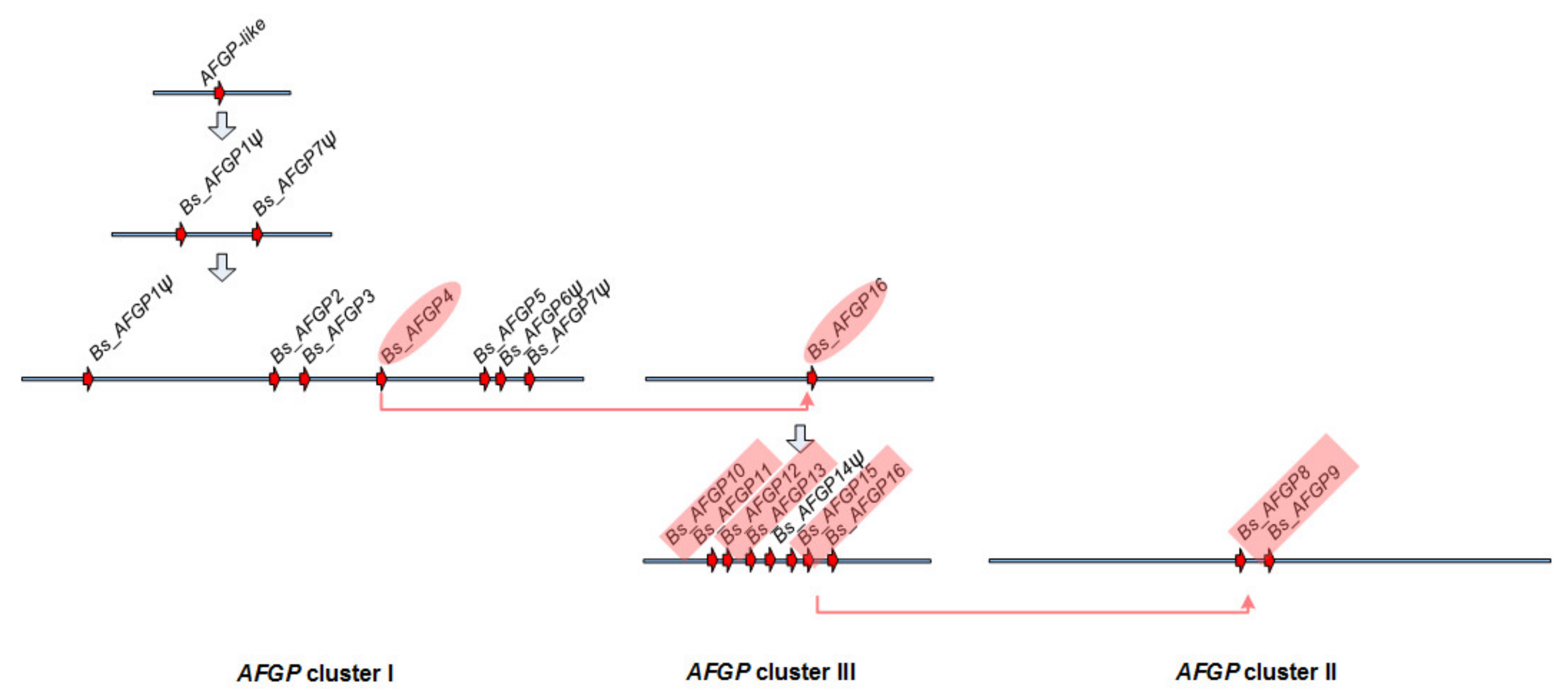
3.6. Expansion of the AFGP Gene Family and Genomic Locus
3.7. AFGP Gene Evolutionary History in Gadid Lineage
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohno, S. Evolution by Gene Duplication; George Alien & Unwin Ltd.: London, UK; Springer: Berlin/Heidelberg, Germany, 1970. [Google Scholar]
- Jacob, F. Evolution and Tinkering. Science 1977, 196, 1161–1166. [Google Scholar] [CrossRef]
- Vakirlis, N.; Carvunis, A.-R.; McLysaght, A. Synteny-based analyses indicate that sequence divergence is not the dominant source of orphan genes. eLife 2020, 9, e53500. [Google Scholar] [CrossRef] [PubMed]
- McLysaght, A.; Hurst, L.D. Open questions in the study of de novo genes: What, how and why. Nat. Rev. Genet. 2016, 17, 567–578. [Google Scholar] [CrossRef]
- Schmitz, J.F.; Bornberg-Bauer, E. Fact or fiction: Updates on how protein-coding genes might emerge de novo from previously non-coding DNA. F1000Research 2017, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Van Oss, S.B.; Carvunis, A.-R. De novo gene birth. PLoS Genet. 2019, 15, e1008160. [Google Scholar] [CrossRef]
- Tautz, D.; Domazet-Lošo, T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011, 12, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, N.; Kosiol, C.; Schlötterer, C. The life cycle of Drosophila orphan genes. eLife 2014, 3, e01311. [Google Scholar] [CrossRef] [PubMed]
- Devries, A.L. Glycoproteins as Biological Antifreeze Agents in Antarctic Fishes. Science 1971, 172, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- DeVries, A.L.; Cheng, C.-H.C. Antifreeze proteins and organismal freezing avoidance in polar fishes. In The Physiology of Polar Fishes; Farrell, A.P., Steffensen, J.F., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2005; Volume 22, pp. 155–201. [Google Scholar]
- Cheng, C.-H.C.; Zhuang, X. Molecular Origins and Mechanisms of Fish Antifreeze Evolution. In Antifreeze Proteins; Ramløv, H., Friis, D., Eds.; Springer: Cham, Switzerland, 2020; Volume 1, pp. 275–313. [Google Scholar]
- Chen, L.; DeVries, A.L.; Cheng, C.-H.C. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proc. Natl. Acad. Sci. USA 1997, 94, 3811–3816. [Google Scholar] [CrossRef]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1994; 600p. [Google Scholar]
- Devries, A.L. Biological antifreeze agents in cold water fishes. Comp. Biochem. Physiol. Part A Physiol. 1982, 73, 627–640. [Google Scholar] [CrossRef]
- O’Grady, S.M.; Schrag, J.D.; Raymond, J.A.; Devries, A.L. Comparison of antifreeze glycopeptides from arctic and antarctic fishes. J. Exp. Zool. 1982, 224, 177–185. [Google Scholar] [CrossRef]
- Cheng, C.-H.C.; Chen, L. Evolution of an antifreeze glycoprotein. Nat. Cell Biol. 1999, 401, 443–444. [Google Scholar] [CrossRef]
- Mortola, E.; Long, M. Turning Junk into Us: How Genes Are Born. Am. Sci. 2021, 109, 174–182. [Google Scholar]
- Matschiner, M.; Hanel, R.; Salzburger, W. On the Origin and Trigger of the Notothenioid Adaptive Radiation. PLoS ONE 2011, 6, e18911. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Yang, C.; Murphy, K.; Cheng, C.-H.C. Molecular mechanism and history of non-sense to sense evolution of antifreeze glycoprotein gene in northern gadids. Proc. Natl. Acad. Sci. USA 2019, 116, 4400–4405. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Chernomor, O.; Von Haeseler, A.; Minh, B.Q. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods); Version 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Mega, X. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Cziko, P.A.; DeVries, A.L.; Evans, C.W.; Cheng, C.-H.C. Antifreeze protein-induced superheating of ice inside Antarctic notothenioid fishes inhibits melting during summer warming. Proc. Natl. Acad. Sci. USA 2014, 111, 14583–14588. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-H.C.; Cziko, P.A.; Evans, C.W. Nonhepatic origin of notothenioid antifreeze reveals pancreatic synthesis as common mechanism in polar fish freezing avoidance. Proc. Natl. Acad. Sci. USA 2006, 103, 10491–10496. [Google Scholar] [CrossRef] [PubMed]
- Miya, T.; Gon, O.; Mwale, M.; Cheng, C.-H.C. The effect of habitat temperature on serum antifreeze glycoprotein (AFGP) activity in Notothenia rossii (Pisces: Nototheniidae) in the Southern Ocean. Polar Biol. 2013, 37, 367–373. [Google Scholar] [CrossRef]
- Zhuang, X.; Murphy, K.; Ghigliotti, L.; Pisano, E.; Cheng, C.-H.C. Reconstruction of the repetitive antifreeze glycoprotein genomic loci in the cold-water gadids Boreogadus saida and Microgadus tomcod. Mar. Genom. 2018, 39, 73–84. [Google Scholar] [CrossRef]
- Frazer, K.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://www.repeatmasker.org (accessed on 20 July 2021).
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Carr, S.M.; Kivlichan, D.S.; Pepin, P.; Crutcher, D.C. Molecular systematics of gadid fishes: Implications for the biogeographic origins of Pacific species. Can. J. Zool. 1999, 77, 19–26. [Google Scholar] [CrossRef]
- Roa-Varón, A.; Ortí, G. Phylogenetic relationships among families of Gadiformes (Teleostei, Paracanthopterygii) based on nuclear and mitochondrial data. Mol. Phylogenet. Evol. 2009, 52, 688–704. [Google Scholar] [CrossRef]
- Roa-Varón, A.; Dikow, R.B.; Carnevale, G.; Tornabene, L.; Baldwin, C.C.; Li, C.; Hilton, E.J. Confronting Sources of Systematic Error to Resolve Historically Contentious Relationships: A Case Study Using Gadiform Fishes (Teleostei, Paracanthopterygii, Gadiformes). Syst. Biol. 2021, 70, 739–755. [Google Scholar] [CrossRef]
- Fletcher, G.L.; Slaughter, D.; Hew, C.L. Seasonal changes in the plasma levels of glycoprotein antifreeze, Na+, Cl−, and glucose in Newfoundland Atlantic cod (Gadus morhua). Can. J. Zool. 1982, 60, 1851–1854. [Google Scholar] [CrossRef]
- DeVries, A.L.; Komatsu, S.K.; Feeney, R.E. Chemical and Physical Properties of Freezing Point-depressing Glycoproteins from Antarctic Fishes. J. Biol. Chem. 1970, 245, 2901–2908. [Google Scholar] [CrossRef]
- Hsiao, K.C.; Cheng, C.H.; Fernandes, I.E.; Detrich, H.W.; DeVries, A.L. An antifreeze glycopeptide gene from the antarctic cod Notothenia coriiceps neglecta encodes a polyprotein of high peptide copy number. Proc. Natl. Acad. Sci. USA 1990, 87, 9265–9269. [Google Scholar] [CrossRef] [PubMed]
- Wise, R.J.; Barr, P.J.; Wong, P.A.; Kiefer, M.C.; Brake, A.J.; Kaufman, R.J. Expression of a human proprotein processing enzyme: Correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc. Natl. Acad. Sci. USA 1990, 87, 9378–9382. [Google Scholar] [CrossRef] [PubMed]
- Douglass, J.; Civelli, O.; Herbert, E. Polyprotein Gene Expression: Generation of Diversity of Neuroendocrine Peptides. Annu. Rev. Biochem. 1984, 53, 665–715. [Google Scholar] [CrossRef]
- Chen, L.; DeVries, A.L.; Cheng, C.-H.C. Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod. Proc. Natl. Acad. Sci. USA 1997, 94, 3817–3822. [Google Scholar] [CrossRef] [PubMed]
- Levinson, G.; Gutman, G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Eichler, E.E.; Holden, J.J.; Popovich, B.W.; Reiss, A.L.; Snow, K.; Thibodeau, S.N.; Richards, C.S.; Ward, P.A.; Nelson, D.L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat. Genet. 1994, 8, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.-Y.; Ranum, L.P.; Duvick, L.A.; Servadio, A.; Zoghbi, H.Y.; Orr, H.T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat. Genet. 1993, 5, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, C.; Sagot, M.-F. A small trip in the untranquil world of genomes: A survey on the detection and analysis of genome rearrangement breakpoints. Theor. Comput. Sci. 2008, 395, 171–192. [Google Scholar] [CrossRef][Green Version]
- Smith, G.P. Evolution of Repeated DNA Sequences by Unequal Crossover. Science 1976, 191, 528–535. [Google Scholar] [CrossRef] [PubMed]
- David, C.; Lange, B.A.; Krumpen, T.; Schaafsma, F.; Van Franeker, J.A.; Flores, H. Under-ice distribution of polar cod Boreogadus saida in the central Arctic Ocean and their association with sea-ice habitat properties. Polar Biol. 2016, 39, 981–994. [Google Scholar] [CrossRef]
- Gradinger, R.R.; Bluhm, B.A. In-situ observations on the distribution and behavior of amphipods and Arctic cod (Boreogadus saida) under the sea ice of the High Arctic Canada Basin. Polar Biol. 2004, 27, 595–603. [Google Scholar] [CrossRef]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmstrøm, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Stransky, C.; Baumann, H.; Fevolden, S.-E.; Harbitz, A.; Høie, H.; Nedreaas, K.H.; Salberg, A.-B.; Skarstein, T.H. Separation of Norwegian coastal cod and Northeast Arctic cod by outer otolith shape analysis. Fish. Res. 2008, 90, 26–35. [Google Scholar] [CrossRef]
- Wassmann, P.; Svendsen, H.; Keck, A.; Reigstad, M. Selected aspects of the physical oceanography and particle fluxes in fjords of northern Norway. J. Mar. Syst. 1996, 8, 53–71. [Google Scholar] [CrossRef]
- Yang, C. Antifreeze Glycoproteins in Northern Cods: Gene Family Sizes, Sequences, Structures, Organizations, and Evolution. Ph.D. Thesis, University of Illinois at Urbana-Champaign, Urbana, IL, USA, 2002. [Google Scholar]
- Tørresen, O.K.; Star, B.; Jentoft, S.; Reinar, W.B.; Grove, H.; Miller, J.R.; Walenz, B.P.; Knight, J.; Ekholm, J.M.; Peluso, P.; et al. An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genom. 2017, 18, 95. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, X.; Cheng, C.-H.C. Propagation of a De Novo Gene under Natural Selection: Antifreeze Glycoprotein Genes and Their Evolutionary History in Codfishes. Genes 2021, 12, 1777. https://doi.org/10.3390/genes12111777
Zhuang X, Cheng C-HC. Propagation of a De Novo Gene under Natural Selection: Antifreeze Glycoprotein Genes and Their Evolutionary History in Codfishes. Genes. 2021; 12(11):1777. https://doi.org/10.3390/genes12111777
Chicago/Turabian StyleZhuang, Xuan, and C.-H. Christina Cheng. 2021. "Propagation of a De Novo Gene under Natural Selection: Antifreeze Glycoprotein Genes and Their Evolutionary History in Codfishes" Genes 12, no. 11: 1777. https://doi.org/10.3390/genes12111777
APA StyleZhuang, X., & Cheng, C.-H. C. (2021). Propagation of a De Novo Gene under Natural Selection: Antifreeze Glycoprotein Genes and Their Evolutionary History in Codfishes. Genes, 12(11), 1777. https://doi.org/10.3390/genes12111777





