Cloning, Expression, and Tobacco Overexpression Analyses of a PISTILLATA/GLOBOSA-like (OfGLO1) Gene from Osmanthus fragrans
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Extraction of Total RNA and Reverse Transcription
2.3. OfGLO1 Gene Cloning from O. fragrans
2.4. Bioinformatics Analysis of Sequences
2.5. Organ-Specific Expression Analysis Using Quantitative Real-Time RT-PCR
2.6. Vector Construction and Tobacco Transformation of OfGLO1
3. Results
3.1. Sequence of the OfGLO1 Gene
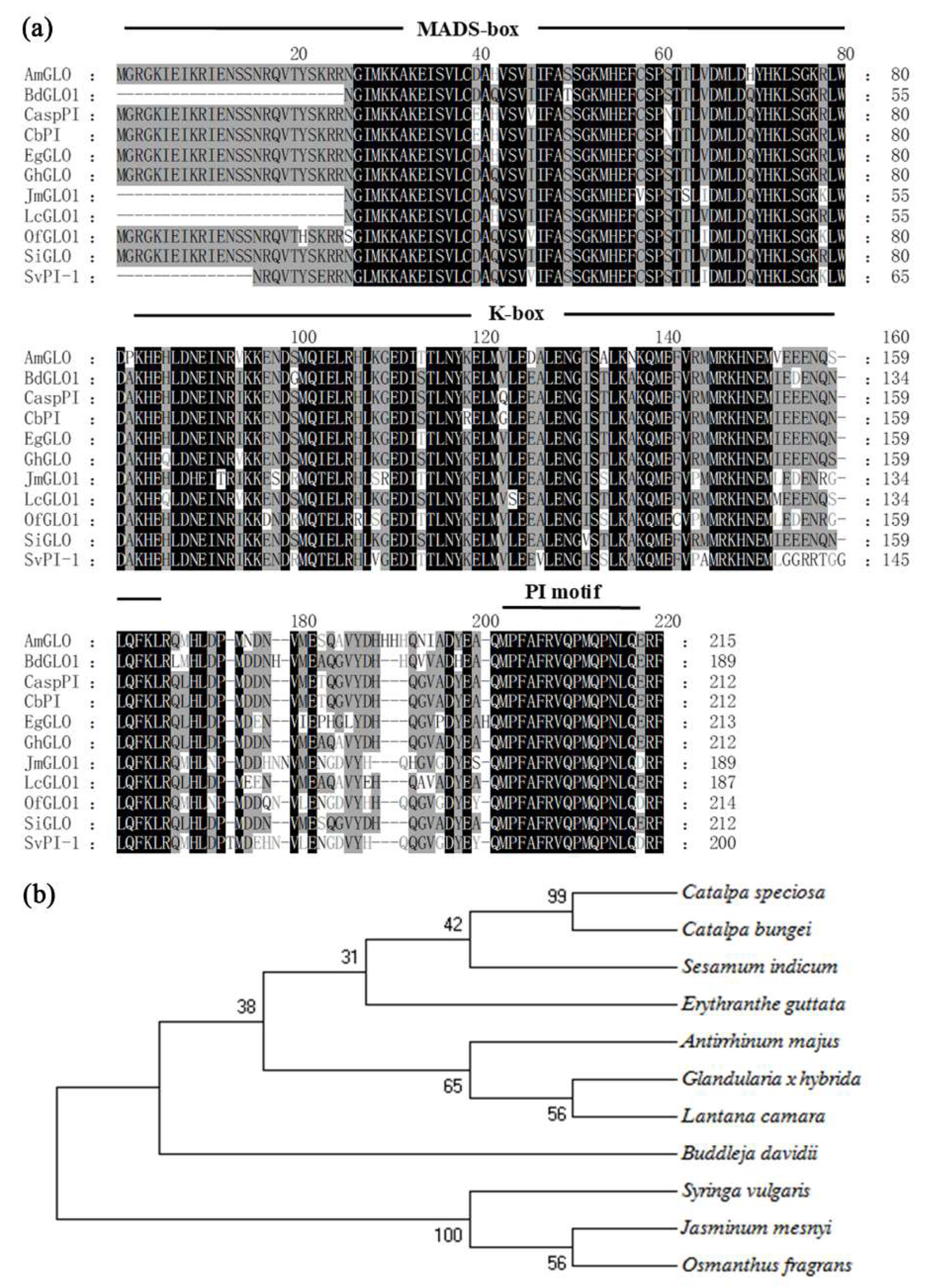
3.2. Sequence Alignment and Phylogenetic Analysis of GLO Homologs among Different Species
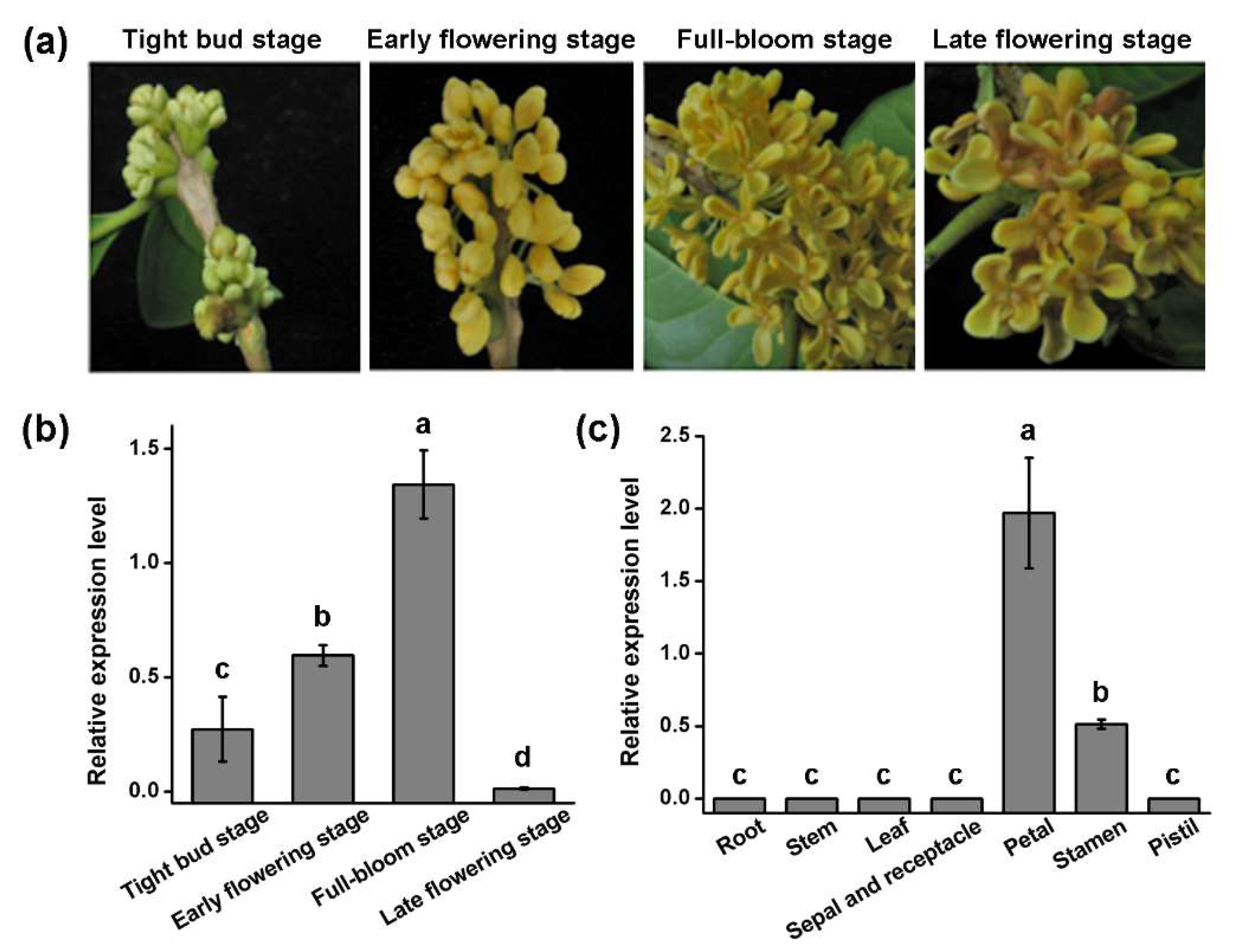
3.3. Organ-Specific Expression Analysis of OfGLO1 Gene
3.4. Phenotypic Analysis of OfGLO1 Gene-Transformed Tobacco Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Soltis, D.E.; Chanderbali, A.S.; Kim, S.; Buzgo, M.; Soltis, P.S. The ABC model and its applicability to basal angiosperms. Ann. Bot. 2007, 100, 155–163. [Google Scholar] [CrossRef]
- Theissen, G.; Saedler, H. Plant biology. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Buzgo, M.; Soltis, P.S.; Soltis, D.E. Floral developmental morphology of Amborella trichopoda (Amborellaceae). Int. J. Plant Sci. 2004, 165, 925–947. [Google Scholar] [CrossRef]
- Bowman, J.L. Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. J. Biosci. 1997, 22, 515–527. [Google Scholar] [CrossRef]
- Kramer, E.M.; Di Stilio, V.S.; Schlüter, P.M. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the ranunculaceae. Int. J. Plant Sci. 2003, 164, 1–11. [Google Scholar] [CrossRef]
- Litt, A. An evaluation of A-function: Evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 2007, 168, 73–91. [Google Scholar] [CrossRef]
- Hernandez-Hernandez, T.; Martinez-Castilla, L.P.; Alvarez-Buylla, E.R. Functional diversification of B MADS-box homeotic regulators of flower development: Adaptive evolution in protein-protein interaction domains after major gene duplication events. Mol. Biol. Evol. 2007, 24, 465–481. [Google Scholar] [CrossRef]
- Urbanus, S.L.; De Folter, S.; Shchennikova, A.V.; Kaufmann, K.; Immink, R.G.; Angenent, G.C. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol. 2009, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Murai, K. Homeotic Genes and the ABCDE Model for Floral Organ Formation in Wheat. Plants 2013, 2, 379–395. [Google Scholar] [CrossRef]
- Abraham-Juarez, M.J.; Schrager-Lavelle, A.; Man, J.; Whipple, C.; Handakumbura, P.; Babbitt, C.; Bartlett, M. Evolutionary variation in MADS box dimerization affects floral development and protein abundance in maize. Plant Cell 2020, 32, 3408–3424. [Google Scholar] [CrossRef]
- Sommer, H.; Beltran, J.P.; Huijser, P.; Pape, H.; Lonnig, W.E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Tröbner, W.; Ramirez, L.; Motte, P.; Hue, I.; Huijser, P.; Lönnig, W.E.; Saedler, H.; Sommer, H.; Schwarz-Sommer, Z. GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992, 11, 4693–4704. [Google Scholar] [CrossRef] [PubMed]
- Jack, T.; Brockman, L.L.; Meyerowitz, E.M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 1992, 68, 683–697. [Google Scholar] [CrossRef]
- Goto, K.; Meyerowitz, E.M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994, 8, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.Y.; Kim, S.R.; Kang, H.G.; Noh, Y.S.; Park, M.C.; Finkel, D.; An, G. Characterization of two rice MADS box genes homologous to GLOBOSA. Plant Sci. 1995, 109, 45–56. [Google Scholar] [CrossRef]
- Munster, T.; Wingen, L.U.; Faigl, W.; Werth, S.; Saedler, H.; Theissen, G. Characterization of three GLOBOSA-like MADS-box genes from maize: Evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene 2001, 262, 1–13. [Google Scholar] [CrossRef]
- Kanno, A.; Saeki, H.; Kameya, T.; Saedler, H.; Theissen, G. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol. Biol. 2003, 52, 831–841. [Google Scholar] [CrossRef]
- Park, J.H.; Ishikawa, Y.; Ochiai, T.; Kanno, A.; Kameya, T. Two GLOBOSA-like genes are expressed in second and third whorls of homochlamydeous flowers in Asparagus officinalis L. Plant Cell Physiol. 2004, 45, 325–332. [Google Scholar] [CrossRef][Green Version]
- Tsai, W.C.; Lee, P.F.; Chen, H.I.; Hsiao, Y.Y.; Wei, W.J.; Pan, Z.J.; Chuang, M.H.; Kuoh, C.S.; Chen, W.H.; Chen, H.H. PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol. 2005, 46, 1125–1139. [Google Scholar] [CrossRef]
- Wu, X.; Shi, J.; Xi, M.; Luo, Z.; Hu, X. A B functional gene cloned from lily encodes an ortholog of Arabidopsis PISTILLATA (PI). Plant Mol. Biol. Rep. 2010, 28, 684–691. [Google Scholar] [CrossRef]
- Xiang, L.; Chen, Y.; Chen, L.; Fu, X.; Zhao, K.; Zhang, J.; Sun, C. B and E MADS-box genes determine the perianth formation in Cymbidium goeringii Rchb.f. Physiol. Plant 2018, 162, 353–369. [Google Scholar] [CrossRef]
- Zhou, K.; Cao, Q.X.; Jin, C.M.; Niu, Y.Y.; Li, G.L.; Zhang, J.J. Identification of two GLOBOSA-Like MADS-Box genes in tea plant (Camellia sinensis [L.] O. Kuntze). Mol. Biol. 2019, 53, 13–23. [Google Scholar] [CrossRef]
- Broholm, S.K.; Pollanen, E.; Ruokolainen, S.; Tahtiharju, S.; Kotilainen, M.; Albert, V.A.; Elomaa, P.; Teeri, T.H. Functional characterization of B class MADS-box transcription factors in Gerbera hybrida. J. Exp. Bot. 2010, 61, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Liu, H.; Almeida, A.M.; Kuang, Y.; Zou, P.; Liao, J. Molecular basis of floral petaloidy: Insights from androecia of Canna indica. AoB Plants 2014, 6, plu015. [Google Scholar] [CrossRef] [PubMed]
- Louati, M.; Salazar-Sarasua, B.; Roque, E.; Beltran, J.P.; Hannachi, A.S.; Gomez-Mena, C. Isolation and functional analysis of a PISTILLATA-like MADS-Box gene from argan tree (Argania spinosa). Plants 2021, 10, 1665. [Google Scholar] [CrossRef]
- Lannenpaa, M.; Parkkinen, S.; Jarvinen, P.; Lemmetyinen, J.; Vepsalainen, S.; Savola, T.; Keinonen, K.; Keinanen, M.; Sopanen, T. The expression and promoter specificity of the birch homologs for PISTILLATA/GLOBOSA and APETALA3/DEFICIENS. Physiol. Plant 2005, 125, 268–280. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, X.; Yang, J.; Cai, X.; Shi, Y.; Zheng, R.; Wang, Z.; Liu, J.; Yi, X.; Xiao, S.; et al. Whole-genome resequencing of Osmanthus fragrans provides insights into flower color evolution. Hortic. Res. 2021, 8, 98. [Google Scholar] [CrossRef]
- Yang, X.; Yue, Y.; Li, H.; Ding, W.; Chen, G.; Shi, T.; Chen, J.; Park, M.S.; Chen, F.; Wang, L. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic. Res. 2018, 5, 72. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Hou, D.; Wang, Y.; Zhang, C.; Bao, Z.; Zhao, H.; Hu, S. Identification of floral aromatic volatile compounds in 29 cultivars from four groups of Osmanthus fragrans by gas chromatography–mass spectrometry. Hortic. Environ. Biotechnol. 2019, 60, 611–623. [Google Scholar] [CrossRef]
- Cai, X.; Mai, R.Z.; Zou, J.J.; Zhang, H.Y.; Zeng, X.L.; Zheng, R.R.; Wang, C.Y. Analysis of aroma-active compounds in three sweet osmanthus (Osmanthus fragrans) cultivars by GC-olfactometry and GC-MS. J. Zhejiang Univ.-Sci. B 2014, 15, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-L.; Irish, V.F. Gene duplication and loss in a MADS box gene transcription factor circuit. Mol. Biol. Evol. 2011, 28, 3367–3380. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Han, N.; Liu, C.; Buerte, B.; Zhou, C.; Chen, J.; Wang, M.; Zhang, Y.; Tang, Y.; Zhu, M.; et al. Functional dissection of HGGT and HPT in barley vitamin E biosynthesis via CRISPR/Cas9-enabled genome editing. Ann. Bot. 2020, 126, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Riechmann, J.L.; Jia, D.; Meyerowitz, E. Minimal regions in the Arabidopsis PISTILLATA promoter responsive to the APETALA3/PISTILLATA feedback control do not contain a CArG box. Sex. Plant Reprod. 2000, 13, 85–94. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Melzer, R.; Theissen, G. Molecular interactions of orthologues of floral homeotic proteins from the gymnosperm Gnetum gnemon provide a clue to the evolutionary origin of ‘floral quartets’. Plant J. 2010, 64, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, J.S.; Zhao, J.; He, C. Distinct subfunctionalization and neofunctionalization of the B-class MADS-box genes in Physalis floridana. Planta 2015, 241, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.M.; Dorit, R.L.; Irish, V.F. Molecular evolution of genes controlling petal and stamen development: Duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 1998, 149, 765–783. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, Z.; Li, X.; Li, J.; Yin, H. The APETALA1 and FRUITFUL homologs in Camellia japonica and their roles in double flower domestication. Mol. Breed. 2014, 33, 821–834. [Google Scholar] [CrossRef]


| Characteristics | Wild Type | OfGLO1-Transformed |
|---|---|---|
| Differentiation rate of floral bud at 70 days | 0.00 | 100.00 * |
| Plant height (cm) | 83.90 | 73.63 * |
| Leaf number | 30.50 | 25.75 * |
| Length–width ratio of leaf | 2.06 | 2.31 * |
| Internode length (cm) | 2.45 | 4.48 * |
| Branching number | 2.25 | 8.38 * |
| Floral bud number | 25.00 | 21.63 * |
| Stamen length (cm) | 4.20 | 3.86 * |
| Pistil length (cm) | 4.16 | 4.18 |
| Corolla diameter (cm) | 2.00 | 1.98 |
| Seed setting rate (%) | 70.00 | 11.25 * |
| Seed germination rate (%) | 96.25 | 28.75 * |
| New Phenotypes of Flower | No. of Total Flowers | Flower No. with New Phenotype | Percentage (%) |
|---|---|---|---|
| No. of petals was less than 5 | 80 | 6 | 7.5 |
| No. of petals was more than 5 | 80 | 12 | 15 |
| Stamen became petal-like | 80 | 10 | 12.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Z.; Chen, S.; Xu, M.; Wang, M.; Chen, Z.; Wang, L.; Pang, J. Cloning, Expression, and Tobacco Overexpression Analyses of a PISTILLATA/GLOBOSA-like (OfGLO1) Gene from Osmanthus fragrans. Genes 2021, 12, 1748. https://doi.org/10.3390/genes12111748
Zeng Z, Chen S, Xu M, Wang M, Chen Z, Wang L, Pang J. Cloning, Expression, and Tobacco Overexpression Analyses of a PISTILLATA/GLOBOSA-like (OfGLO1) Gene from Osmanthus fragrans. Genes. 2021; 12(11):1748. https://doi.org/10.3390/genes12111748
Chicago/Turabian StyleZeng, Zhanghui, Si Chen, Mingrui Xu, Min Wang, Zhehao Chen, Lilin Wang, and Jiliang Pang. 2021. "Cloning, Expression, and Tobacco Overexpression Analyses of a PISTILLATA/GLOBOSA-like (OfGLO1) Gene from Osmanthus fragrans" Genes 12, no. 11: 1748. https://doi.org/10.3390/genes12111748
APA StyleZeng, Z., Chen, S., Xu, M., Wang, M., Chen, Z., Wang, L., & Pang, J. (2021). Cloning, Expression, and Tobacco Overexpression Analyses of a PISTILLATA/GLOBOSA-like (OfGLO1) Gene from Osmanthus fragrans. Genes, 12(11), 1748. https://doi.org/10.3390/genes12111748






